Abstract
The transcription map of simian parvovirus (SPV), an Erythrovirus similar to Parvovirus B19, was investigated. RNA was extracted from tissues of experimentally infected cynomolgus macaques and subjected to reverse transcription-PCR with SPV-specific primers. The PCR products were cloned and sequenced to identify splice junctions. A total of 14 distinct sequences were identified as putative partial transcripts. Of these, 13 were spliced; a single unspliced transcript putatively encoded NS1. Sequence analysis revealed that spliced partial transcripts may encode portions of open reading frames for the major capsid proteins VP1 and VP2 and smaller, unknown proteins. These unspliced and spliced transcripts and putative proteins encoded by SPV were similar to those of B19. Initial splice junctions at nucleotides 279 and 333 were analogous to those at nucleotides 406 and 441, respectively, in B19. Seven of the 10 splices identified had typical GT/AG donor/acceptor junctions. The splice sites were confirmed by Northern blotting and autoradiography. In contrast to B19, which has a maximum of two splices per transcript, up to three splices were observed in SPV transcripts. A spliced transcript putatively encoding a truncated version of NS1, as seen with minute virus of mice and adeno-associated virus 2, was also observed. The findings indicate that that the splicing pattern of transcripts of SPV and B19 is similar, but SPV also has coding strategies in common with other parvoviruses.
Simian parvovirus (SPV) was first identified in cynomolgus macaques suffering from severe anemia as a consequence of erythroid cell destruction (20). These animals were immunosuppressed due to a concurrent infection with type D simian retrovirus (22). Clinically, this was similar to cases of human infection with parvovirus B19, which is responsible for causing transient aplastic crisis in individuals with underlying hemolysis and red cell aplasia in immunosuppressed individuals, such as AIDS patients (9, 22). B19 infection also results in hydrops fetalis and fetal death in pregnant women (9), sequelae also observed with experimental SPV infection in pregnant female cynomolgus macaques (M. G. O’Sullivan, J. C. Veille, W. A. Block, A. R. Turner, N. S. Young, and K. E. Brown, Abstr. 17th Annu. Meet. Am. Soc. Virol., abstr. W22-9, 1998).
SPV was classified as a parvovirus considering molecular similarities to other parvoviruses, i.e., adeno-associated virus 2 (prototype dependovirus), the autonomous parvoviruses (the prototype being minute virus of mice), and parvovirus B19 (prototype erythrovirus). In general, members of the subfamily Parvovirinae have a small genome of about 5 kb composed of single-stranded DNA with terminal palindromic sequences that serve as primers for the synthesis of the complementary strand. Only the positive strand of these viruses encodes proteins, suggesting that the positive-sense mRNA is transcribed from the minus-strand genomic DNA (3). To maximize the coding capacity, a number of promoters and all three reading frames are used to generate multiple RNAs by splicing (3). Also, for efficient utilization of the genome, capsid proteins are encoded by overlapping in-frame DNA sequences, implying that the coding sequences share the carboxyl terminus and that the smaller capsid proteins (VP2 and VP3) are truncated versions of the large capsid protein (VP1) (3).
The genus Erythrovirus was formed considering the predilection for the prototype parvovirus B19 to infect erythroid cells and significant differences between B19 and other members of the subfamily Parvovirinae (3, 11). All nine B19 transcripts observed utilize the same P6 promoter (5, 12, 23), and all transcripts initiate from the 5′ end of the genome. In contrast, minute virus of mice and adeno-associated virus 2 use two and three promoters, respectively (13, 18, 25). All parvovirus transcripts studied to date terminate at the 3′ end of the genome. The only known exception is B19, which uses two polyadenylation signals. B19 forms two types of small, relatively abundant RNAs which are absent in minute virus of mice and adeno-associated virus (2, 23). Also, B19 transcripts have multiple large introns not seen in either adeno-associated virus or minute virus of mice (1, 16, 19, 27).
SPV has 50% homology to B19 at the DNA level. At the amino acid level, there is 70% homology with B19 capsid proteins and 50% homology with B19 nonstructural protein (7). SPV thus became the second virus to be considered a member of the genus Erythrovirus. The goal of this study was to investigate the transcripts produced by SPV. The working hypothesis was that SPV transcription would resemble that of B19. Our findings confirmed this hypothesis but also indicated that SPV, unlike B19, has coding strategies in common with other parvoviruses.
MATERIALS AND METHODS
Experimental infection.
SPV was purified from a viremic cynomolgus monkey in the original outbreak investigated by O’Sullivan et al. (20). Purification was achieved by ultracentrifugation of viremic serum through a sucrose gradient, followed by resuspension in phosphate-buffered saline (20). This material was used to inoculate adult cynomolgus monkeys by the intravenous route (21) and the fetuses of pregnant females by the intramuscular route (M. G. OSullivan, J. C. Veille, W. A. Block, A. R. Turner, N. S. Young, and K. E. Brown, Abstr. 17th Annu. Meet. Am. Soc. Virol., abstr. W22-9, 1998). Bone marrow was collected from two viremic adult females, and the liver (an important organ for erythropoiesis in the fetus) was obtained from two viremic fetuses. These tissues contained numerous intranuclear inclusions within erythroid cells and, for this reason, were used for isolation of RNA.
The research performed complied with all relevant federal guidelines and institutional policies on animal use (with the protocol approval by the Institutional Animal Care and Use Committee, University of Minnesota).
Reverse transcription.
Total RNA was extracted from adult bone marrow and fetal liver recovered at necropsy from experimentally infected macaques (RNeasy, Qiagen Inc., Valencia, Calif.). RNA was quantified both before and after performing DNase treatment (DNA-free; Ambion Inc., Austin, Tex.). The absence of SPV DNA in the DNase-treated RNA was confirmed by the inability to amplify products with PCR with SPV-specific primers SP3 and SP5 (7). This RNA was then reverse transcribed into cDNA with random decamers as primers (Retroscript; Ambion Inc.).
PCR.
We amplified 5 μl of reverse transcription products (cDNAs) by PCR with Taq polymerase (Retroscript). Cycling conditions were as follows: 95°C for 3 min and 59°C for 2 min, 35 cycles of 72°C for 3.30 min, 95°C for 45 s, and 59°C for 45 s; and 72°C for 10 min. All primers used were approximately 20 nucleotides in length and were synthesized at the Advanced Genetic Analysis Center at the University of Minnesota.
The nucleotide positions referred to in this manuscript indicate positions on the positive-sense DNA strand of the SPV genome (excluding the terminal repeats) cloned previously (GenBank accession number U26342) (7). A forward primer located at nucleotide position 230 (F230) was primarily used in conjunction with four reverse (R) primers at nucleotide positions 2346, 2568, 3369, and 4854. Two other primers used were F1545 and R1598 (see Fig. 2).
FIG. 2.
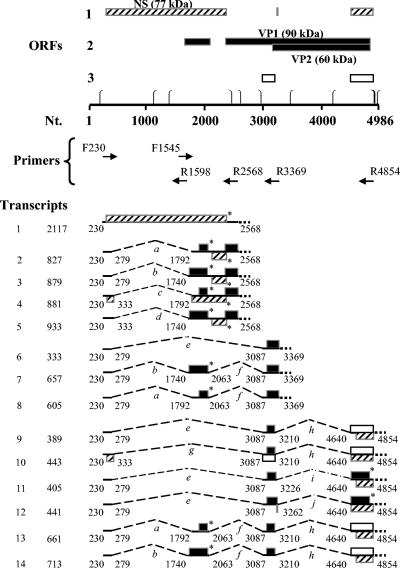
Splice junction map of SPV. Open reading frames (with ATG as the start codon) greater than 40 amino acids are indicated with boxes of different shades. Nt., nucleotide position in simian parvovirus genome (GenBank Accession U26342). ⎧, promoter site (TATAA). ⎫, site for polyadenylation (AATAAA). F, forward primer. R, reverse primer. On the left-hand side are shown the designated numbers and total lengths in nucleotides of cloned partial transcripts. The ends of these transcripts correspond to the forward and reverse primers used. Lines indicate exons, and brackets indicate spliced introns (designated alphabetically in italics). Asterisks indicate stop codons. ORFs and transcripts ending with dotted lines indicate the ends of partial transcripts corresponding to the position of the reverse primer; NS = 307 to 2370, VP1 = 2363 to 4819, and VP2 = 3149 to 4819 (6).
Mouse liver RNA amplified with mouse liver-specific primers (Retroscript) was used as a positive control for the PCR. SPV-infected macaque RNA to which reverse transcriptase was not added during reverse transcription and amplified with F230 and R2346 primers served as a negative control to check DNA contamination during reverse transcription. Water (5 μl) used in place of DNA in the PCR and amplified with the F230 and R2346 primers served as a negative control to rule out DNA contamination during PCR.
PCR products (concentrations averaging 0.6 μg/μl) were used directly for ligation, or PCR products were used for ligation after separation by gel electrophoresis (1% agarose in 0.5× Tris-boric acid-EDTA buffer). Bands were visualized after 30 min of staining with ethidium bromide, followed by 10 min of destaining with distilled water on a platform shaker. Different-sized products were mechanically separated and gel purified with silica gel membrane assembly for ligation (QIAquick; Qiagen Inc., Valencia, Calif.). The concentrations of gel-extracted products averaged 20 ng/μl.
Cloning.
PCR products were ligated overnight into the pCR 2.1 vector (original TA cloning; Invitrogen Corporaticon, Carlsbad, Calif.) with T4 DNA ligase in a total volume of 10 μl at 14°C. Ligated products were cloned in transformed Escherichia coli cells and plasmids were isolated (QIAprep). Plasmid samples were restriction enzyme digested with EcoRI (New England BioLabs Inc., Beverly, Mass.) to select clones for sequencing.
Sequence analysis.
Plasmids (1 to 2 μg) were sequenced at the Advanced Genetic Analysis Center at the University of Minnesota in a total reaction volume of 12 μl with the T7P and M13R primers (3.2 pmol). The software programs Seqman, Megalign, and Editseq (version 4.0; Lasergene; DNAStar Inc., Madison, Wis.) were used to align trace files of sequences of cloned PCR products with SPV sequence (GenBank accession no. U26342). This identified splice junctions. Putative open reading frames (ORFs) were identified with the program Mapdraw (version 4.0; DNAStar).
BLAST search.
Standard nucleotide-nucleotide BLAST and standard protein-protein BLAST searches were performed on the NCBI website.
Northern blotting and autoradiography.
RNA samples were electrophoresed in sets in a denaturing 1% agarose-formaldehyde gel. Each set consisted of DNase-treated sample RNA extracted from tissues of infected macaques (referred to hereafter as SPV RNA) (7 μg), negative control (in most instances RNA from tissues of noninfected macaques, and occasionally MA104 or CHO cell RNA), and the radiolabeled marker synthesized by in vitro transcription (Ribomark labeling system; Promega Corporation). An in vitro-transcribed product was also used as positive control (see below).
In vitro-transcribed positive control.
One of the commonly observed partial transcripts (transcript number 6 encoding the ORF for VP2; Fig. 2), 333 bases in length, was selected. Plasmids containing the PCR products corresponding to this partial transcript (in an orientation that would give a ribonucleotide of positive sense) were used for in vitro transcription. A stock solution of such plasmids was purified from a culture of previously transformed cells maintained in 20% glycerol at −86°C (QIAprep; Qiagen Inc., Valencia, Calif.). For a single reaction, 10 μg of plasmid was linearized with BamHI (New England BioLabs Inc.) to obtain a 5′ overhang. After RNase treatment with proteinase K and ethanol precipitation, the product was suspended in a total volume of 16 μl. This was suitable for a 40-μl reaction of in vitro transcription with T7 reaction components (Ribomax large scale RNA production system). Synthesized RNA was separated from the DNA template by DNase treatment followed by phenol-chloroform extraction and ethanol precipitation. Unincorporated nucleotides were removed with chromatography columns (Micro Bio-Spin; Bio-Rad Laboratories, Hercules, Calif.). The yield of RNA was typically approximately 500 μg (10 μg/μl).
After denaturing gel electrophoresis, RNA was transferred (downward transfer) by blotting onto a nylon filter and cross-linked with a UV cross-linker (Stratalinker; Stratagene, Cedar Creek, Tex.) set at auto-cross-link (12,000 mJ/cm2 for 90 s). Northern blots were blocked with salmon sperm DNA (Stratagene) and probed with one of the following probes. One was a splice junction-specific antisense 40-mer oligonucleotide (spanning 20 bases on either side of the 10 different splices identified) tailed with 32P-labeled dATP (Redivue; Amersham Pharmacia Biotech Inc., Piscataway, N.J.) with terminal deoxynucleotidyl transferase (New England BioLabs Inc.). Splice junction-specific oligonucleotides were obtained from the Advanced Genetic Analysis Center at the University of Minnesota or from Integrated DNA Technologies Inc., Coralville, Iowa. The other was a cloned 333-bp partial transcript radiolabeled with random primer extension with Klenow fragment of polymerase I (Prime-It II; Stratagene).
The radioactive probes were purified with chromatography columns (Micro Bio-Spin; Bio-Rad Laboratories, Hercules, Calif.). The specific activity of the probes was approximately 2 × 108 cpm. The temperature conditions used for hybridization and washing were optimized after calculating the melting temperatures (Tm) of various oligonucleotides used as probes. Hybridization was performed at 68°C for 12 to 18 h and three 10-min washes were performed at 68°C with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate.
Autoradiography results.
The AssayZap (version 2.50; Biosoft, Ferguson, Mo. [http://www.biosoft.com/w/assayzap.htm]) and GraphPad (version 2.0C, GraphPad Software, Inc., San Diego, Calif. [http://www.graphpad.com/welcome.htm]) programs were used to determine the sizes of the bands observed after autoraudiography. The results were checked on both the PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and imaging films (Eastman Kodak Company, Rochester, N.Y.).
Nucleotide sequence accession number.
The GenBank accession number for the SPV sequence used to analyze the sequences of the observed transcripts is U26342.
RESULTS
PCR amplification products.
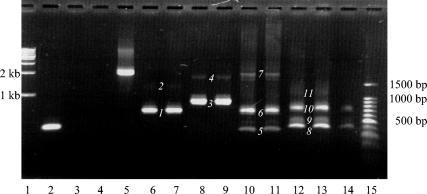
PCR products were visualized by ethidium bromide staining after gel electrophoresis. As indicated in Fig. 1, 11 bands were detected after reverse transcription-PCR was performed on infected macaque sample RNA with F230 and R2346, R2568, R3369, and R4854 primers. The 11 bands observed indicated that SPV DNA is transcribed to produce several mRNA species in infected cells.
FIG. 1.
PCR products obtained from the four main primer sets. Lanes 1 and 15, 1-kb and 100-bp markers. Lane 2, mouse liver RNA amplified with mouse liver-specific primers (positive control for PCR of size 361 bp). Lane 3, SPV-infected macaque RNA to which reverse transcriptase was not added during reverse transcription, amplified with the F230 and R2346 primers (negative control). Lane 4, 5 μl of water used for the PCR amplified with the F230 and R2346 primers (negative control). Lane 5, 1 μl of the complete SPV genome amplified with the F230 and R2346 primers. PCR products from amplification of 5 μl of reverse transcription product of infected macaque sample RNA with primers F230 and R2346 (lanes 6 and 7); primers F230 and R2568 (lanes 8 and 9); primers F230 and R3369 (lanes 10 and 11); and primers F230 and R4854 (lanes 12, 13, and 14). PCR products (bands) obtained with the different primer sets (see above) and used in subsequent cloning reactions are numbered 1 through 11.
Cloning.
Initially, complete PCR products from different primer sets were used for ligation into the pCR2.1 vector. With this approach, products corresponding to 4 out of the 11 bands (numbered 1, 3, 6, and 8, Fig. 1) were cloned. Gel purification of bands prior to cloning resulted in the cloning of an additional three bands (numbered 5, 9, and 10). Figure 1 illustrates that PCR products were run in sets to provide enough DNA for gel extraction. Cloning of the remaining four PCR products larger than 1 kb in size (bands numbered 2, 4, 7, and 11) was unsuccessful with this technique. This observation may suggest that strong intramolecular secondary structures are present in the larger SPV reverse transcription-PCR generated fragments.
cDNA sequence analysis of tissue derived RNA transcripts.
Thirteen spliced, partial transcripts (Fig. 2) were found by nucleotide sequence analysis of seven cloned PCR products (Fig. 1, as described above). The exact nucleotide positions where these transcripts begin and end were not mapped, and the numbers refer to the nucleotide position of the primers used to detect them (positions correspond to the previously cloned SPV genome, GenBank accession number U26342).
Thirteen different transcripts were observed because bands seen with ethidium bromide staining on agarose gel (Fig. 1) represented one or more transcripts, which had a narrow range of molecular sizes. For example, band 3 (Fig. 1, lanes 8 and 9), observed with primers F230 and R2568, represented transcripts numbered 2, 3, 4, and 5 in Fig. 2. Similarly, band 5 (Fig. 1, lanes 10 and 11), observed with primers F230 and R3369, represented transcript number 6 in Fig. 2, while band 6 (lanes 10 and 11) observed with the same primer set represented transcripts numbered 7 and 8 in Fig. 2. The transcripts detected with primers F230 and R4854 (numbers 9 to 14, Fig. 2) can similarly be correlated to bands 8, 9, and 10 in Fig. 1. Band 8 represented transcripts 9 and 11, band 9 represented transcripts 10 and 12, and band 10 represented transcripts 13 and 14, respectively. Transcripts identified with primers F230 and R2346 (Fig. 1, lanes 6 and 7, band number 1) were shorter versions (not shown in Fig. 2) of transcripts observed with F230 and R2568 (described above).
The large number of transcripts (13 from seven bands) was explained by the identification of 10 alternatively used splices (Table 1), indicated by letters a to j in Fig. 2. Several clones of similar sized inserts were sequenced to ensure that all possible species of mRNAs in these were included. It is surmised that the more abundant transcripts/splices were detected more often (Table 1).
TABLE 1.
Splice junctions observed with different primer setsa
| Primer set | Splice junction(s) | Splice site(s) | Transcript no. | No. of times observed in sequencing | No. of PCR in which observed |
|---|---|---|---|---|---|
| F230, R1598 | None | 1 | 4 | 1 | |
| F1545, R2568 | None | 1 | 3 | 1 | |
| F230, R2346 | 279/1792 | a | 2 | 3 | 2 |
| 279/1740 | b | 3 | 4 | 2 | |
| 333/1792 | c | 4 | 1 | 1 | |
| 333/1740 | d | 5 | 1 | 1 | |
| F230, R2568 | 279/1792 | a | 2 | 11 | 3 |
| 279/1740 | b | 3 | 14 | 3 | |
| 333/1792 | c | 4 | 1 | 1 | |
| 333/1740 | d | 5 | 3 | 2 | |
| F230, R3369 | 279/3087 | e | 6 | 18 | 4 |
| 279/1740, 2063/3087 | b, f | 7 | 2 | 1 | |
| 279/1792, 2063/3087 | a, f | 8 | 3 | 1 | |
| F230, R4854 | 279/3087, 3210/4640 | e, h | 9 | 3 | 2 |
| 333/3087, 3210/4640 | g, h | 10 | 1 | 1 | |
| 279/3087, 3226/4640 | e, i | 11 | 3 | 1 | |
| 279/3087, 3262/4640 | e, j | 12 | 2 | 1 | |
| 279/1792, 2063/3087, 3210/4640 | a, f, h | 13 | 1 | 1 | |
| 279/1740, 2063/3087, 3210/4640 | b, f, h | 14 | 2 | 1 |
A slash indicates a splice. Transcript numbers correspond to the numbers used in Fig. 2.
An unspliced transcript (number 1, Fig. 2) was detected with two sets of primers (F230 andR1598 and F1545 and R2568). Although R2568 revealed a PCR product (band number 4, lanes 8 and 9, Fig. 1) approximately equal to 2 kb when used with the F230 primer, cloning of this band was unsuccessful. However, PCR with primers F1545 and R2568 yielded an unspliced PCR product. When aligned with the unspliced PCR product generated from primers F230 and R1598, a complete, unspliced PCR product from 230 to 2568 was identified.
Splice sites.
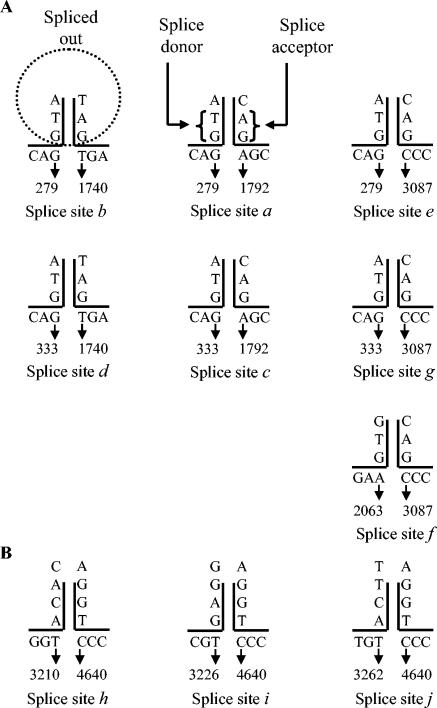
Seven out of 10 splice sites (Fig. 3A) had typical GT-AG donor/acceptor junctions (6, 26). Nonconventional splice donor sites (AC and GA) observed in the case of SPV were at positions 3211, 3227, and 3263, and their common, nonconventional, splice acceptor site (GT) was at position 4638 (Fig. 3B). Peculiarly, these nonconventional donor sites had GT sequences preceding them by two positions (at nucleotide positions 3209, 3225, and 3261) and the nonconventional acceptor site had AG sequences preceding it by two positions (at nucleotide 4636). With evidence of nonconventional splice donors in minute virus of mice and B19 (10, 14), the three nonconventional splice donors and one nonconventional acceptor in SPV are not unique.
FIG. 3.
Conventional (A) and nonconventional (B) splice donor and acceptor sites. Sequences in the proximity of the splice junctions are shown.
Deviations from predictions.
Based on the SPV sequence and similarity with parvovirus B19, Brown et al. predicted a splice acceptor site at nucleotide 3068 for the spliced VP2 coding transcript (7). We observed that the predicted site is spliced out and the actual site utilized is at nucleotide position 3085 (Fig. 2, transcripts numbered 6 to 14, splice sites e, f, and g). In addition, when sequencing the cDNAs obtained from tissue-derived RNA transcripts, nucleotide position 4727 was always found to be cytosine instead of adenine, as was reported in GenBank (accession no. U26342). This change in nucleotide would cause the incorporation of the amino acid proline instead of threonine in the second reading frame (capsid proteins).
Promoter site.
Six consensus RNA transcript promoter sites (TATAA), at nucleotide positions 195, 1140, 1387, 3487, 4189, and 4968 (Fig. 2), are present in the SPV sequence (7). The last site (at nucleotide position 4968) should be nonfunctional unless the virus utilizes a circular replicative form. All transcripts detected in our study begin at the 5′ end of the genome, because the predominant forward primer used was F230. This indicates the use of the promoter at position 195. It is supported by the observation of a highly GC-rich sequence, two SP1 sites upstream of the first TATA box at position 195 (7), promoter activity in the analogous region in B19 (4), and in vitro confirmation with reporter assays (S. W. Green and K. E. Brown, unpublished data). Whether SPV utilizes downstream promoters requires additional study.
Polyadenylation site.
Five sites with the sequence AATAAA were identified in the SPV sequence beginning at nucleotide positions 2449, 2618, 2959, 4948, and 4958 (7). Considering these positions, all spliced partial transcripts seen with primer sets F230 and R3369 and F230 and R4854 (Fig. 2, transcripts numbered 6 to 14) would end at the 3′ end of the genome, utilizing either of the two polyadenylation sites starting at nucleotide 4948 or 4958. Other spliced partial transcripts seen with F230 and R2568 (Fig. 2, transcripts numbered 2 to 5) could use any of the polyadenylation sites after nucleotide position 2568 (nucleotide position 2618, 2959, 4948, or 4958). The unspliced transcript coding for NS1 may end in the middle of the genome, with one of the two polyadenylation signals after nucleotide 2449 (nucleotide 2618 or 2959). The polyadenylation site utilized by these transcripts cannot be accurately deduced from the results obtained to date.
Potential classes of full-length transcripts.
To predict the sizes of bands that would be observed with Northern blotting and autoradiography, the full lengths of the spliced transcripts were estimated (Table 2). It was assumed that partial transcripts would terminate utilizing the polyadenylation signals at the 3′ end of the genome (discussed above) to derive the maximum coding potential of SPV. This step was taken with the understanding that potential variations may be realized when SPV transcripts were investigated with Northern blot analysis, as completed below. The transcripts observed with primer R4854 span almost the entire genome (Fig. 2, transcripts numbered 9 to 14), and hence there would be little variation in the actual size of these full-length transcripts, when compared to the determined estimates. However, as mentioned before and discussed later, variations are possible in transcripts observed with primers R2568 and R3369 (Fig. 2, transcripts numbered 2 to 8).
TABLE 2.
Estimated full-length spliced transcripts and predicted proteinsa
| Transcript no. | Size of partial transcript (bases) | Estimated full length (kb) | Sizes of predicted proteinsb (kDa) |
|---|---|---|---|
| 2 | 827 | 3.37 | 5.4, 9.6, VP1 |
| 3 | 879 | 3.43 | 10.4, 9.6, VP1 |
| 4 | 881 | 3.43 | NS* (22.4), 5.4, VP1 |
| 5 | 933 | 3.48 | 10.4, 9.6, VP1 |
| 6 | 333 | 2.01 | VP2 |
| 7 | 657 | 2.4 | 10.4, VP2 |
| 8 | 605 | 2.35 | 5.4, VP2 |
| 9 | 389 | 0.69 | 13.2, 7.2 |
| 10 | 443 | 0.75 | 13.5, 13.2 |
| 11 | 405 | 0.7 | VP2* (9.6), 7.2 |
| 12 | 441 | 0.74 | VP2* (10.2), 8 |
| 13 | 661 | 0.96 | 5.4, 13.2, 7.2 |
| 14 | 713 | 1 | 10.4, 13.2, 7.2 |
The size of each transcript (numbers used to designate partial transcripts are the same as in Fig. 2) was calculated as the sum of the lengths of exons, leader (estimated 60 bases), trailer (estimated 40 bases), and poly(A) tail (estimated 200 bases). *, truncated protein.
NS = 77 kDa, VP1 = 90 kDa, and VP2 = 60 kDa.
SPV specificity of the amplified, cloned and sequenced cDNA.
The observed sequences of partial transcripts aligned to the SPV sequence. BLAST searches revealed significant alignments to SPV and other Erythrovirus sequences. Repeated observation of a particular sequence (Table 1) supported the existence of the corresponding transcript. With the estimates of full-length transcripts (Table 2) and the splice junctions shared by each of these products (Fig. 2), the number and molecular sizes of bands that would be observed after Northern blot analysis and autoradiography utilizing 32P-labeled, splice junction-specific oligonucleotide probes were predicted (Table 3). Northern blots were interpreted as described below.
TABLE 3.
Summary of splice site analysisa
| Splice site | Transcript no(s). | Band predicted (kb) | Band observed |
|---|---|---|---|
| a | 2 | 3.4 | 3.4 |
| 8 | 2.4 | 2.3 | |
| 13 | 1 | 0.8 | |
| b | 3 | 3.4 | 3.4 |
| 7 | 2.4 | 2.2 | |
| 14 | 1 | 0.9 | |
| c | 4 | 3.4 | 3.4 |
| 2.3 | |||
| 0.8 | |||
| d | 5 | 3.4 | None |
| e | 6 | 2 | 2.2 |
| 9, 11, 12 | 0.7 | 0.8 | |
| f | 7, 8 | 2.4 | 2.4 |
| 13, 14 | 1 | 0.9 | |
| g | 10 | 0.7 | None |
| h | 2.4 | ||
| 13, 14 | 1 | 0.9 | |
| 9, 10 | 0.7 | 0.6 | |
| i | 11 | 0.7 | 0.6 |
| j | 2.2 | ||
| 12 | 0.7 | 0.8 |
Transcript(s) indicates the numbers designated to different transcripts in which splice sites are present (Fig. 2). Bands predicted are the sizes (in kilobases) of bands based on the calculations described in Table 2, footnote a. Bands observed are the sizes (in kilobases) of the bands seen with Northern blot analysis with splice junction-specific probes (Fig. 4). Unpredicted observations are underlined.
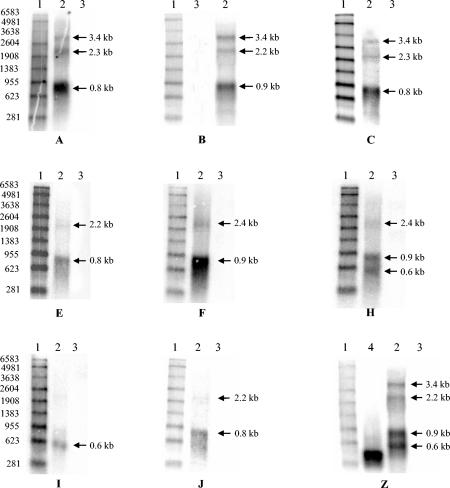
The in vitro-transcribed positive control lane of the blot labeled Z when probed with the reverse complement radiolabeled oligonucleotide detected only a single size-specific band, corresponding to 333 bases (Fig. 4, blot Z, lane 4). However, four transcript classes were detected in the lane with SPV RNA (Fig. 4, lane 2) with the same radiolabeled oligonucleotide probe. These results are consistent with the radiolabeled PCR-amplified DNA copy of the 333-bp partial transcript hybridizing to the four different transcript classes, predicted to have sizes of 3.4, 2.4, 1.0, and 0.7 kb when full length (Fig. 2, Table 2). The hybridization to so many classes is explained by the presence of probe complementary regions in all transcripts.
FIG. 4.
Northern blots. Blots A, B, C, E, F, H, I, and J were probed with splice junction-specific 40-mer probes spanning 20 bases on either side of the splices a, b, c, e, f, h, i, and j, respectively (Fig. 2 and Table 3). Lane 1, radiolabeled markers (sizes mentioned on the left). Lane 2, DNase-treated RNA from SPV-infected tissues. Lane 3, negative control RNA, in most instances from tissues of noninfected macaques and occasionally MA104 or CHO cell RNA. Blot Z was probed with radiolabeled antisense strands of the cloned partial transcript of 333 bases. Lane 4, positive control sense strands of RNA transcribed in vitro from the cloned partial transcript of 333 bases. Arrows indicate predicted band sizes.
When SPV RNA was probed with a radiolabeled, antisense, 40-mer probe spanning 20 bases on either side of the splice site a (Fig. 2), bands of 0.8, 2.3, and 3.4 kb were detected (Fig. 4, blot A, lane 2). These correlated to the predictions as splice site a is shared in partial transcripts numbered 2, 8, and 13 (Fig. 2), the predicted full-length sizes of which approximately equal 1, 2.4 and 3.4 kb (Tables 2 and 3).
Similarly, splice site b is shared in the partial transcripts numbered 3, 7 and 14 (Fig. 2), the predicted full-length sizes of which approximately equal 1, 2.4, and 3.4 kb. When SPV RNA was probed with a radiolabeled, antisense, 40-mer probe spanning 20 bases on either side of the splice site b, bands of sizes 1, 2.4 and 3.4 kb were detected (Fig. 4, blot B, lane 2). The predictions and observations for all probes are summarized in Table 3.
The predicted number and sizes of bands were observed with five of the 10 splice junction-specific probes (correlating to splice sites a, b, e, f, and i, Table 3). Our observations did not correlate to predictions for probes corresponding to the remaining 5 splice sites. These are discussed in some detail below.
DISCUSSION
SPV and other members of the genus Erythrovirus replicate in erythroid precursor cells. Their replication is difficult to study in vitro because of the need for explant bone culture. Also, analyses with explant cultures of bone marrow or fetal liver cells do not provide a real account of host-viral interaction. Hence, the findings of this study, in which transcripts formed in vivo by SPV were investigated, are significant.
The transcription maps for B19 and minute virus of mice were originally produced with S1 nuclease mapping (14, 23). Nuclease protection assays have a detection limit of 4,000 to 5,000 copies of mRNA. Reverse transcription-PCR is presently the most sensitive method available for mRNA detection. The procedure is somewhat tolerant of degraded RNA, i.e., as long as the RNA is intact within the region spanned by the primers, the target will be amplified. A major disadvantage with reverse transcription-PCR is observation of artifacts. It was used considering the high sensitivity and limited amount of macaque tissue available. The combination of cDNA sequencing and Northern blot analysis helped us better analyze our results.
No bands were detected with probes correlating to the splice sites d and g (Table 3 and data not shown). Splice sites d and g appear to be utilized by less abundant transcripts among those observed (numbered 5 and 10, Fig. 2, observations summarized in Table 1). It is possible that their abundance is so low that they could not be visualized with Northern blot analysis. Another possibility is that they are PCR artifacts.
The partial transcript with splice site d putatively encodes VP1. The corresponding RNA species encoding VP1 in B19 was the least abundant among those observed, and this was noted to be consistent with the relative expression of the protein (23). This may be explained by the fact that VP1 comprises only about 5% of the capsid proteins (9). Since RNA was extracted from in vivo-derived tissues from infected macaques in this study, it is possible that transcripts encoding only VP1 were of such low abundance that they could not be detected. Alternatively, the VP1 encoding transcript could be utilizing a downstream promoter.
When estimating the full-lengths of spliced partial transcripts observed by Northern blot analysis, it was assumed that the transcripts observed would terminate at the 3′ end of the genome. However, transcripts observed with primer R2568 (Fig. 2) might only code for small ORFs and end before coding for the capsid proteins by utilizing an earlier polyadenylation signal. These would then be similar to B19 transcripts ending in the middle of the genome (Fig. 5). This hypothesis is supported by the observation of strong bands of 0.8 or 0.9 kb with the probes spanning splice sites a and c (Fig. 4), and could explain the observation of smaller, unpredicted bands, as seen with the probe corresponding to splice site c (Table 3).
FIG. 5.
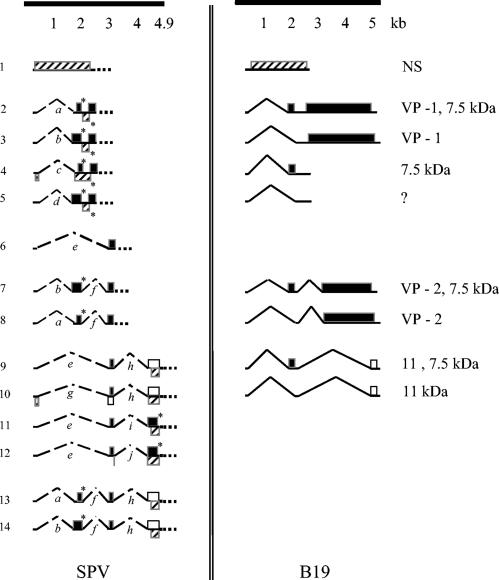
Comparison of the messages encoded by SPV and B19 based on the splice junctions observed. The method of representation of transcripts and ORFs putatively encoded by SPV is the same as in Fig. 2. The transcription map of B19 on the right is based on the data from Luo et al. (17) and Ozawa et al. (23), with all ORFs indicating completely translated proteins (no asterisks).
Alternatively, additional splice sites could shorten the length of the transcripts. This is easily understood in case of transcripts observed with primer R2568 and could also be true for transcripts observed with primer R3369 (Fig. 2). However, because the exons of transcripts observed with primer R3369 include the splice donor sites of splice site h or i or j, it appears to be less likely that additional splice sites would be detected with primer R3369.
The observation of faint, unpredicted bands with probes corresponding to splice sites h and j may be a result of non-splice-specific binding of the probes. Northern blots with each probe were repeated two to three times to minimize procedural errors, but the possibility cannot be negated.
Some features of the SPV transcripts detected are analogous to those of B19. The initial splice junctions for SPV are at nucleotides 279 and 333 (Fig. 2). These are similar to nucleotide 406 and nucleotide 441 for B19 (10) as described by Brunstein et al. (variant splice sites of B19 not shown in Fig. 5). Brunstein et al. had noted this variation in splicing-pattern with reverse transcription-PCR. This variant has not been detected in case of B19 in studies with other techniques. Hence, it is possible that such splice variants are low in abundance and hence, only detected with a technique as sensitive as reverse transcription-PCR.
As shown in Fig. 5, and in agreement with the predictions of Brown et al. (7), an unspliced SPV message putatively coding for NS (transcript number 1) is present in the 5′ end of the genome. Also, depending on the use of the polyadenylation signal, partial transcripts observed with primer R2568 (numbers 2 to 5) may terminate in the middle of the genome. These would then be analogous to smaller transcripts of B19, such as one encoding the ORF for a 7.5-kDa protein (Fig. 5). Smaller putative ORFs (for 5.4- and 10.4-kDa proteins) are also encoded by transcripts numbered 13 and 14, respectively in the left half of the genome. These may represent small nonstructural proteins as seen with B19 (17). Putative ORFs for the capsid proteins (incomplete ORFs followed by dotted lines in transcripts numbered 2 to 8, Fig. 5) are in the 3′ end of the genome. Both SPV and B19 have a small overlap of sequences encoding the large 5′ ORF for NS (ORF ends at nucleotide position 2370 in SPV and 2448 in B19) and the large 3′ ORF for capsid proteins (ORF begins at nucleotide position 2363 in SPV and at 2444 in B19) (7, 23).
Considering the above-mentioned observations, SPV and B19 appear closely related evolutionarily. However, some differences were also noted. SPV has zero to three splices per transcript, compared to zero to two reported for B19 (23) (Fig. 5). The spliced transcript putatively encoding ORF for the truncated NS protein (transcript number 4) is more like the transcripts encoding ORFs for Rep 68/40 of adeno-associated virus 2 and NS2 of minute virus of mice (14, 16). An identical spliced transcript was observed in studies of SPV infection of human bone marrow (8), suggesting that this is not an artifact. Antibodies that bind to the carboxyl end of the SPV NS protein are currently not available, and we were unable to confirm production of this truncated NS protein by Western blotting. In case of the capsid protein encoding transcripts, B19 has a double-spliced message with an ORF only for VP2 (Fig. 5). The SPV transcript putatively encoding only for VP2 was single-spliced (transcript number 6). Also contrary to B19, SPV did not appear to utilize a transcript encoding VP1 alone, although the reverse transcription-PCR product for such a transcript may have been too low in abundance for detection in this study.
Sequence analysis of observed transcripts with Mapdraw also revealed additional ORFs encoding unidentified proteins (possible truncated versions of capsid proteins and the nonstructural protein, and small, unknown proteins) (Fig. 2 and 5). Interestingly, SPV switches frame after splice junctions in many cases. Putative ORFs of unidentified proteins that did not end before nucleotide position 4854 for transcripts numbered 9 to 14 (Fig. 2) could utilize the stop codon at nucleotide position 4870 in the first reading frame, or the stop codon at nucleotide position 4950 in the third reading frame. Molecular sizes for proteins have been calculated accordingly (Table 2).
Predicted amino acid analysis of the deduced complete transcripts indicated that potential translational units begin at nuleotide positions 307, 1781, 1916, 2113, 2363, 3149, 3261, and 4678 (Fig. 2). Brown et al. predicted the sequences encoding major SPV proteins NS = 307 to 2370 (≈2.1 kb), VP1 = 2363 to 4819 (≈2.5 kb), and VP2 = 3149 to 4819 (≈1.7 kb), and confirmed the expression of capsid proteins with Western blot analysis (7). Western blotting of SPV-infected fetal liver with rabbit polyclonal antibody raised against SPV V1, SPV VP2, and a B19 NS peptide confirmed the presence of 90 kDa, 60 kDa and 77 kDa proteins, respectively (data not shown). In addition, the VP2 encoding sequence was recently also used to express the protein in a baculovirus system. Virus-like particles that agglutinated red blood cells were successfully produced (8). This again supports the previous predictions and the use of ATG as the start codon by SPV.
The Kozak consensus sequence for initiation of translation in vertebrates is (GCC) GCCRCCATGG, where R is a purine (A or G) (15). Ozawa et al. indicated a part of the sequence (purine-NNATG) when they determined the transcription map of B19 (23). Only two of the putative ORFs observed in this study on SPV have the sequence “purine-NN-ATG-G”, i.e., have a second G. These two ORFs putatively encode NS1 and VP2. None of the other putative ORFs have the second G. The initial portion of the sequence as quoted by Kozak [(GCC) GCCRCC] is absent in all putative ORFs. On the other hand, 6 out of 8 putative ORFs (beginning at nucleotide positions 307, 1781, 2363, 3149, 3261, and 4678) have the sequence “purine-NN-ATG”. This may be explained by a recent analysis in which it was observed that a large number of start codons used for translation initiation deviate significantly from Kozak's sequence (24). It was suggested that other means used to translate proteins, including leaky scanning, reinitiation, or internal initiation of translation may have greater roles than previously imagined.
SPV seems to utilize a diversified splicing mechanism for its transcripts. Variation in transcripts and their coding potential could be due to splicing, promoter strength and differential termination. The reverse transcription-PCR approach used in this study was successful in amplifying SPV RNA, but the larger PCR products could not be cloned/sequenced. The cloning approach favored the identification of smaller, highly spliced products. Compared to B19, which was shown to have 9 transcripts (23), 14 putative messages were observed (Fig. 5). Most of the work on transcription of the prototype parvoviruses has to date not employed this technique and hence the use of various splice sites had only been predicted. As was suggested by Morgan et al. and Jongeneel et al. based on their work on minute virus of mice, the use of alternate nearby splice donor and acceptor sites can generate large numbers of transcripts that can be detected by cDNA cloning only (14, 19). This map of splice junctions is in agreement with their prediction. Even though functionality of these junctions needs confirmation with further studies, SPV may be efficiently switching reading frames in vivo with extensive use of alternative splicing to make different proteins in infected cells and exemplifies the elegant splicing pattern that parvoviruses may use.
Although incomplete, this splice junction map provides substantial information on the splice junctions in SPV transcripts. The splice junction map suggests that the general transcription pattern for SPV is similar to that of B19 with transcripts originating from 5′ end of the genome (Fig. 5). The abundance of transcripts with ORFs for putative capsid proteins indicates permissive infection of cells from which the RNA was extracted.
Acknowledgments
This work was supported by Public Health Service grants RR14098 and HD34364 from the National Center for Research Resources and the National Institute of Child Health and Human Development, respectively, of the National Institutes of Health, Bethesda, Md.
REFERENCES
- 1.Astell, C. R., M. Thomson, M. Merchlinsky, and D. C. Ward. 1983. The complete DNA sequence of minute virus of mice, an autonomous parvovirus. Nucleic Acids Res. 11:999-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard, C., J. St Amand, and C. R. Astell. 1989. Transient expression of B19 parvovirus gene products in COS-7 cells transfected with B19-SV40 hybrid vectors. Virology 172:659-664. [DOI] [PubMed] [Google Scholar]
- 3.Berns, K. I., M. Bergoin, M. Bloom, M. Lederman, N. Muzyczka, G. Siegl, J. Tal, and P. Tattersall. 2000. Family Parvoviridae, p. 311-323. In M. H. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses: seventh report of the International Committee on Taxonomy of Viruses. Virology Division, International Union of Microbiological Societies, San Diego, Calif.
- 4.Blundell, M. C., and C. R. Astell. 1989. A GC-box motif upstream of the B19 parvovirus unique promoter is important for in vitro transcription. J. Virol. 63:4814-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blundell, M. C., C. Beard, and C. R. Astell. 1987. In vitro identification of a B19 parvovirus promoter. Virology 157:534-538. [DOI] [PubMed] [Google Scholar]
- 6.Breathnach, R., C. Benoist, K. O’Hare, F. Gannon, and P. Chambon. 1978. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. USA 75:4853-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, K. E., S. W. Green, M. G. OSullivan, and N. S. Young. 1995. Cloning and sequencing of the simian parvovirus genome. Virology 210:314-322. [DOI] [PubMed] [Google Scholar]
- 8.Brown, K. E., Z. Liu, G. Gallinella, S. Wong, I. P. Mills, and M. G. O’Sullivan. Simian parvovirus (SPV) infection: a potential zoonosis. J. Infect. Dis., in press. [DOI] [PubMed]
- 9.Brown, K. E., N. S. Young, and J. M. Liu. 1994. Molecular, cellular and clinical aspects of parvovirus B19 infection. Crit. Rev. Oncol. Hematol. 16:1-31. [DOI] [PubMed] [Google Scholar]
- 10.Brunstein, J., M. Söderlund-Venermo, and K. Hedman. 2000. Identification of a novel RNA splicing pattern as a basis of restricted cell tropism of erythrovirus B19. Virology 274:284-291. [DOI] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., and P. Tattersall. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 226:1161-1165. [DOI] [PubMed] [Google Scholar]
- 12.Doerig, C., P. Beard, and B. Hirt. 1987. A transcriptional promoter of the human parvovirus B19 active in vitro and in vivo. Virology 157:539-542. [DOI] [PubMed] [Google Scholar]
- 13.Green, M. R., and R. G. Roeder. 1980. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell 22:231-242. [DOI] [PubMed] [Google Scholar]
- 14.Jongeneel, C. V., R. Sahli, G. K. McMaster, and B. Hirt. 1986. A precise map of splice junctions in the mRNAs of minute virus of mice, an autonomous parvovirus. J. Virol. 59:564-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, M. 1984. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12:857-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laughlin, C. A., H. Westphal, and B. J. Carter. 1979. Spliced adenovirus-associated virus RNA. Proc. Natl. Acad. Sci. USA 76:5567-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, W., and C. R. Astell. 1993. A novel protein encoded by small RNAs of parvovirus B19. Virology 195:448-455. [DOI] [PubMed] [Google Scholar]
- 18.Lusby, E. W., and K. I. Berns. 1982. Mapping of the 5′ termini of two adeno-associated virus 2 RNAs in the left half of the genome. J. Virol. 41:518-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, W. R., and D. C. Ward. 1986. Three splicing patterns are used to excise the small intron common to all minute virus of mice RNAs. J. Virol. 60:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Sullivan, M. G., D. C. Anderson, J. D. Fikes, F. T. Bain, C. S. Carlson, S. W. Green, N. S. Young, and K. E. Brown. 1994. Identification of a novel simian parvovirus in cynomolgus monkeys with severe anemia. A paradigm of human B19 parvovirus infection. J. Clin. Investig. 93:1571-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Sullivan, M. G., D. K. Anderson, J. A. Goodrich, H. Tulli, S. W. Green, N. S. Young, and K. E. Brown. 1997. Experimental infection of cynomolgus monkeys with simian parvovirus. J. Virol. 71:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Sullivan, M. G., D. K. Anderson, J. E. Lund, W. P. Brown, S. W. Green, N. S. Young, and K. E. Brown. 1996. Clinical and epidemiological features of simian parvovirus infection in cynomolgus macaques with severe anemia. Lab. Anim. Sci. 46:291-297. [PubMed] [Google Scholar]
- 23.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 61:2395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peri, S., and A. Pandey. 2001. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 17:685-687. [DOI] [PubMed] [Google Scholar]
- 25.Pintel, D., D. Dadachanji, C. R. Astell, and D. C. Ward. 1983. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 11:1019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seif, I., G. Khoury, and R. Dhar. 1979. BKV splice sequences based on analysis of preferred donor and acceptor sites. Nucleic Acids Res. 6:3387-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]