Abstract
We previously demonstrated that after vaccination of BALB/c mice with DNA encoding murine cytomegalovirus (MCMV) IE1 or M84, a similar level of protection against MCMV infection was achieved. However, the percentage of antigen-specific CD8+ T cells elicited by IE1 was higher than that by M84 as measured by intracellular cytokine staining when splenocytes were stimulated with an epitope peptide (M. Ye at al., J. Virol. 76:2100-2112, 2002). We show here that after DNA vaccination with M84, a higher percentage of M84-specific CD8+ T cells was detected when splenocytes were stimulated with J774 cells expressing full-length M84. When the defined M84 epitope 297-305 was deleted, the mutant DNA vaccine was still protective against MCMV replication and induced strong M84-specific CD8+-T-cell responses. The M84 gene was subsequently subcloned into three fragments encoding overlapping protein fragments. When mice were immunized with each of the M84 subfragment DNAs, at least two additional protective CD8+-T-cell epitopes were detected. In contrast to strong responses after DNA vaccination, M84-specific CD8+-T-cell responses were poorly induced during MCMV infection. The weak M84-specific response after MCMV infection was not due to poor antigen presentation in antigen-presenting cells, since both J774 macrophages and primary peritoneal macrophages infected with MCMV in vitro were able to efficiently and constitutively present M84-specific epitopes starting at the early phase of infection. These results indicate that antigen presentation by macrophages is not sufficient for M84-specific CD8+-T-cell responses during MCMV infection.
The cytomegaloviruses (CMVs) are double-stranded DNA viruses that can establish persistent and latent infection in the host. Although infection in immunocompetent individual is usually benign, human CMV (HCMV) is responsible for significant morbidity and mortality in immunosuppressed individuals (28). Because CMV is highly species specific, there are no animal models available to study directly the pathogenesis of HCMV and to test preventive strategies. Murine CMV (MCMV) greatly resembles its human counterpart with respect to the organization and expression of their genomes and host immune response to the viral gene products. After acute infection, it also establishes latency and latent virus can be reactivated under various conditions of immunosuppression. Therefore, this animal model has been very useful for the development and testing of strategies for prevention and treatment of CMV infection.
Like HCMV, MCMV encodes several proteins that are very important in controlling the infection of the virus through the induction of adaptive immune responses in its host. Some of the gene products have homologs in HCMV, whereas others are unique to MCMV. Although both humoral and cell-mediated immune responses are induced during MCMV infection, the cell-mediated immune responses play a central role in the control of the CMV infection. The importance of the CD8+ T cells has been aptly documented in a mouse model of bone marrow transplantation in which adoptive transfer of CD8+ cytotoxic T lymphocytes (CTLs) from MCMV-infected mice into immunocompromised infected recipients protects against lethal disease (29, 31, 36). To date, seven gene products, namely, IE1 (pp89), M83, M84, m164, m18, m04 (gp34), and M45, have been reported to elicit cell-mediated immune responses during MCMV infection (5, 9, 10, 14-17). Of particular interest, MCMV IE1 is a functional homolog and M84 and M83 are sequence homologs of HCMV dominant antigens IE1 and pp65, respectively.
The MCMV immediate-early (IE) protein IE1 was the first gene product identified to elicit CTL responses in mice (32). The nonapeptide 168YPHFMPTNL176 of IE1 is the dominant CTL epitope in infected BALB/c mice and plays a significant role in controlling the replication of the virus (4, 11, 15, 33). The MCMV M83 and M84 genes are both homologs of the HCMV UL83 gene, which encodes pp65 (2). The M83 protein is expressed with early-late gene kinetics and is present in the virion, whereas the M84 gene product is a nonstructural protein that is expressed at early times in the infection (2, 25). Interestingly, the M84 open reading frame (ORF) of MCMV possesses greater homology to the HCMV UL83 ORF than does the positional homolog M83 (2). Although HCMV pp65 is a dominant immunogen, both MCMV M83 and M84 are less immunogenic during MCMV infection. One Ld-restricted epitope in the 105-kDa M83 gene product (761YPSKEPFNF769) and one Kd-restricted epitope of the 65-kDa M84 gene product (297AYAGLFTPL305) have been identified (12, 16). Polyclonal cell lines generated against either M83 epitope 761-769 or M84 epitope 297-305 have been shown to confer protection against MCMV replication in adoptive-transfer experiments (12). The M83-specific T-cell lines appeared to be more protective than the M84-specific T-cell lines. This observation is in contrast to our previous study showing that immunization of mice with DNA encoding M84, but not M83, provided strong protection against viral replication in the spleen when the mice were challenged with MCMV (26).
In past years, our laboratory has been using MCMV infection as a model to explore vaccine strategies against CMV replication. By using DNA-based vaccines, we demonstrated that BALB/c mice immunized intradermally (i.d.) with plasmids expressing IE1 and M84 were similarly protected against subsequent MCMV challenge, as measured by the reduction of virus titers in the spleen (6, 26). Moreover, a synergistic effect was observed when mice were coimmunized with IE1 and M84. Since both IE1 and M84 are nonstructural proteins, the cellular immune response is primarily responsible for the protection provided by DNA vaccine immunizations. Using the technology of intracellular cytokine staining (ICCS) assay, as well as traditional CTL lysis assay, a strong IE1 epitope (168-176)-specific CTL response was determined (6, 37). Approximately 4 to 5% of the spleen CD8+ T cells were IE1 specific at 10 days after the last immunization. When the known M84 epitope (297-305) was used in the ICCS assay to measure the CTL response after DNA-based vaccine immunization, a significantly lower percentage (1 to 2%) of M84 epitope-specific CD8+ T cells was detected. These results raised the question of why there were major differences in the CTL responses between the two vaccines, whereas both displayed similar protective effects against a MCMV challenge.
In the present study, we further investigated the M84-specific CTL response generated after i.d. DNA immunization by using a modified ICCS assay in which antigen-presenting cells expressing full-length M84 were used as a stimulator. Using this modified ICCS assay, a much higher percentage of M84-specific CD8+ T cells were detected compared to the response to the single defined epitope. By immunizing mice with plasmids expressing subfragments of the M84 gene, we also demonstrated that there are at least two additional CD8+-T-cell epitopes in M84. These results provide an explanation for the correlation between the strength of immune responses and the viral protection following DNA-based vaccine immunization. We also reinvestigated the IE1- and M84-specific CD8+-T-cell responses after MCMV infection of BALB/c mice with the modified ICCS assay. Although we were able to detect a modest increase in the percentage of M84-specific CD8+ T cells when the additional epitopes were used in the assay, indicating that the additional epitopes contributed to the response to M84 during infection, the overall response remains low relative to that after M84 DNA immunization. The lack of response after infection was not due to a defect in presentation, since we found that the M84 epitopes were efficiently presented by BALB/c primary macrophage at early times in the MCMV infection.
MATERIALS AND METHODS
Mice.
Female BALB/c mice that were 3 to 4 weeks old were purchased from Harlan Sprague-Dawley, Inc. All mice were housed in microisolator-covered cages in the Pacific Hall vivarium at the University of California, San Diego. After arrival, mice were allowed to acclimate to the facility for 1 to 2 weeks before any experiments were performed.
Cell culture.
Culture and maintenance of NIH 3T3 cells (ATCC CRL-1658) and COS-7 cells (ATCC CRL-1651) were described previously (37). HeLa cells (ATCC CCL-2) were maintained at 37°C in 7% CO2 in minimal essential medium (MEM) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine. J774A.1 (ATCC CRL TIB-67) macrophages (H-2d) and BALB/c mouse embryo fibroblasts (MEFs) were maintained as described previously (27).
BALB/c peritoneal macrophages were prepared as described previously (21). Briefly, each of four mice per experiment was injected with 2 ml of thioglycolate intraperitoneally (i.p.). Three days after injection, mice were sacrificed and 8 ml of cold phosphate-buffered saline was injected i.p. with a 25-G needle. The peritoneal skin was subsequently opened, and the intact peritoneal cavity was exposed. The peritoneal fluid was recovered with a 20-G needle. The peritoneal macrophage containing fluid from four mice was pooled and pelleted at 400 × g at 4°C for 10 min. After one wash with 10 ml of RP-10 medium (34), macrophages were resuspended in RP-10 and counted. The resulting cells were seeded at 6 × 106 per 10-cm tissue culture dishes for the antigen presentation assay.
Virus propagation.
MCMV strain K181 was grown in MEFs or NIH 3T3 cells (tissue culture virus) or in vivo in the salivary glands of BALB/c mice (salivary gland-derived virus) and stored at −80°C as described previously (37).
The recombinant vaccinia viruses pp89-vacc and M84-vacc, which express MCMV IE1 and M84, respectively, were constructed as described previously (26). Wild-type and recombinant vaccinia viruses were propagated in HeLa cells as described previously with minor modifications (2, 18). Briefly, 107 HeLa cells were seeded in each T185 flask 1 day before infection such that each flask contained 1.4 × 107 cells the next day. For infection, 0.5 ml of 0.25% trypsin was added to 3 × 108 PFU of virus stock, and the virus mixture was incubated at 37°C for 30 min with vortexing every 10 to 15 min. The virus was then diluted to 10 ml with MEM containing 5% FBS and 2 mM l-glutamine, and 5 ml of diluted virus was added to each of two flasks such that a multiplicity of infection (MOI) of 10 was attained. After incubation at 37°C for 1 h with rocking every 20 min, the inoculum was removed, and the cells were washed once with MEM and refed with 15 ml of HeLa cell medium. When 80% of the cells displayed a cytopathic effect (typically 18 to 20 h p.i.), the medium was removed, and the cells were washed once with MEM containing 5% FBS (MEM-5). The washed cells were dislodged into 5 ml of fresh MEM-5 by using a rubber policeman, and the cells from two T-185 flasks were combined into 15 ml of medium and pelleted. After 10 ml of the supernatant was set aside, the cell pellet was resuspended in the remaining 5 ml of supernatant and subjected to three freeze-thaw cycles in a dry ice-ethanol bath. The cell suspension was then homogenized with 20 to 30 strokes in a glass Dounce homogenizer and added to the 10 ml of supernatant that had been set aside. The virus preparation usually had a titer on HeLa cells of >108 PFU/ml and was stored at −80°C in 1-ml aliquots.
Construction of pc3-pp89ΔLd, pc3-M84ΔKd, pc3-NFrg, pc3-MFrg, pc3-ΔKdMFrg, and pc3-CFrg plasmids.
A recombinant plasmid expressing a mutant IE1 with a deletion of the known Ld-restricted epitope 168-176 sequence was derived from pc3-pp89 (37) by QuikChange (Stratagene) site-directed mutagenesis according to the manufacturer's recommendations. The primers consisted of nucleotides immediately upstream and downstream of the epitope sequence. The sequences of all complementary primer sets used for mutagenesis are listed in Table 1. A silent mutation of G to A in the primer pp89ΔLd Sense (underlined in the sequence in Table 1) resulted in elimination of an ApaI site. This was done to facilitate the screening of clones. The resulting plasmid expressing mutant IE1 without the 168-176 epitope was named pc3-pp89ΔLd. Similarly, the defined Kd-restricted epitope 297-305 in M84 was deleted from pcDNA3-M84 (2) by site-directed mutagenesis with another complementary primer pair (Table 1) to yield pc3-M84ΔKd. To construct a recombinant plasmid expressing the amino-terminal one-third of M84, a stop codon was inserted into the M84 gene at the end of amino acid (aa) 194 by site-directed mutagenesis. A ClaI restriction site immediately downstream of the stop codon was introduced to assist in clone selection and is underlined in the sequence (Table 1). The plasmid pc3-M84 (37) was used as a template, and the resulting plasmid was named pc3-NFrg. Similarly, pc3-MFrg was constructed to express the middle one-third of the M84 gene from aa 187 to 396, pc3-CFrg was constructed to express the carboxy-terminal one third of the M84 gene from aa 389 to 587, and pc3-ΔKdMFrg was constructed to express the middle one third of the M84 gene from aa 187 to 396 that has a deletion of the defined epitope 297-305. The corresponding DNA fragments were derived from pc3-M84 or pc3-M84ΔKd by PCR and subcloned into HindIII and EcoRI sites (underlined in Table 1) of the vector pc3Δneo (37). The complete nucleotide sequences of the mutant IE1 and M84 genes, as well as genes coding for the subfragments of M84, were verified by sequencing (University of California at San Diego Cancer Center DNA Sequencing Shared Resource). Of particular note, the putative amino acid sequence of the M84 gene of strain K181 was identical to that published for Smith strain (30), except for a Gly (Smith)-to-Glu (K181) mutation at position 44, and a repeated Leu-Gln in the K181 sequence after Smith position 564.
TABLE 1.
Nucleotide sequences of primers used for construction of recombinant plasmids
| Primer | Nucleotide sequence |
|---|---|
| pp89ΔLd sense | 5′-CTCATGTATGACATGGGACCCTCAGAAAAGAGAG-3′ |
| pp89ΔLd antisense | 5′-CTCTCTTTTCTGAGGGTCCCATGTCATACATGAG-3′ |
| M84ΔKd sense | 5′-CTACTTCTCGAGGGCCTACCGTCCGGGGTTGAGC-3′ |
| M84ΔKd antisense | 5′-GCTCAACCCCGGACGGTAGGCCCTCGAGAAGTAG-3′ |
| M84 NFrg sense | 5′-CATAGCGCACGTCCTCTCC TGA ATCGAT TGCTCGCACTCGGACATC-3′ |
| M84 NFrg antisense | 5′-GATGTCCGAGTGCGAGCA ATCGAT TCA GGAGAGGACGTGCGCTATG-3′ |
| M84 MFrg sense | 5′-CCAAGCTT GGTACCGAGCTCGGATATGTTCCACATAGCGCACGTCCTC-3′ |
| M84 MFrg antisense | 5′-CTGCA GAATTC CAGCACACTGGCG TCA CGAGGGTCTCCGGTGTGCCGG-3′ |
| M84 CFrg sense | 5′-CCAAGCTTGGTACCGAGCTCGGAT ATG CCGCCGGCACACCGGAGACC-3′ |
| M84 CFrg antisense | 5′-CTGCAGAATTCCAGCACACTGGCG TCA GATGTTCTGCTGCAGCC-3′ |
The expression of gene products from the above constructs was confirmed by Effectene-mediated transfection of plasmids into COS-7 cells according to the manufacturer's recommendations (Qiagen). Two days after transfection, cells were lysed in 20 μl of loading buffer per 40,000 cells and then sonicated. Cell lysate proteins were resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) and transferred onto a nitrocellulose membrane. The expression of pp89ΔLd was confirmed by Western blotting with an anti-MCMV hyperimmune serum (data not shown), and expression of M84ΔKd, M84 NFrg, M84 MFrg, M84ΔKdMFrg and M84 CFrg was similarly confirmed by Western blotting with an affinity-purified rabbit anti-GST-M84 serum (25, 37). Bound antibodies were detected by using the SuperSignal West Pico kit (Pierce), followed by fluorography according to the manufacturer's recommendations.
Immunization and challenge of mice with MCMV and plaque assay.
Mice were DNA immunized i.d. and challenged i.p. with MCMV as described previously (37). Virus titers in the spleen were measured by plaque assay on NIH 3T3 cells in 24-well or 10-cm dishes as described previously (27).
ICCS assay.
An ICCS assay in which epitope peptides were used to stimulate splenocytes prior to fluorescence-activated cell sorting (FACS)-based enumeration of gamma interferon (IFN-γ)-synthesizing CD8+ T cells was performed as described previously (37). To perform a cell-mediated ICCS assay, J774 cells in 6-cm dishes were infected at an MOI of 10 with recombinant vaccinia virus expressing M84 or IE1 for 10 h. The infected cells were then placed at 4°C for 30 min and dislodged by using a cell scraper. After counting, 100 μl of responder splenocytes were mixed with 100 μl of vaccinia virus infected J774 cells at an optimal ratio of 5:1 (splenocyte/J774 cell). The cell mixture was incubated at 37°C for 7 h in the presence of brefeldin A (GolgiPlug; BD Bioscience catalog no. 2301kz). Background cytokine staining was determined by mixing splenocytes with J774 cells that had been infected similarly with wild-type vaccinia virus (strain WR). The cells were then pelleted and resuspended in FACS buffer (phosphate-buffered saline supplemented with 2% FBS and 0.09% sodium azide) containing 1:100-diluted Fc Block (Pharmingen catalog no. 553142). To eliminate nonspecific binding of antibodies, the cells were incubated with Fc Block (Pharmingen) on ice for 15 min and then pelleted and resuspended in FACS buffer containing 1:250-diluted fluorescein isothiocyanate-conjugated anti-CD8a monoclonal antibody (Pharmingen catalog no. 553031). Cells were incubated on ice for 30 min, washed twice with FACS buffer, and then fixed and permeabilized by using the Cytofix/Cytoperm kit as suggested by the manufacturer (Pharmingen catalog no. 554714). The intracellular IFN-γ was detected by incubating the cells on ice for 30 min with a 1:100 dilution of R-phycoerythrin-conjugated rat monoclonal antibody to mouse IFN-γ (Pharmingen catalog no. 18115A). After the cells were washed with Cytoperm/Cytowash and FACS buffer, they were resuspended in FACS buffer. The stained cells were analyzed by Elite flow cytometer (Beckman Coulter) at the Flow Cytometry Core, VA Medical Center, La Jolla, Calif. Typically, 300,000 cell events of the small lymphocyte population were analyzed. IFN-γ-antigen positive CD8+ T cells were enumerated by using Beckman Coulter cytometer software and were expressed as a percentage of the total CD8+ T cells.
Antigen presentation assay.
To perform the antigen presentation assay, responder splenocytes were isolated from a group of three mice immunized i.d. three times with pc3-M84 or pc3-pp89 at 10-day intervals. Stimulator cells consisted of MEF, J774, or freshly isolated BALB/c peritoneal macrophages. Stimulator cells were infected with MCMV under various conditions and used for the cell-mediated ICCS assay as described above. Cells presenting IE1 or M84 epitopes on their major histocompatibility complex (MHC) class I molecules stimulate responder splenocytes. In the presence of brefeldin A, the latter synthesize and accumulate IFN-γ inside the cells. Therefore, the level of stimulation can be evaluated by measuring the percentage of responder cells that are IFN-γ positive. To prepare stimulator cells, 6 × 106 J774 or peritoneal macrophages or 5 × 106 MEFs were seeded into 6-cm dishes. For stimulator cells infected with MCMV for 24 h, one set of dishes was infected with MCMV at an MOI of 5 with centrifugal enhancement at 800 × g for 30 min (8). At 8 h postinfection (p.i.), two other plates were infected with MCMV at an MOI of 5 with centrifugal enhancement for the16-h time point. One plate contained 250 μg of phosphonoacetic acid (PAA) per ml to inhibit viral DNA replication and the expression of late viral genes. At 15 to 16 h p.i., another dish was infected with MCMV at an MOI of 5 and served as an 8- or 9-h p.i. time point. For the rest of the time points under the various infection conditions, the infection order was staggered to avoid the simultaneous processing of an excessive number of samples. For infection of stimulator cells with UV-inactivated MCMV, MCMV was diluted in 5 ml of medium with 5% FBS in a 6-cm dish and exposed with 9,999 J of UV light by using a Stratalinker (Stratagene). The resulting virus was used to infect a dish of stimulator cells at a pre-UV inactivation MOI of 5. To prepare stimulator cells expressing enhanced levels of immediate-early gene only, cells were infected with MCMV at an MOI of 5 in the presence of 50 μg of cycloheximide (ChX) per ml, incubated for 2.5 h, washed three times with medium, fed with fresh medium containing 5 μg of actinomycin D (ActD) per ml, and incubated an additional 3 h. At the end of each time point, cells were placed on ice and then dislodged with a cell scraper. After the cells were washed and counted, they were resuspended in 4°C medium containing GolgiPlug and used in the 7-h cell-mediated ICCS assay described above.
Statistical analysis.
One-way analysis of variance was performed on MCMV titer and CD8+-T-cell data that had three or more groups, and a Fisher protected-least-significant-difference (PLSD) test was used as the post hoc test for pairwise comparisons. Similarly, repeated-measures analysis of variance was performed for paired data in three or more groups, and the subsequent post hoc tests used were pairwise, two-tailed paired t tests with the Bonferroni correction for multiple comparisons. In the cases of a single pairwise comparison, paired or unpaired, two-tailed Student t tests were performed. Analyses were performed by using StatView 4.5 software for Macintosh and statistical significance was achieved when the P value was <0.05.
RESULTS
Strong M84-specific CD8+-T-cell responses after DNA immunization.
We previously demonstrated that BALB/c mice that were immunized with pc3-pp89 or -M84 were similarly protected against MCMV replication after sublethal viral challenge (37). When the levels of IE1- and M84-specific CD8+ T cells were examined by an ICCS assay, a significantly higher percentage of IE1-specific CD8+ T cells was detected. In that report, the percentages of IE1- and M84-specific CD8+ T cells were determined by enumerating CD8+ T cells synthesizing IFN-γ after incubation of splenocytes with the known CD8+ T-cell epitope peptides of IE1 (i.e., 168-176) or M84 (i.e., 297-305) (16, 33). It has been demonstrated that there exists only a single CD8+-T-cell epitope in IE1 (presented by MHC class I molecules of the H-2d haplotype) that is responsible for the protection observed following immunization with a recombinant vaccinia virus expressing IE1 (4). M84-specific CD8+-T-cell responses vary significantly from mouse to mouse after immunization and are generally far lower than that elicited by IE1 DNA immunization. The protective CD8+-T-cell epitope in M84 was also found to be only weakly immunogenic in MCMV-infected mice (12, 16, 37). Therefore, we reasoned that there were probably other protective CD8+-T-cell epitope(s) that contributed to the strong protection observed following M84 DNA immunization.
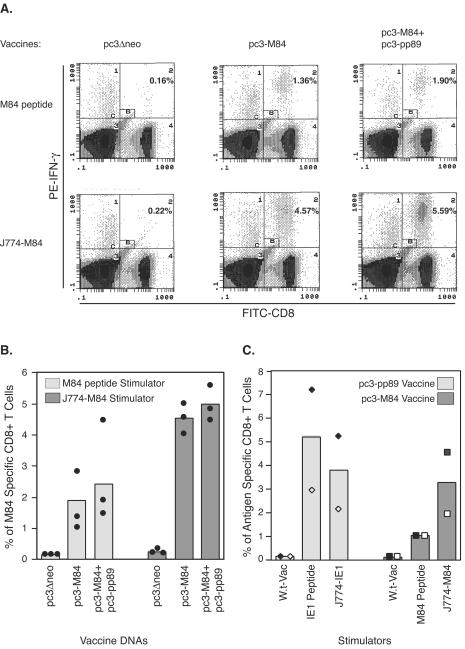
To explore this possibility, we modified our previously established ICCS assay in order to detect all epitopes derived from the M84 protein. In addition to using the known epitope peptide to stimulate splenocytes from mice immunized with pc3-M84, the macrophage cell line J774 (which presents H-2d haplotype-specific antigens) was infected for 10 h with a recombinant vaccinia virus expressing full-length M84. The M84-expressing J774 cells were subsequently incubated with mouse splenocytes in the presence of brefeldin A. The CD8+ T cells expressing IFN-γ after stimulation with the M84 peptide or M84-expressing J774 cell were detected by fluorescent staining with anti-IFN-γ antibody, followed by flow cytometry analysis. Figure 1A is one set of the representative dot plot graphs after flow cytometry analyses. In mice immunized with vector DNA (pc3Δneo) only, similar background levels of M84-specific CD8+ T cells were detected in ICCS assays with either M84 peptide or M84-expressing J774 cell as a stimulator. In mice immunized with either pc3-M84 or pc3-M84+pc3-pp89, stimulation in the ICCS assay with M84-expressing J774 cells resulted in the detection of a higher percentage of M84-specific CD8+ T cells than with M84 peptide. When peptide was used as the stimulator, about 1.8 and 2.5% of M84-specific CD8+ T cells were detected in mice immunized with pc3-M84 or coimmunized with pc3-M84 and pc3-pp89, respectively (Fig. 1B, left). When these same splenocytes were stimulated with J774 cells infected with recombinant vaccinia virus expressing M84, significantly higher levels of ca. 4.5% (P = 0.001) or 5% (P = 0.004) of CD8+ T cells from the pc3-M84- or pc3-M84 plus pc3-pp89-coimmunized mice, respectively, were specific for M84—levels ca. 1.5- to 2.5-fold higher than those detected with epitope peptide stimulation (Fig. 1B, right).
FIG. 1.
Detection of M84-specific CD8+ T cells with a J774 cell-mediated ICCS assay. (A and B) Three BALB/c mice per group were immunized i.d. three times with 10 μg of pc3Δneo, pc3-M84, or pc3-M84+pc3-pp89 at 10-day intervals. Ten days after the last immunization, splenocytes from individual mice were stimulated with 1 μM M84 epitope 297-305 peptide or J774 macrophage cells infected with an M84-expressing vaccinia virus at an MOI of 10 for 10 h. The resulting cells were processed for the ICCS as described in Materials and Methods. Fluorescein isothiocyanate-anti-CD8 and phycoerythrin-anti-IFN-γ antibodies were used to stain the cells. CD8+ T cells that had accumulated intracellular IFN-γ were detected and enumerated by flow cytometry. Panel A shows representative dot plots displaying IFN-γ-positive CD8+ T cells detected from various mouse groups by using either M84 peptide or M84-expressing J774 cells (J774-M84) as stimulators in the ICCS assay. The numbers in quadrant C2 represent the percentages of IFN-γ positive CD8+ T cells after exclusion of the nonspecific cells in gate B (small rectangle in quadrant C2). The symbols around the columns in panel B represent the percentages of IFN-γ-positive CD8+ T cells in individual mice. (C) Two mice per group were immunized i.d. three times with 10 μg of pc3-M84 or pc3-pp89 plasmids at 10-day intervals. Ten days after the last immunization, mouse splenocytes were incubated with the IE1 epitope 168-176 peptide, the M84 epitope 297-305 peptide, or J774 cells infected for 10 h with recombinant vaccinia virus expressing IE1 or M84 and then analyzed by ICCS assay. Percentages of antigen-specific CD8+ T cells from a mouse immunized with pc3-pp89 (⧫), from the other pc3-pp89-immunized mouse (◊), from a mouse immunized with pc3-M84 (▪), and for the other M84-immunized mouse (□) are shown.
To exclude the possibility that these results were due to a higher efficiency of detection with the M84-expressing J774 cells than with the epitope peptide, a separate experiment was conducted to compare the percentages of M84- and IE1-specific CD8+ T cells detected with the two different ICCS assays (Fig. 1C). In mice immunized with pc3-pp89, approximately 5% of IE1-specific CD8+ T cells were detected by using the IE1 peptide 168-176. When the same splenocytes were examined with J774 cells expressing full-length IE1 after infection with a recombinant vaccinia virus, the levels of IE1-specific CD8+ T cells dropped slightly (P > 0.05 [paired t test]). These results are in contrast to what was observed for the mice immunized with M84, in which a slightly higher percentage of M84-specific CD8+ T cells was detected with the M84-expressing J774 cell-mediated ICCS assay (Fig. 1C, right). However, with only two mice tested per group in this experiment, the difference was not statistically significant (P > 0.05 [paired t test]). Therefore, in the case of the single epitope-containing IE1, antigen-expressing J774 cell stimulation in the ICCS assay was actually slightly less efficient than epitope peptide stimulation. These trends suggested that CD8+ T cells with specificities other than the known Kd-restricted M84 epitope 297-305 may contribute to the higher observed response to M84.
M84-specific immune responses and protection provided by DNA immunization with an M84 mutant lacking the defined 297-305 epitope.
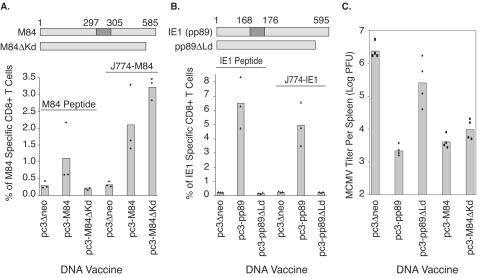
To investigate whether additional CD8+-T-cell epitopes were present in M84, we deleted the nucleotide sequence coding for the previously defined Kd epitope (that is, 297-305) in the M84 gene (Fig. 2A, top). The resulting recombinant plasmid coding for the mutant M84 (pc3-M84ΔKd) was subsequently used to vaccinate BALB/c mice. We also confirmed the presence of the previously defined 297-305 epitope of M84 in the M84 gene of MCMV strain K181 by sequencing of pc3-M84 (data not shown). As a control, the known Ld-restricted epitope of IE1, 168-176, was similarly deleted to yield pc3-pp89ΔLd and used to immunize BALB/c mice, along with pc3-M84, pc3-M84ΔKd, and pc3-pp89. Ten days after the last immunization, three mice from each immunization group were examined for antigen-specific CD8+-T-cell responses. The remaining four mice per group were challenged i.p with 3.5 × 105 PFU (0.5 50% lethal dose [LD50]) of salivary gland-derived MCMV. Five days postchallenge, virus titers in the spleen were measured by using a highly sensitive plaque assay as previously described (27).
FIG. 2.
Detection of an additional protective CD8+-T-cell epitope in M84. Diagrams above panels A and B represent wild-type M84, IE1, and their mutant proteins, M84ΔKd and pp89ΔLd. The known CD8+-T-cell epitopes 297-305 (M84) or 168-176 (IE1) are indicated. They were deleted in M84ΔKd and pp89ΔLd. The numbers represent the amino acid position in the protein. BALB/c mice were immunized i.d. three times with DNA vaccines pc3Δneo (vector), pc3-M84, pc3-M84ΔKd, pc3-pp89, or pc3-pp89ΔLd. Ten days after the last immunization, three mice per group were examined for M84-specific CD8+ T cells by using a peptide-mediated or M84-expressing-J774 cell-mediated ICCS assay. The other four mice per group were challenged i.p. with 3.5 × 105 PFU (0.5 LD50) of salivary gland-derived MCMV (A and B). Five days later, the virus titer in the spleen was examined (C). (A) M84-specific CD8+-T-cell responses measured with ICCS assay by using M84 297-305 peptide (left three columns) or M84-expressing-J774 cells (right three columns) as a stimulator. (B) The IE1-specific CD8+-T-cell responses measured with ICCS with IE1 168-176 peptide (left three columns) or IE1-expressing-J774 cells as a stimulator. (C) Titer of MCMV per spleen after challenge of immunized mice. Solid symbols stand for measurement of CD8+-T-cell responses or virus titers in individual mice.
Figure 2A shows the levels of M84-specific CD8+ T cells detected by the ICCS assay using the M84 297-305 epitope peptide (M84 peptide) or J774 cells infected with recombinant vaccinia virus expressing M84 (J774-M84) as stimulators. When the M84 peptide 297-305 was used as a stimulator, an average of 1.2% of M84-specific CD8+ T cells was detected in mice immunized with plasmid coding for wild-type M84 (pc3-M84). The level of antigen-specific CD8+ T cells detected in mice immunized with the mutant M84 lacking the 297-305 epitope (pc3-M84ΔKd) was very similar to that detected in mice immunized with vector only (pc3Δneo), indicating that M84 epitope (i.e., 297-305)-specific CD8+ T cells were not elicited by mutant M84 DNA (Fig. 2A, left 3 columns). When the same splenocytes were examined for M84-specific CD8+ T cells by ICCS assay with J774 cells infected with recombinant vaccinia virus expressing M84 as stimulators, a different profile of M84-specific CD8+-T-cell responses was detected (Fig. 2A, right 3 columns). An average of 2.2% of M84-specific CD8+ T cells were detected in mice immunized with the wild-type M84 plasmid (pc3-M84): a level nearly twofold higher than that detected with the epitope peptide. More specifically, the level of M84-specific CD8+ T cells detected by J774 cell-mediated ICCS assay in each individual mouse was higher than that measured with epitope peptide as a stimulator (P = 0.01 [paired t test]). When the splenocytes from mice immunized with the plasmid coding for the M84 deletion mutant (M84ΔKd) were examined by using J774 cells infected with M84-expressing recombinant vaccinia virus as stimulators in the ICCS assay, an average of 3.3% of M84-specific CD8+ T cells were detected. The level of M84-specific CD8+ T cells detected in mice immunized with vector alone was comparable to that detected with epitope peptide as a stimulator. The experiment-to-experiment variability inherent in the absolute percentages of specific CD8+ T cells should be noted, since the mean percentages of M84-specific CD8+ T cells after immunization with pc3-M84 and stimulation with M84-expressing vaccinia virus-infected cells were 4.5 and 3.2% in the two experiments in Fig. 1 (Fig. 1B and C) and 2.2% in this third independent experiment. Thus, the higher percentages of M84-specific T cells measured by stimulation with M84-expressing J774 cells instead of the M84 peptide suggest that at least one CD8+-T-cell epitope is present in M84, in addition to the previously defined epitope 297-305.
In a control experiment shown in Fig. 2B, the previously defined IE1-specific CD8+-T-cell epitope 168-176 was deleted, and the resulting mutant gene (pp89ΔLd) was used to immunize mice along with wild-type IE1 as diagramed on top of Fig. 2B. When the IE1 epitope peptide 168-176 was used as a stimulator in the ICCS assay, antigen-specific CD8+ T cells were detected only in mice immunized with the wild-type IE1 gene. When the same splenocytes were examined by using J774 cells expressing full-length IE1 as a stimulator, no IE1-specific CD8+ T cells were detected in mice immunized with the mutant IE1 (pc3-pp89ΔLd). In addition, in mice immunized with pc3-pp89, a slightly lower percentage of IE1-specific CD8+ T cells was detected by using IE1-expressing J774 cells as stimulators compared to peptide stimulation (P > 0.05 [Fisher PLSD]) (Fig. 2B). This result is consistent with a previous report that demonstrated the presence of only a single CD8+-T-cell epitope in the IE1 protein (4). Taken together, these observations suggest that the increased percentage of M84-specific CD8+ T cells detected by J774 cell-mediated ICCS assay resulted from immune responses to additional epitope(s).
Finally, the protective effects of additional CD8+-T-cell epitopes in M84 were investigated by challenging the mice immunized with the wild-type or mutant IE1 or M84 plasmids with a sublethal dose of salivary gland derived MCMV, as shown in Fig. 2C. In mice immunized with vector alone, titers of >106 PFU were detected in the spleen. Immunization with IE1 reduced viral titers in the spleen ∼1,000-fold. When the epitope 168-176 in IE1 was deleted, however, the DNA vaccine encoding mutant IE1 (pc3-pp89ΔKd) was only modestly protective, with variable levels of protection and a mean titer reduction of ninefold being within the range of the experimental variability of the assay. Most notably, the viral titers after immunization full-length IE1 were significantly lower than those after immunization with the mutant IE1 (P < 0.001 [Fisher PLSD]). This result is consistent with the previous report showing that the epitope 168-176 is likely the single protective epitope of IE1 (4). In contrast, deletion of the previously defined epitope in M84 (297-305) only slightly reduced the protective effects. The reduction in spleen virus titer was ∼600-fold in mice immunized with DNA vaccine coding for wild-type M84 and 250-fold in mice immunized with the mutant M84 (pc3-M84ΔKd) (P = 0.19 [Fisher PLSD]), indicating that at least one additional epitope in M84 confers protection (Fig. 2C).
Multiple CD8+-T-cell epitopes are involved in M84-mediated immune protection.
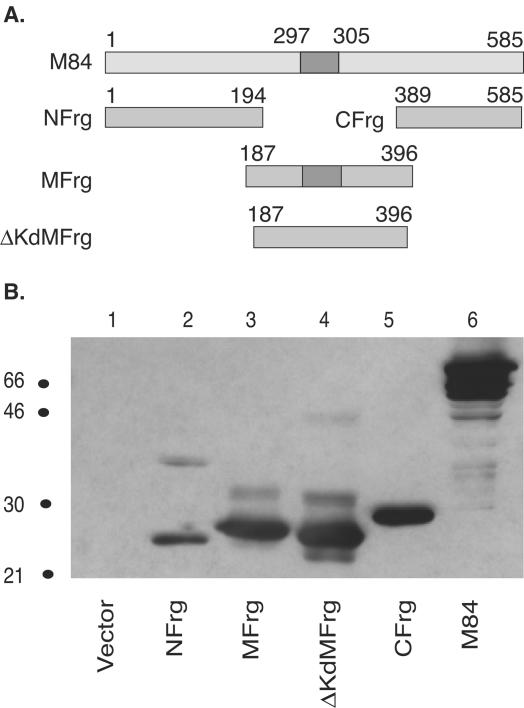
To further characterize the nature of additional epitope(s) in M84, we subcloned the M84 gene into three overlapping minigenes as shown in Fig. 3A. They were designated N fragment (NFrg), M fragment (MFrg), and C fragment (CFrg). In order to minimize the loss of potential epitopes, an overlapping sequence coding for 8 aa was shared between adjacent fragments. In addition, a minigene that contained the M fragment lacking the defined Kd-restricted epitope 297-305 was constructed and named ΔKd M fragment (ΔKdMFrg). The expression of each minigene was confirmed by transient transfection of COS-7 cells, followed by Western blotting analysis with a rabbit polyclonal antibody to M84 (26, 37). As shown in Fig. 3B, all four minigenes expressed gene products that migrated with the expected mobility. In addition, faint higher-molecular-weight bands were observed in lanes 2, 3, and 4, and these bands are likely aggregates of the minigene products and their respective degradation products, since the full-length M84 gene product has been found to readily aggregate if expressed at very high levels or overheated prior to SDS-PAGE analysis (25).
FIG. 3.
Expression of M84 subfragments in COS-7 cells. (A) Schematic diagram of M84 and its overlapping subfragments. The numbers mark the amino acid position in the protein. The known M84 297-305 epitope is indicated with dark shading. NFrg, amino-terminal fragment of M84; MFrg, middle fragment; CFrg, carboxy-terminal fragment; and ΔKdMFrg, middle fragment without the M84 297-305 epitope. (B) COS-7 cells were transfected with pc3Δneo (Vector), pc3-NFrg, pc3-MFrg, pc3-ΔKdMFrg, pc3-CFrg, or pc3-M84. Two days posttransfection, cells were lysed and cell lysates from an equal number of cells were subjected to SDS-PAGE. After transfer of the proteins to nitrocellulose, M84 protein or individual subfragments were examined by Western blotting with a rabbit polyclonal antibody against bacterially expressed GST-M84. Size markers (in kilodaltons) are on the left side of the figure.
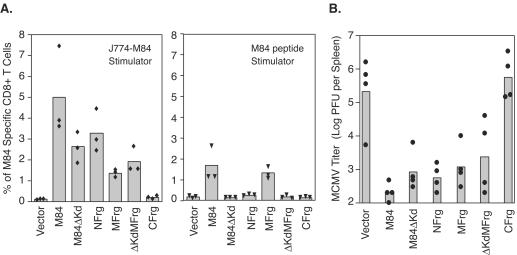
Subsequently, seven groups of BALB/c mice were immunized i.d. three times with either pc3Δneo (Vector), pc3-M84, pc3-M84ΔKd, pc3-NFrg, pc3-MFrg, pc3-ΔKdMFrg, or pc3-CFrg. Ten days after the last immunization, three mice from each group were examined for M84-specific CD8+-T-cell responses by using both recombinant vaccinia virus-infected J774-mediated (Fig. 4A, left panel) and peptide-mediated (Fig. 4A, right panel) ICCS assays. Another four mice from each group were challenged i.p. with a sublethal dose of salivary gland-derived MCMV. At 5 days postchallenge, virus titers in the spleen were determined as described in Materials and Methods.
FIG. 4.
M84-specific CD8+-T-cell responses and immune protection after vaccination of mice with M84 DNA or M84 DNA subfragments. BALB/c mice were immunized three times i.d. with pc3Δneo (Vector), pc3-M84, pc3-M84Δ Kd, pc3-NFrg, pc3-MFrg, pc3-ΔKdMFrg, or pc3-CFrg DNA vaccines. Ten days after the last immunization, three mice per group were examined for M84-specific CD8+-T-cell responses by using the ICCS assay with either M84 297-305 peptide or M84-expressing J774 cells as stimulators. The other four mice per group were challenged with 3.5 × 105 PFU (0.5 LD50) of salivary gland-derived MCMV. Five days later, the virus titer in the spleen was determined. (A) M84-specific CD8+-T-cell responses measured by ICCS assay with M84-expressing J774 cells (left) or M84 297-305 peptide (right) as a stimulator. (B) Titer of MCMV per spleen after viral challenge. The limit of virus detection was 200 PFU per spleen, and virus titers below the detection limit were arbitrarily set to 100 PFU for display purposes and mean calculation. Each individual mouse is represented by a solid symbol.
The left panel of Fig. 4A displays the M84-specific CD8+-T-cell responses measured by ICCS assay with M84-expressing J774 cells as a stimulator. As shown above, immunization of mice with plasmids expressing either wild-type M84 or M84ΔKd induced strong CD8+-T-cell responses (5 and 2.7%). It is important to note that the variability inherent in the plasmid DNA immunization from experiment to experiment precludes the comparison of absolute percentages of antigen-specific CD8+ T cells between the different immunization experiments but permits instead the comparisons within experiments of T-cell levels resulting from stimulation of the same splenocytes with different stimulators. For example, when the splenocytes of pc3-M84-immunized mice were stimulated with the M84-expressing J774 cells as a stimulator in the independent experiments shown in Fig. 1B and C, 2A, and 4A, the mean percentages of M84-specific T cells measured were 4.5, 3.3, 2.1, and 5.0%, respectively, whereas within each experiment, the T-cell levels measured after peptide stimulation were lower. In addition to the CD8+-T-cell responses observed after immunization with wild-type M84 or M84ΔKd, strong M84-specific CD8+-T-cell responses were also elicited by immunization with plasmids expressing NFrg, MFrg, and ΔKdMFrg 3.4, 1.5, and 2%, respectively. In contrast, no CD8+-T-cell response was detected after immunization with CFrg. When the same splenocytes were examined by ICCS with epitope 297-305 peptide as a stimulator (Fig. 4A, right panel), M84-specific CD8+-T-cell responses were detected only in mice immunized with plasmids encoding wild-type M84 and MFrg of the protein. Of note, when mice are immunized with the wild-type M84 or the various M84 mutants and fragments, the inability to predict the overall T-cell responses to the remaining epitopes in the absence of the deleted epitope(s) precludes comparing the total antigen-specific T-cell levels elicited by the different M84 fragments or deducing the relative contributions of each fragment or epitope to the overall T-cell response. For example, immunization with MFrg elicited the same mean percentage of M84-specific T cells when measured by stimulation with J774-M84 or M84 peptide, whereas ΔKdMFrg immunization resulted in a slightly higher level of M84-specific T cells than the wild-type MFrg when measured by stimulation with J774-M84 (Fig. 4A). Taken together, these results demonstrate that M84-specific CD8+-T-cell responses were elicited by epitopes in the NFrg and the ΔKdMFrg, in addition to the defined epitope 297-305 in the MFrg.
In order to investigate the biological significance of the CD8+-T-cell responses measured above, protection in the remaining immunized mice was determined after sublethal i.p. challenge with MCMV. As shown in Fig. 4B, protection was observed in mice immunized with plasmids expressing NFrg, ΔKdMFrg, and M84ΔKd, in addition to wild-type M84 and MFrg, the latter two of which contain the 297-305 epitope (P < 0.01 [Fisher PLSD for these groups compared to the vector group]). Consistent with the lack of detectable M84-specific CD8+-T-cell responses, no protection was observed in mice immunized with plasmid expressing CFrg of M84 (P > 0.05). These experiments confirm that at least two CD8+-T-cell epitopes in addition to the 297-305 epitope are present in M84—one in the NFrg and one in the ΔKdMFrg—and that they are protective against MCMV replication when they are used individually to immunize mice.
M84-specific CD8+-T-cell responses in MCMV-infected mice.
Previously, we and others demonstrated that MCMV infection in mice elicits weak M84-specific CD8+-T-cell responses (12, 16, 37). In those reports, CD8+-T-cell responses were measured by an ICCS assay with the M84 epitope 297-305 as a stimulator. In the present study, we found that a higher percentage of CD8+-T-cell responses in M84 DNA-immunized mice could be detected with J774 cells expressing full-length M84 as a stimulator. Therefore, we reassessed these M84-specific CD8+-T-cell responses in MCMV-infected mice by using the modified assay.
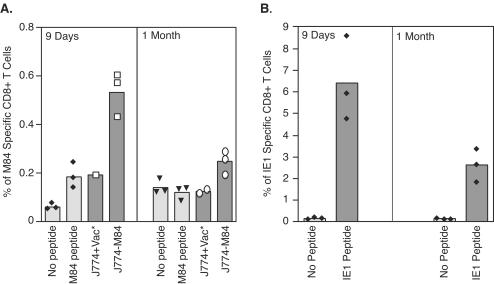
To this end, 4- to 5-week-old female BALB/c mice were infected with 1.2 × 105 PFU of tissue culture-derived MCMV. At both 9 days and 1 month p.i., M84-specific CD8+ T cells were measured by ICCS assay with either epitope 297-305 peptide or M84-expressing J774 cells as stimulators. The background levels with either no peptide or J774 cells infected with only vaccinia virus are shown (0.04 and 0.2%, respectively). As shown in Fig. 5A, at 9 days p.i., only low levels (0.14% above background) of the CD8+ T cells were directed against the M84 epitope 297-305. When J774 cells were infected with recombinant vaccinia virus expressing M84 and used as stimulators, there was an ∼2-fold increase in M84-specific CD8+ T cells detected. Although the ICCS assay with the M84-expressing J774 cells revealed a significantly higher level of M84-specific CD8+ T cells (P < 0.001 [Fisher PLSD]), the percentage is still much lower than that detected in mice immunized with the DNA vaccine expressing M84, which is typically 2 to 4%. It is also significantly lower than the percentage of IE1-specific CD8+ T cells (Fig. 5B), although the absolute percentages of specific T cells are variable between individual immunization experiments. One month after infection, the percentage of M84 epitope 297-305-specific CD8+ T cells detected was at the background level when peptide was used as a stimulator (Fig. 5A, right panel). However, by using the J774 cell-mediated ICCS assay, low levels of M84-specific CD8+ T cells could still be detected (mean of 0.12% above background) (P < 0.01 [Fisher PLSD compared to J774+Vac]). Although this percentage is low compared to IE1-specific CD8+-T-cell responses (Fig. 5B), these data clearly demonstrated that a low level of M84-specific memory CD8+ T cells was present 1 month after infection. Since epitope 297-305-specific CD8+ T cells were barely detectable 1 month after MCMV infection, other epitopes in the NFrg and ΔKdMFrg may have contributed to the M84-specific memory T cells. Alternatively, the contribution to stimulation provided by the additional M84 epitopes may have increased the number of IFN-γ-positive cells to a level over the threshold needed for detection.
FIG. 5.
M84- and IE1-specific CD8+-T-cell responses after MCMV infection. Three BALB/c mice per group were infected with 1.2 × 105 PFU of tissue culture-derived MCMV. Nine days or 1 month after infection, M84- or IE1-specific CD8+-T-cell responses were measured by ICCS assay after either peptide-based stimulation with the M84 297-305 or IE1 168-176 epitope peptides or cell-based stimulation with J774 cells infected with either the wild-type or M84-expressing vaccinia virus. Open or solid symbols represent the percentage of antigen-specific CD8+ T cells in each individual mouse except when indicated by an asterisk. In these cases, the splenocytes were pooled.
Antigen presentation of CD8+-T-cell restricted M84 epitopes in macrophages following MCMV infection.
Although M84 is a very potent vaccine candidate, especially when coimmunized with IE1, immune responses to M84 are very low when mice are infected with MCMV (Fig. 5A) (12, 16). It has been demonstrated that MCMV encodes several immune modulators, such as m04, m06, and m152 (5, 19, 20, 22, 34, 38). The m04 gene product forms a complex with MHC class I molecules in the endoplasmic reticulum that is transported to the cell surface, whereas the m06 and m152 gene products play a role in downregulating antigen presentation by diverting MHC class I molecules to the lysosome for degradation (m06) or by retaining the MHC molecules in the endoplasmic reticulum (m152). Therefore, one possible explanation for the weak immune responses elicited against M84 during MCMV infection was that there was poor M84-specific antigen presentation. MEFs have been shown to be fully permissive for MCMV replication and have been used extensively to document the early-phase downregulation of class I-mediated antigen presentation (3, 7). However, recent studies suggest that this downregulation occurs more readily in primary fibroblasts than in professional antigen-presenting cells, such as the J774 macrophage line (8). We thus measured the presentation of M84-derived CD8+-T-cell epitopes in MEFs and macrophages. To exclude potential cell line-specific effects when the J774 macrophages are used, we also infected primary peritoneal macrophages, a readily obtainable macrophage that is infected early in vivo after i.p. infection (24).
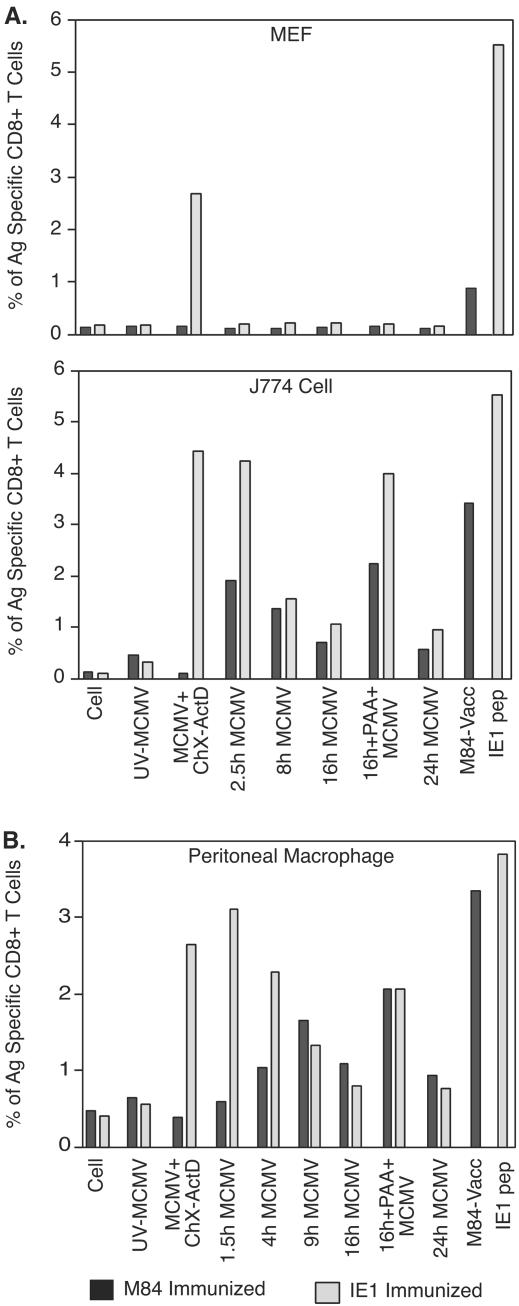
Since we developed and verified an ICCS assay by using the J774 macrophage line infected with recombinant vaccinia virus expressing M84 as a stimulator (Fig. 1), we modified this cell mediated ICCS assay to examine the presentation of M84 epitopes during the MCMV infection. In this ICCS assay, splenocytes from BALB/c mice immunized with either pc3-M84 or pc3-pp89 were used as effector cells. BALB/c MEFs, the macrophage cell line J774, and BALB/c primary peritoneal macrophage were either mock infected or infected with MCMV under various conditions to regulate viral gene expression. Figure 6A shows the percentage of M84- and IE1-specific CD8+ T cells that are detected when M84 and IE1 antigens are presented by MEFs. Generally, MEFs present M84 and IE1 antigens poorly during MCMV infection, which is consistent with the findings of others regarding IE1 presentation in MEFs (3, 8). Antigen presentation of IE1 was seen only when cells were infected with MCMV for 3 h in the presence of ChX; the ChX was removed, and infection proceeded for an additional 2.5 h in the presence of ActD. For herpesviruses, ChX treatment allows accumulation of immediate-early transcripts (such as IE1) without concomitant protein synthesis, whereas the removal of ChX and subsequent ActD treatment allows the accumulated transcripts to be translated but inhibits additional transcription. Therefore, ChX, followed by ActD treatment, significantly increased the level of IE gene products such as IE1, which subsequently resulted in increased IE1 antigen presentation by the MEFs. In contrast, stimulation of splenocytes from pc3-M84 immunized mice by MEFs was not detectable under any of the infection conditions tested. However, we also found that M84 can be presented by this cell type, since MEFs infected with the M84 expressing vaccinia virus were capable of simulating low levels of M84-specific splenocytes (Fig. 6A). As a control, MEFs infected with UV-inactivated MCMV (UV-MCMV) were only able to provide background levels of stimulation of M84-or IE1-specific effectors, a finding consistent with these antigens being nonstructural.
FIG. 6.
Presentation of M84 and IE1 antigens in MCMV-infected cells. Three BALB/c mice per group were immunized three times (A) or four times (B) i.d. with 10 μg of pc3-M84 or pc3-pp89 DNA vaccine at 7-day intervals. At 10 days (A) or 6 days (B) after the last immunization, mouse splenocytes were prepared and pooled as effector cells in the ICCS assay. BALB/c MEFs (A, top panel), J774 cells (A, lower panel), or freshly isolated primary BALB/c peritoneal macrophages (B) were infected with tissue culture-derived MCMV at an MOI of 5 for the indicated periods or conditions. The infected cells were subsequently used as stimulators in the ICCS assay to measure the level of antigen presentation as described in Materials and Methods. Uninfected cells were used as a negative control. The IE1 168-176 peptide or J774 cells infected with the M84-expressing recombinant vaccinia virus served as positive controls to measure the maximum level of M84- or IE1-specific CD8+-T-cell responses. Darkly shaded columns represent the measurement of M84-specific CD8+ T cells in splenocytes of M84-immunized mice. Lightly shaded columns stand for the measurement of IE1-specific CD8+ T cells in the splenocytes of IE1-immunized mice.
The inability of infected MEFs to efficiently present viral antigen has been attributed to the expression of MCMV-encoded immune modulators (3, 22). Whether antigen presentation is inhibited in J774 macrophages or primary macrophages depends upon the antigen (8, 22). A study by Hengel et al. showed that IE1 antigen presentation is not inhibited in primary macrophages from BALB/c mice that have an H-2d MHC class I haplotype. In contrast, LoPiccolo et al. have recently shown that there is restriction of M45 antigen presentation in primary macrophages from C57BL/6 mice that have an H-2b MHC class I haplotype. Since only recognition of the IE1 protein was studied by Hengel et al., it was possible that M84, an early gene product, would not be presented by the infected primary macrophage. Therefore, we tested M84 and IE1 antigen presentation by primary peritoneal macrophage and the macrophage cell line J774. Similar to the results observed in MEFs, MCMV-infected J774 cells treated with ChX, followed by ActD, were able to stimulate IE1-specific effector cells efficiently, and a high percentage of IE1-specific CD8+ T cells was detected (Fig. 6B). As expected, M84-specific CD8+ T cells were not stimulated by the ChX- and ActD-treated J774 cells because M84 is an early gene product and no early RNA is transcribed (25). At 2.5 h p.i., however, significant numbers of both M84- and IE1-specific CD8 T cells were stimulated by the infected J774 cells, indicating that M84 epitopes were presented at very early times of infection. At this time, IE1 antigen presentation is also at a very high level. Presentation of both IE1 and M84 decreased at 8 h p.i. but still was above background at 24 h p.i. PAA has been used as an inhibitor of late gene expression, and we found that J774 cells infected with MCMV in the presence of PAA were able to stimulate a high number of M84- and IE1-specific CD8+ T cells, suggesting that late viral gene products may provide some inhibition of antigen presentation. In contrast, J774 cells infected with UV-MCMV did not stimulate either group of effector cells. These results indicate that M84 and IE1 are presented on MHC class I in J774 macrophages throughout the viral replication cycle.
Although J774 macrophages have been shown to be fully permissive for MCMV infection (8), we performed a separate experiment to measure the presentation of M84 and IE1 in primary macrophages, since these may more accurately reflect antigen presentation after MCMV infection. Freshly isolated BALB/c murine peritoneal macrophages were infected with MCMV in vitro and used as described above as stimulator cells to confirm the above observation. As shown in Fig. 6C, we found patterns of antigen presentation very similar to those observed in J774 macrophages, although statistical analysis between simulator groups was not possible due to the need for pooling of splenocytes from the immunized mice resulting in a single measure for each combination of immunization and stimulation. Antigen presentation of IE1 was highest at 1.5 h p.i. At that time, M84 antigen presentation was very low or absent, a finding consistent with early gene expression (12, 25). Similar to the results observed in MCMV-infected J774 cells, antigen presentation of IE1 decreased during the period from 1.5 to 16 h p.i. In contrast, M84 presentation was maintained from early (4 h p.i.) to late (24 h p.i.) times after infection. Treatment with PAA also markedly increased the presentation of both IE1 and M84. Taken together, these experiments demonstrate that the macrophage cell line J774 and freshly isolated peritoneal macrophages from BALB/c mice are able to efficiently present both IE1 and M84 antigens.
DISCUSSION
In previous reports, we demonstrated that an M84 DNA vaccine was as protective against viral replication in the spleen as IE1 DNA (26). However, when the immune responses after vaccination were investigated by using an epitope peptide based ICCS assay, a much lower percentage of M84-specific CD8+ T cells were detected compared to the IE1-specific CD8+ T cells (37). In the study presented here, we sought to elucidate the basis of this discrepancy.
Our experiments showed that when a macrophage cell line J774 was infected with a recombinant vaccinia virus expressing full-length M84 and used as a stimulator in an ICCS assay, higher percentages of M84-specific CD8+ T cells were detected in mice immunized with M84 DNA. More specifically, when the percentages of the M84-specific T cells that were detected following either peptide or J774 stimulation from all of the pc3-M84 immunization experiments are combined (Fig. 1B and C, 2A, and 4A), J774-M84 stimulation resulted in significantly higher percentages of T cells than after peptide stimulation (P < 0.001 [Fisher PLSD]). Even when the known Kd-restricted M84 epitope 297-305 was deleted from the M84 gene, a high percentage of M84-specific CD8+ T cells was detected in mice immunized with the mutant M84 DNA (pc3-M84ΔKd). These results provide evidence that there are additional CD8+ T cell epitope(s) that contribute to the overall protection elicited after M84 DNA immunization.
The presence of additional CD8+-T-cell epitope(s) in M84 was further studied by immunizing mice with subfragments of M84 and measuring M84-specific responses by using the J774 mediated ICCS assay. Our results clearly demonstrate that there are at least three CD8+-T-cell epitopes in M84. One is present in the amino-terminal one-third of M84. Immunization of mice with a minigene encoding this fragment, from aa 1 to 194, elicited strong M84-specific CD8+-T-cell responses and provided significant protection against sublethal MCMV challenge (Fig. 4B). The other two epitopes are present in the middle one-third of the protein from aa 187 to 396. Previously, one of the epitopes was identified in this region and mapped to aa 297 to 305, and a CTL line was established based on this epitope (16). However, our experiments show the existence of another epitope in this fragment. When the known epitope 297-305 in the fragment was deleted, immunization of mice with the resulting minigene (ΔKdMFrg) resulted in strong M84-specific immune responses and significant protection against MCMV replication. Therefore, the current study detected at least two additional M84-specific CD8+-T-cell epitopes in addition to the previously mapped epitope 297-305. The amino acid sequences of the two additional epitopes remain to be determined. Two epitope candidates, M84 33-41 (FGPYETKFL) and M84 47-55 (LSPQAPCVL), with high scores based on computer program analysis as described by Holtappels et al. (16) were investigated in the epitope peptide-based ICCS assay. However, no significant levels of M84-specific CD8+ T cells from mice immunized with M84 DNA were detected by using these two peptides (data not shown). Therefore, the new epitopes may consist of amino acid sequences divergent from the MHC peptide consensus sequences described by Holtappels et al. (16). It is likely that an exhaustive mapping of the M84 amino acid sequence will be necessary to define the two minimal peptide epitopes targeted by protective M84-specific CD8+ T cells, and these experiments are beyond the scope of this report.
In a study conducted with lymphocytic choriomeningitis virus (LCMV), Rodriguez et al. (35) found that a cryptic epitope which is not immunogenic in the viral N protein was able to elicit CD8+-T-cell responses when it is present in a subfragment minigene without the dominant epitope. Our current study differs from the LCMV study at least in two aspects. First, higher percentages of M84-specific CD8+ T cells were consistently detected by the J774-mediated ICCS assay in splenocytes from mice immunized with full-length M84 gene. In contrast, no LCMV-specific CD8+-T-cell responses corresponding to the cryptic epitope were detected in mice immunized with full-length N protein. The cryptic epitope-specific CD8+-T-cell responses were strictly associated with the minigene of LCMV nucleoprotein in the absence of the dominant epitope. The second difference is that in our studies, a higher percentage of M84-specific CD8+ T cells were also detected in mice infected with MCMV, although the percent increase was not as great as that observed in mice immunized with the DNA vaccine. Therefore, additional epitopes detected by M84 minigene immunization represent true unidentified immunogenic epitopes in M84. Once these minimal epitopes are mapped, it will be possible to determine the levels of CD8+ T cells specific for each epitope after M84 DNA immunization or MCMV infection.
In a previous study, we investigated the immune responses to IE1 and M84 after MCMV infection of BALB/c mice by peptide-mediated ICCS assay (37). Although the immune responses to IE1 were vigorous and peaked between days 7 to 9 p.i., the immune responses against M84 were very weak (5.5% versus 0.14%). Based upon our new evidence that M84 may contain at least three CD8+-T-cell epitopes, we reinvestigated M84-specific CD8+-T-cell responses via J774-mediated ICCS assay. To our surprise, the percentage of M84-specific CD8+ T cells were still very low compared to IE1-specific CD8+-T-cell responses (6.5% versus 0.38% over background), although the percentage detected was ∼2-fold higher than that with the M84 epitope 297-305 peptide (Fig. 5). One possible explanation for the weak immune responses to M84 during MCMV infection is that there may be insufficient priming of naive CD8+ T cells to recognize M84. This may be due to M84, which is an early gene product, not being optimally presented on the surface of the infected cell due to the MHC class I downregulation of proteins that are expressed at early times of infection. There is also increasing evidence that CTL responses may be generated by cross-priming that is mediated by professional antigen-presenting cells, most likely dendritic cells, following the uptake and processing of exogenous proteins from other cells (for a review, see reference 23). Thus, inefficient uptake and processing of M84 (produced during the infection of other cells) by dendritic cells might prevent the induction of strong CTL responses. This might be due either to low levels of available M84 as a result of slow biosynthesis or rapid turnover of the protein or to some impairment of the dendritic cells by the infection. Alternatively, there may be insufficient help from the CD4+ T cells to generate a strong CTL response. In this regard, it has been shown that CD4-T-cell help is required during the priming phase to elicit functional CD8-T-cell memory cells that can mount robust recall responses (for a review, see reference 1).
It has been well documented that MCMV contains at least three immune suppressive genes (m04, m06, and m152) and that all are expressed at the early phase of the virus replication cycle, coinciding with the expression of M84. Therefore, it was possible that the presentation of M84 antigen was suppressed by these gene products through the inhibition of MHC expression on the cell surface. To explore this possibility, we examined M84 antigen presentation in three types of cells: MEFs, the macrophage cell line J774, and BALB/c peritoneal macrophages. Consistent with other reports (3, 8) investigating IE1 antigen presentation, the presentation of IE1 in MEFs was not detected in our assay unless the expression of the IE1 protein was amplified by incubating the cells in the presence of ChX that was then removed and replaced by ActD, which inhibited further viral gene transcription. M84 was not detected under any of the conditions, indicating that IE1 and M84 antigen presentation was likely blocked by the inhibition of MHC class I expression on the cell surface. When the J774 cell line and primary BALB/c peritoneal macrophages were similarly examined, the presentation of both IE1 and M84 was detected. These results are consistent with the findings of Hengel et al. that IE1 antigen presentation in J774 cells was not suppressed by immune suppressive genes during early and late phases of virus replication (8). Since macrophages are important antigen-presenting cells and play a central role in the induction of immune responses against invading infectious agents, efficient presentation of M84 antigen by these cells strongly suggests that this presentation is not sufficient for priming of M84-specific CD8+ T cells.
In the present study, we describe the MHC class I presentation of pM84, an MCMV early antigen that is considered subdominant due to the low levels of M84-specific CD8+ T cells elicited during acute viral infection. Although the levels of M84-specific CD8+ T cells detectable after stimulation with the full-length pM84 protein were approximately two- to threefold higher than those measured with a single defined M84 epitope peptide, these increased levels were still very low relative to the IE1-specific responses during infection. In contrast, peak levels of pM84-specific CD8+ T cells were five- to eightfold higher after M84 DNA vaccination than after viral infection. Importantly, the resultant M84-specific CD8+ T cells following vaccination were found to be as highly protective against viral challenge as those primed by IE1 DNA. This feature highlights an important difference between these two protective antigens: while IE1-specific CD8+ T cells might be predicted to be protective due to their high levels after the acute viral infection, the low levels of M84-specific CD8+ T cells do not reflect a diminished potential of these cells to contribute to viral clearance. Further support for the potential antiviral function of the M84-specific CD8+ T cells comes from these studies that have shown that clonally expanded, M84 peptide-specific cytolytic-T-cell lines can help resolve infection in a murine model of adoptive cytoimmunotherapy (12). Interestingly, the converse also has been observed. In this case, it was found that whereas MCMV infection of C57BL/6 mice elicited high levels of M45-specific CD8+ T cells, adoptive transfer of these antivirally active cells did not protect the recipients against the wild-type virus (13). The design of candidate vaccines must therefore focus on the specific immune responses generated in the context of vaccination and the resultant levels of protection. By these criteria, the MCMV vaccine candidate must be functionally evaluated not only for its efficacy in priming CD8+ T cells but also for the ability of these immune cells to recognize the cognate antigen in infected cells that also express the interfering viral immune-evasive genes.
Acknowledgments
This study was supported by research grants 1-FY02-199 from the March of Dimes Birth Defects Foundation and AI51557 from the National Institutes of Health and by National Institutes of Health training grant T32 AI07036.
REFERENCES
- 1.Behrens, G., M. Li, C. M. Smith, G. T. Belz, J. Mintern, F. R. Carbone, and W. R. Heath. 2004. Helper T cells, dendritic cells, and CTL immunity. Immunol. Cell. Biol. 82:84-90. [DOI] [PubMed] [Google Scholar]
- 2.Cranmer, L. D., C. L. Clark, C. S. Morello, H. E. Farrell, W. D. Rawlinson, and D. H. Spector. 1996. Identification, analysis, and evolutionary relationship of the putative murine cytomegalovirus homologues of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J. Virol. 70:7929-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Val, M., K. Munch, M. J. Reddehase, and U. H. Koszinowski. 1989. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell 58:305-315. [DOI] [PubMed] [Google Scholar]
- 4.Del Val, M., H. J. Schlicht, H. Volkmer, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 1991. Protection against lethal cytomegalovirus infection by a recombinant vaccine containing a single nonameric T-cell epitope. J. Virol. 65:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold, M. C., M. W. Munks, M. Wagner, U. H. Koszinowski, A. B. Hill, and S. P. Fling. 2002. The murine cytomegalovirus immunomodulatory gene m152 prevents recognition of infected cells by M45-specific CTL but does not alter the immunodominance of the M45-specific CD8 T cell response in vivo. J. Immunol. 169:359-365. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez Armas, J. C., C. S. Morello, L. D. Cranmer, and D. H. Spector. 1996. DNA immunization confers protection against murine cytomegalovirus infection. J. Virol. 70:7921-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hengel, H., P. Lucin, S. Jonjic, T. Ruppert, and U. H. Koszinowski. 1994. Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J. Virol. 68:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hengel, H., U. Reusch, G. Geginat, R. Holtappels, T. Ruppert, E. Hellebrand, and U. H. Koszinowski. 2000. Macrophages escape inhibition of major histocompatibility complex class I-dependent antigen presentation by cytomegalovirus. J. Virol. 74:7861-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtappels, R., N. K. Grzimek, C. O. Simon, D. Thomas, D. Dreis, and M. J. Reddehase. 2002. Processing and presentation of murine cytomegalovirus pORFm164-derived peptide in fibroblasts in the face of all viral immunosubversive early gene functions. J. Virol. 76:6044-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtappels, R., N. K. Grzimek, D. Thomas, and M. J. Reddehase. 2002. Early gene m18, a novel player in the immune response to murine cytomegalovirus. J. Gen. Virol. 83:311-316. [DOI] [PubMed] [Google Scholar]
- 11.Holtappels, R., J. Podlech, G. Geginat, H. P. Steffens, D. Thomas, and M. J. Reddehase. 1998. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J. Virol. 72:7201-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holtappels, R., J. Podlech, N. K. Grzimek, D. Thomas, M. F. Pahl-Seibert, and M. J. Reddehase. 2001. Experimental preemptive immunotherapy of murine cytomegalovirus disease with CD8 T-cell lines specific for ppM83 and pM84, the two homologs of human cytomegalovirus tegument protein ppUL83 (pp65). J. Virol. 75:6584-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtappels, R., J. Podlech, M. F. Pahl-Seibert, M. Julch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtappels, R., D. Thomas, J. Podlech, G. Geginat, H. P. Steffens, and M. J. Reddehase. 2000. The putative natural killer decoy early gene m04 (gp34) of murine cytomegalovirus encodes an antigenic peptide recognized by protective antiviral CD8 T cells. J. Virol. 74:1871-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtappels, R., D. Thomas, J. Podlech, and M. J. Reddehase. 2002. Two antigenic peptides from genes m123 and m164 of murine cytomegalovirus quantitatively dominate CD8 T-cell memory in the H-2d haplotype. J. Virol. 76:151-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtappels, R., D. Thomas, and M. J. Reddehase. 2000. Identification of a Kd-restricted antigenic peptide encoded by murine cytomegalovirus early gene M84. J. Gen. Virol. 81:3037-3042. [DOI] [PubMed] [Google Scholar]
- 17.Jonjic, S., M. del Val, G. M. Keil, M. J. Reddehase, and U. H. Koszinowski. 1988. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J. Virol. 62:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenyon, T. K., J. I. Cohen, and C. Grose. 2002. Phosphorylation by the varicella-zoster virus ORF47 protein serine kinase determines whether endocytosed viral gE traffics to the trans-Golgi network or recycles to the cell membrane. J. Virol. 76:10980-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleijnen, M. F., J. B. Hupp, P. Lucin, S. Mukherjee, H. Farrell, A. E. Campbell, U. H. Koszinowski, A. B. Hill, and H. L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER is not retained but is transported to the cell surface. EMBO J. 16:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krmpotic, A., M. Messerle, I. Crnkovic-Mertens, B. Polic, S. Jonjic, and U. H. Koszinowski. 1999. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T-cell control in vivo. J. Exp. Med. 190:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruisbeek, A. M., and S. N. Vogel. 1994. Isolation of murine macrophages, p. 14.11.11-14.11.13. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology, vol. 3. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 22.LoPiccolo, D. M., M. C. Gold, D. G. Kavanagh, M. Wagner, U. H. Koszinowski, and A. B. Hill. 2003. Effective inhibition of Kb- and Db-restricted antigen presentation in primary macrophages by murine cytomegalovirus. J. Virol. 77:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melief, C. J. 2003. Regulation of cytotoxic T lymphocyte responses by dendritic cells: peaceful coexistence of cross-priming and direct priming? Eur. J. Immunol. 33:2645-2654. [DOI] [PubMed] [Google Scholar]
- 24.Mercer, J. A., and D. H. Spector. 1986. Pathogenesis of acute murine cytomegalovirus infection in resistant and susceptible strains of mice. J. Virol. 57:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morello, C. S., L. D. Cranmer, and D. H. Spector. 2000. Suppression of murine cytomegalovirus (MCMV) replication with a DNA vaccine encoding MCMV M84 (a homolog of human cytomegalovirus pp65). J. Virol. 74:3696-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morello, C. S., M. Ye, and D. H. Spector. 2002. Development of a vaccine against murine cytomegalovirus (MCMV), consisting of plasmid DNA and formalin-inactivated MCMV, that provides long-term, complete protection against viral replication. J. Virol. 76:4822-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pass, R. F. 2001. Cytomegaloviruses, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 29.Podlech, J., R. Holtappels, N. Wirtz, H. P. Steffens, and M. J. Reddehase. 1998. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J. Gen. Virol. 79:2099-2104. [DOI] [PubMed] [Google Scholar]
- 30.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddehase, M. J., S. Jonjic, F. Weiland, W. Mutter, and U. H. Koszinowski. 1988. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J. Virol. 62:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddehase, M. J., and U. H. Koszinowski. 1984. Significance of herpesvirus immediate-early gene expression in cellular immunity to cytomegalovirus infection. Nature 312:369-371. [DOI] [PubMed] [Google Scholar]
- 33.Reddehase, M. J., J. B. Rothbard, and U. H. Koszinowski. 1989. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature 337:651-653. [DOI] [PubMed] [Google Scholar]
- 34.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez, F., M. K. Slifka, S. Harkins, and J. L. Whitton. 2001. Two overlapping subdominant epitopes identified by DNA immunization induce protective CD8+ T-cell populations with differing cytolytic activities. J. Virol. 75:7399-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steffens, H. P., S. Kurz, R. Holtappels, and M. J. Reddehase. 1998. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J. Virol. 72:1797-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye, M., C. S. Morello, and D. H. Spector. 2002. Strong CD8 T-cell responses following coimmunization with plasmids expressing the dominant pp89 and subdominant M84 antigens of murine cytomegalovirus correlate with long-term protection against subsequent viral challenge. J. Virol. 76:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]