Abstract
Hepatitis C virus (HCV) is a major cause of viral hepatitis. There is no effective therapy for most patients. We have identified a nucleotide binding motif (NBM) in one of the virus's nonstructural proteins, NS4B. This structural motif binds and hydrolyzes GTP and is conserved across HCV isolates. Genetically disrupting the NBM impairs GTP binding and hydrolysis and dramatically inhibits HCV RNA replication. These results have exciting implications for the HCV life cycle and novel antiviral strategies.
Over 150 million people are infected with hepatitis C virus (HCV) worldwide (1). Current therapies are inadequate for most of these individuals (18). HCV is a positive single-stranded RNA virus. Its 9.6-kb genome encodes a single ∼3,000-amino-acid polyprotein, which is proteolytically processed into structural proteins, which are components of the mature virus, and nonstructural proteins, which are involved in replicating the viral genome (26). A characteristic feature of positive-strand RNA viruses is their use of cytoplasmic membranes as platforms for replication (27). These membranes can either be preexisting host cell compartments or novel structures induced by the virus (2, 4, 8, 17, 27). HCV is also believed to replicate in association with intracellular membranes, although how the RNA replication complex is assembled and maintained remains unknown. Recently the HCV NS4B protein has been shown to induce the formation of a distinct membranous structure designated the membranous web (5), which represents the candidate site for HCV RNA replication (12). The mechanism whereby NS4B mediates its function(s) in membrane-associated RNA replication, however, remains to be elucidated and may offer insights for the development of novel antiviral strategies. Here we report the identification of a nucleotide binding motif (NBM) within NS4B and show that this motif mediates both binding and hydrolysis of GTP and HCV RNA replication.
MATERIALS AND METHODS
Cell cultures.
Cell monolayers of the human hepatoma cell line Huh-7 were routinely grown in complete medium consisting of equal volumes of Dulbecco's modified minimal essential medium (Gibco) and RPMI 1640 (Gibco), supplemented with 1% l-glutamine (Gibco), 1% penicillin, 1% streptomycin, and 10% fetal bovine serum. Cell lines were passaged twice weekly after treatment with 0.05% trypsin-0.02% EDTA and seeding at a dilution of 1:10.
Antibodies.
A rabbit polyclonal antibody against green fluorescent protein (GFP) and an anti-rabbit secondary antibody were purchased from Molecular Probes. A monoclonal antibody against glutathione S-transferase (GST) was purchased from Cell Signaling Technology.
Plasmids.
Standard recombinant DNA technology was used to construct and purify all plasmids. All regions that were amplified by PCR were analyzed by automated DNA sequencing. Plasmid DNAs were prepared from large-scale bacterial cultures and purified by a Maxiprep kit (Marligen Biosciences). Restriction enzymes were purchased from New England Biolabs.
The Bart79I plasmid was described previously. Briefly, it was made by PCR mutagenesis (9) of HCVrep1bBartMan/AvaII (3) such that nucleotide 5336 was changed from a G to T, resulting in a change in NS5A amino acid 1179 from serine to isoleucine. This mutation results in a dramatic increase in replication efficiency of the HCV subgenomic replicon (3). The NS4B NBM mutations of Bart79I (numbers represent the amino acid positions relative to amino acid 1 of NS4B), G129V, I131N, K135S, and K135R, were generated by site-directed mutagenesis using a PCR-based method. Briefly, complementary primers (forward primer 1, 3, 5, or 7 with primer 10 or reverse primer 2, 4, 6, or 8 with primer 9; Table 1) and the enzyme Platinum Pfx (Invitrogen) were used to generate by PCR two DNA fragments with overlapping ends containing the mutation. These ends were annealed to allow 3′ extension of the complementary strand with the 3′ overlap of each strand as a primer. The product was then further amplified by PCR using primers 9 and 10 (Table 1). The PCR products and the Bart79I vector were cut with SspI and MluI, followed by ligation with T4 DNA ligase (Invitrogen) and transformation into chemically competent Escherichia coli (One Shot Top10 competent cells; Invitrogen).
TABLE 1.
Sequences of the oligonucleotides used in this study
| Primer no. | Primer namea | Sequence (5′→3′) |
|---|---|---|
| 1 | G129V-for | CGCTGGAGCGGCTGTTGTCAGCATAGGCCTTGGGAAGG |
| 2 | G129V-rev | CCTTCCCAAGGCCTATGCTGACAACAGCCGCTCCAGCG |
| 3 | I131N-for | GCGGCTGTTGGCAGCAACGGCCTTGGGAAGGTGC |
| 4 | I131N-rev | GCACCTTCCCAAGGCCGTTGCTGCCAACAGCCGC |
| 5 | K135S-for | GCAGCATAGGCCTTGGGAGTGTGCTTGTGGATATTTTGG |
| 6 | K135S-rev | CCAAAATATCCACAAGCACACTCCCAAGGCCTATGCTGC |
| 7 | K135R-for | GCAGCATAGGCCTTGGGAGGGTGCTTGTGGATATTTTGG |
| 8 | K135R-rev | CCAAAATATCCACAAGCACCCTCCCAAGGCCTATGCTGC |
| 9 | 3800sp-for | GTCATTGTGGGCAGGATCATCTTGTCCGGAAAGCC |
| 10 | 5Right-rev | GTGACCCAACCAGGTATATTGATTGAGCCCGACCAGGAATGTGACC |
| 11 | Ncol-4B-for | CAGCCATGGCCTCACACCTCCCTTACATCG |
| 12 | 4B-NcoI-rev | CATGCCATGGCGCATGGCGTGGAGCAGTCCTCG |
| 13 | Attb-TEV-4B-for | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAAACCTGTATTTTCAGGGCGCCTCACACCTCCCTTACATCGAAC |
| 14 | 4B-stop-attb-rev | GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGCATGGCGTGGAGCAGTCCTCG |
| 15 | 4Left-for | AGAGCGTCTTTACAGGCCTCACCCACATAGACGCCCATTTCTTGTCCCAG |
| 16 | 4Right-rev | AGGGCGCCAGGGGAGAGGATAGCAGGGAGTAGGTTAACCAGGTCCTCGG |
for, forward primers; rev, reverse primers.
The plasmid PEF-NS4B-GFP was constructed in a two-step cloning procedure as follows. A PCR fragment of the NS4B gene amplified from the Bart79I plasmid with forward and reverse primers containing NcoI restriction sites (primers 11 and 12; Table 1) was digested with NcoI and ligated with the NcoI-digested T7GFP plasmid (6) to generate the plasmid T7NS4BGFP. The plasmid T7NS4BGFP was digested with BglII-KpnI, and the fragment corresponding to NS4B-GFP was inserted into BamHI-KpnI-digested PEF6myc-HisA (Invitrogen) to yield PEF-NS4B-GFP. To obtain the plasmids encoding mutations in the NBM in NS4B, coding sequences for proteins with G129V, I131N, K135S, and K135R mutations in the NBM (see above) were digested with NdeI-HpaI and the fragment corresponding to the mutated NBM was inserted into NdeI-HpaI-digested PEF-NS4B-GFP.
The GST-NS4B plasmid and those encoding the corresponding NBM mutant proteins were generated by using the Gateway technology (Invitrogen) according to the manufacturer's protocol. In brief, a forward primer introducing a recombination site and a TEV protease cleavage site (primer 13; Table 1) and a reverse primer introducing a second recombination site and a stop codon (primer 14; Table 1) were used to generate a PCR product encoding wild-type or mutant NS4B flanked by the two recombination sites. This product was first introduced into a donor vector (pDonor 201), from which it was transferred to the destination vector, pDEST15, to yield the GST-NS4B plasmid by a two-step recombination procedure. The 5A-GFP plasmid was described previously (6).
Infection and transfection.
A vaccinia virus that expresses the T7 RNA polymerase (T7RNAP) was used to infect Huh-7 cells. Following a 45-min incubation at 37°C the cells were washed twice with Optimem (Invitrogen) and subjected to transfection with the appropriate construct by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The cells were supplemented with growth medium and incubated for 5 h at 37°C.
GTP binding assay.
Photoaffinity labeling of NS4B-GFP in membrane preparations with 32P-labeled GTP-γ-4-azidoanilide ([γ-32P]GTPγAA; 38 Ci/mmol; Affinity Labeling Technologies, Inc.) was carried out essentially as described previously (13). Cellular membrane preparations were prepared from vaccinia virus-infected and transfected Huh-7 cells. Following infection and transfection the cells were collected by trypsinization, washed once with phosphate-buffered saline (PBS), and resuspended in HME buffer (20 mM HEPES [pH 7.4], 1 mM EDTA, 2 mM MgCl2), which was supplemented with phenylmethylsulfonyl fluoride to a final concentration of 1 mM and a protease inhibitor cocktail (Sigma). The cells were lysed by two cycles of freeze-thaw in dry ice-ethanol and then passaged through a 27.5-gauge needle 10 times. Nuclei were removed by centrifugation at 250 × g for 10 min, and the postnuclear supernatant was subjected to ultracentrifugation at 100,000 × g for 30 min to obtain the membrane preparation. All steps were done at 4°C. One hundred and fifty micrograms of total membrane protein was resuspended in 20 mM Na-HEPES, pH 7.4. The assay mixture containing a 30-μl membrane preparation, 30 μl of 3× binding buffer (30 mM Na-HEPES [pH 7.4], 100 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2), and 30 μl of [γ-32P]GTPγAA (total of 15 μCi) was incubated for 1 h at 30°C in the dark. Samples were then irradiated with UV light at a 3-cm distance for 1 min (2,000 μW, 254 nm; UVS-28; UV Products) to allow covalent attachment of the bound radiolabeled guanine nucleotide. Unbound nucleotides were removed by ultracentrifugation for 10 min at 100,000 × g, and the membranes were resuspended in 1× binding buffer containing 2 mM dithiothreitol (for inactivation of the unbound material) and irradiated on ice for an additional 3 min with UV light.
Immunoprecipitation of labeled NS4B-GFP.
To identify the [γ-32P]GTPγAA-labeled NS4B-GFP, membrane preparations were incubated in 1 ml of TDB buffer (2.5% Triton X-100, 25 mM triethanolamine-Cl [pH 8.6], 20 mM NaCl, 0.5 M EDTA, 0.2% NaN3), followed by ultracentrifugation at 100,000 × g for 10 min. The supernatants were incubated overnight with a rabbit polyclonal antibody directed against GFP (Molecular Probes) and protein A-Sepharose (Amersham Biosciences). Following three washes in NET buffer (150 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl [pH 8.0]) immunoprecipitates were solubilized in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Nitrocellulose membranes were also subjected to Western analysis with mouse anti-GFP antibodies (Roche) and horseradish peroxidase-conjugated donkey anti-mouse immunoglobulin G, followed by chemiluminescence (Amersham) development.
Transfection.
DNA constructs were transfected into Huh-7 cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Fluorescence microscopy.
Cells expressing GFP fusion proteins were fixed in 4% formaldehyde 18 h posttransfection and mounted with polyvinyl alcohol (Mowiol) mounting medium. Fluorescence images were captured with a Nikon E600 fluorescence microscope equipped with a SPOT digital camera and the Openlab (Improvision) image acquisition software.
Expression and purification of wild-type and mutant GST-NS4B.
Proteins were expressed and purified as previously reported (30). Overnight cultures of E. coli transformed with parental or recombinant pDEST15 plasmids were diluted 1:100 in 400 ml of fresh medium and grown at 37°C to an optical density of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG; Invitrogen) was then added to a final concentration of 0.1 mM. After 2 h of growth at room temperature, cells were pelleted and resuspended in 25 ml of lysis buffer (PBS [pH 7.3], 1% Triton X-100 [J. T. Baker], 100 U of DNase [Sigma]/ml, 100 μg of Lysozyme [Sigma]/ml, protease inhibitor cocktail [Sigma], 1 mM phenylmethylsulfonyl fluoride [Sigma], 2 mM MgCl2). After 15 min of incubation on ice, cells were lysed by one cycle in a French press at a pressure of 10,000 lb/in2 for 1 min, followed by centrifugation at 12,000 × g for 5 min at 4°C. The supernatant was mixed at 4°C on a rotating platform with 200 μl of 50% glutathione-agarose beads (Sigma). Beads were then washed three times with PBS. GST-NS4B was eluted by a 10-min incubation at room temperature in 100 μl of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione, 0.1% Triton X-100). Elution was repeated twice. Glycerol was added to the pooled eluates at a final concentration of 20% and stored at −20°C until use as described below. Expression and purification were monitored by SDS-PAGE, followed by Coomassie staining or Western blot analysis with an anti-GST antibody. We estimate the maximum amount of non-GST-containing protein to be <5%. In addition to the expected full-length GST-NS4B band at 58 kDa, some faster-migrating GST-containing bands were detected. The latter appeared to be the result of premature termination, as their size correlated with the positions of codons poorly recognized by standard E. coli strains, and they were found to significantly decrease following the addition of appropriate tRNAs in an in vitro expression system (Rapid Translation system; Roche) (T. Danieli, unpublished data). Typical final yields were 5 μg of total protein per 100-ml bacterial culture. Of note, there were no differences in yield or purity between the NBM mutant proteins and wild-type GST-NS4B.
GTPase assays.
The standard GTPase assay was performed as previously described (25). One-half microgram of purified protein was incubated in a 30-μl reaction mixture containing 20 mM HEPES-KOH (pH 6.8), 10 mM MgCl2, 2 mM dithiothreitol, 40 μM cold GTP (Promega), and 15 μCi of [γ-32P]GTP/ml (5,000 Ci/mmol; Amersham Biosciences). Serial aliquots were collected at different incubation time intervals (5, 15, 30, 45, and 60 min), while the reaction was performed at 37°C. The reaction was terminated on ice by the addition of EDTA to a final concentration of 5 mM. Aliquots (0.5 μl) were then spotted onto polyethyleneimine cellulose-coated thin-layer chromatography (TLC) plates (Merck). Plates were developed in 0.15 M LiCl-0.15 M formic acid (pH 3.5) in a TLC chamber, dried, and subjected to autoradiography and quantitative phosphorimager analysis.
In vitro RNA transcription.
Plasmid DNA of the wild-type HCV (Bart79I) replicon and replicons encoding the various NS4B NBM mutations were linearized with ScaI and treated with proteinase K, followed by phenol-chloroform extraction and precipitation with ethanol. The DNA was resuspended in RNase-free water to a final concentration of 1 μg/μl. Four micrograms of DNA was used as a template for transcription with the Ribomax RNA production kit (Promega) according to the manufacturer's protocol. The template DNA was digested by the addition of 5 U of RQ1 DNase (Promega) and a 15-min incubation at 37°C. The unincorporated ribonucleotides were removed by size exclusion with a Micro Bio-Spin P-30 column (Bio-Rad), and the transcribed RNA was extracted with phenol-chloroform, followed by precipitation in ethanol. The RNA pellet was washed with 70% ethanol and resuspended in H2O. Determination of the RNA concentration was performed by measurement of the optical density at 260 nm. The integrity of the RNA and its concentration were confirmed by 1% agarose gel electrophoresis and ethidium bromide staining.
Colony formation assays.
The standard replicon colony formation assay was performed as previously described (3, 6). Briefly, subconfluent Huh-7 cells were trypsinized and collected by centrifugation at 700 × g for 5 min. The cells were then washed three times in ice-cold RNase-free PBS (BioWhittaker) and resuspended at 107 cells/ml in PBS. Five micrograms of in vitro-transcribed RNA was mixed with 0.4 ml of washed Huh-7 cells in a 2-mm-gap cuvette (BTX) and immediately pulsed (0.68 kV, five 99-μs pulses) with a BTX-830 electroporator. After a 10-min recovery at room temperature, pulsed cells were diluted into 10 ml of prewarmed growth medium. Cells were plated in 10-cm3 tissue culture dishes at different densities (4 × 106, 4 × 105, 8 × 104, and 4 × 104 cells per dish) to permit accurate colony counting. Twenty-four hours postelectroporation, the cells were supplemented with plain Huh-7 cells to a final density of 106 cells/plate. Following an additional 24 h, the selecting drug, G418 (Invitrogen), was added to the medium to a final concentration of 1 mg/ml. Growth medium supplemented with G418 was replaced every 4 days for 3 weeks. The plates were then washed twice with PBS and incubated in 1% crystal violet made in 20% ethanol for 5 min, followed by three washes with H2O to facilitate colony counting. The G418 transduction efficiency was calculated based on the number of G418-resistant colonies relative to the number of Huh-7 cells plated after electroporation. Results were expressed as number of colonies per microgram of transfected RNA of each mutant relative to the wild-type replicon.
RNA extraction, RT-PCR amplification, and sequencing.
Several G418-resistant clones were isolated from colony formation assays performed with the replicon harboring the G129V mutation. Total cellular RNA of individual clones was extracted with TRIZOL reagent (Invitrogen) according to the manufacturer's protocol. The reverse transcriptase reaction and PCR amplification were performed with the Superscript One-Step reverse transcriptase PCR (RT-PCR) kit (Invitrogen) according to the kit's protocol. Briefly, the amplification reaction mixture included 1 μg of total RNA as the template and 10 pmol of each primer. Two sets of primers (4Left-for with 4Right-rev and 3800sp-for with 4Right-rev; Table 1) were used to amplify 1-kb and 650-bp segments, respectively, each containing the NBM region coding sequence. Performing two independent amplification reactions for each clone provided further confirmation of the sequencing results. The RT reaction was performed at 50°C for 30 min and was followed by incubation at 95°C for 2 min. The DNA was amplified by 28 cycles of 95°C for 15 s, 60°C for 30 s, and 68°C for 1 min. A final elongation step was performed at 68°C for 10 min. The PCR products were purified from agarose gels with the Ultra Clean 15 DNA purification kit (MoBio) and sent for automatic sequencing on an ABI Prism 377 DNA sequencer (Sequetech).
Protein assays.
Concentrations of purified protein and protein content in membrane preparations were determined by the Bradford dye binding procedure using a Bio-Rad (Richmond, Calif.) protein assay kit.
RESULTS AND DISCUSSION
NS4B contains an NBM.
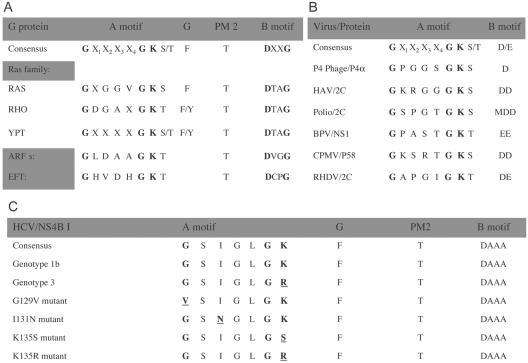
Inspection of the NS4B primary sequence reveals the presence of an NBM in the middle of NS4B. This motif consists of a set of conserved amino acids found in both the GTP-binding members of the G protein superfamily, as well as several viral proteins with nucleotide-binding domains (10, 11, 31, 32). The most highly conserved elements within these nucleotide-binding domains are the so-called A motif and B motif (Fig. 1A and B). Because binding and hydrolysis of nucleotides mediate a variety of critical signaling, membrane trafficking, and membrane fusion events (7, 23, 24), we hypothesized that the NBM within NS4B may similarly be important for NS4B's role in HCV RNA replication.
FIG. 1.
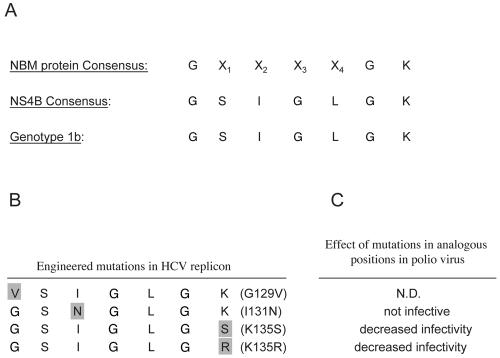
Conserved sequence elements in NBM-containing proteins (10, 11, 31, 32). The most conserved element of the NBM is the so-called A motif. Other conserved elements known from crystal structures to participate in nucleotide binding (G, PM2, and B motif) are also indicated. (A and B) Consensus sequences of the NBM from some representative family members of the G protein superfamily of GTP-binding proteins (A) and selected viruses with the indicated NBM-containing protein (B). (C) An NBM was located in HCV NS4B. The consensus amino acid sequences of all HCV isolates available for examination, the genotype 1b clone used in this study, genotype 3, and the engineered G129V, I131N, K135S, and K135R mutant NS4Bs are indicated. X, any amino acid. See text for details.
NS4B binds GTP.
To determine the properties associated with the wild-type and mutated versions of NS4B's NBM, we first constructed a plasmid, termed PEF-NS4B-GFP, which encodes a NS4B protein with a C-terminal, in-frame GFP tag. This tag allows for visualization in live cells and provides a convenient epitope outside of any future field of mutagenesis within NS4B. Importantly, GFP fusions to NS4B have been previously reported to have no difference in intracellular localization patterns from wild-type NS4B (14, 19).
To test the hypothesis that NS4B can bind GTP, we performed GTP-binding experiments using Huh-7 cells infected with a T7RNAP-expressing vaccinia virus and transfected with plasmids encoding NS4B-GFP or GFP or mock transfected. Membrane preparations were prepared, and aliquots were incubated with [γ-32P]GTPγAA (a UV-photoactivatable nonhydrolyzable GTP analog) essentially as described previously (13). Following a brief pulse of UV irradiation to activate covalent attachment of any bound GTP, pelleted membranes were washed and subjected to immunoprecipitation with a rabbit anti-GFP antibody, SDS-PAGE, transfer to nitrocellulose, autoradiography, and Western blotting.
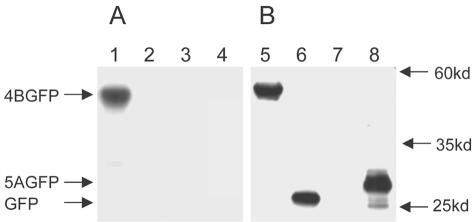
As shown in Fig. 2A, NS4B-GFP, but not GFP, was specifically labeled with GTP (Fig. 2A, lane 1 versus 2). Western analysis with an antibody against GFP of the immunoprecipitates revealed comparable expression levels of the two proteins (Fig. 2B, lane 5 versus 6). To provide another measure of the specificity of the observed labeling, we performed experiments with the 5A-GFP plasmid, which encodes the first 31 amino acids of HCV NS5A fused in frame to the N terminus of GFP (6). The resulting fusion protein thus contains, like NS4B (6a), a potent membrane-targeting N-terminal amphipathic helix yet does not include a known nucleotide-binding element. Essentially no GTP labeling of 5A-GFP was observed (Fig. 2A, lane 4) in spite of a larger amount of expressed protein (Fig. 2B, lane 8 versus 5). Labeling of 5A-GFP, detectable only after extensive film exposure, was used for background subtraction purposes in subsequent quantitative analyses. To our knowledge, this is the first demonstration that NS4B has GTP-binding activity. In addition, these results indicate that such binding activity is preserved when NS4B is expressed in the form of a fusion protein. The latter is a convenient although not surprising finding, as nucleoside triphosphate binding activity remains intact in fusion proteins made with other nucleotide-binding proteins (21, 28).
FIG. 2.
HCV NS4B binds GTP. Membrane preparations from Huh-7 cells transfected with plasmids encoding NS4B-GFP (lanes 1 and 5) or GFP (lanes 2 and 6), mock-transfected cells (lanes 3 and 7), and cells transfected with the plasmid encoding 5A-GFP (lanes 4 and 8) were incubated with 32P-labeled photoactivatable GTP. (A) Following 1 min of UV irradiation to activate covalent attachment of any bound GTP, samples were washed and subjected to immunoprecipitation with a rabbit anti-GFP antibody, SDS-PAGE, and autoradiography. (B) Aliquots of the immunoprecipitates were also analyzed by Western blotting. The blot was probed with a mouse anti-GFP antibody, followed by chemiluminescence detection. Molecular mass markers are indicated on the right.
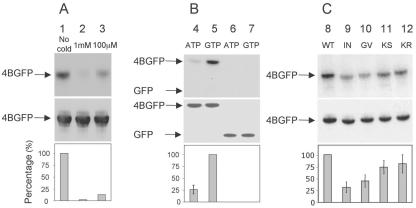
We further evaluated the specificity of NS4B's GTP binding by performing binding experiments in the presence of excess unlabeled ligand. Membrane aliquots of Huh-7 cells expressing NS4B-GFP were incubated with a 32P-labeled GTP analog as described above, except that cold guanosine 5′-O-(3-thiotriphosphate) (GTPγS) was added to the incubation mixture. As shown in Fig. 3A, the binding of labeled GTP progressively decreased in the presence of increasing concentrations of the cold competitor nucleotide (Fig. 3A, top, lane 1 versus 3 and 2).
FIG. 3.
The NS4B NBM is specific for GTP and sensitive to genetic mutation. Each panel shows an autoradiograph (top), a Western blot analysis with an anti-GFP antibody (middle), and a graph quantifying nucleotide binding relative to wild-type control (bottom). (A) Binding of labeled GTP is progressively decreased in the presence of increasing concentrations of cold competitor nucleotide. Huh-7 cells were transfected with a plasmid encoding NS4B-GFP. Membrane preparations were incubated with 10 μM labeled GTP compound in the absence of cold GTPγS (lane 1) or presence of 1 mM (lane 2) or 100 μM (lane 3) competing cold GTPγS, followed by immunoprecipitation as in Fig. 2A. (B) NS4B-GFP binds ATP significantly less efficiently than GTP. Membrane preparations prepared from Huh-7 cells transfected with plasmids encoding NS4B-GFP (lanes 4 and 5) or GFP (lanes 6 and 7) were incubated with equal concentrations of labeled ATP (lanes 4 and 6) or GTP (lanes 5 and 7), followed by immunoprecipitation as in Fig. 2A. (C) Mutations within the NBM impair GTP binding. Huh-7 cells were transfected with plasmids encoding wild-type NS4B-GFP (lane 8) or NS4B-GFP with one of the following NBM mutations: Ile131Asn (lane 9), Gly129Val (lane 10), Lys135Ser (lane 11), or Lys135Arg (lane 12). As above, membrane fractions were incubated with labeled GTP, followed by immunoprecipitation. Experiments were repeated between two and four times. When present, any detectable binding of GTP to the 5A-GFP negative control protein was used for background subtraction purposes. Representative gels are shown. Mean values are plotted in the graphs, and error bars represent standard errors.
Although the NBM of poliovirus 2C can bind GTP, its preference is for ATP (25). We therefore asked whether NS4B behaves similarly. Binding assays were performed using Huh-7 cells expressing NS4B-GFP as in Fig. 2. Membrane aliquots were incubated with equal concentrations of 32P-labeled ATP-γ-4-azidoanilide or the GTPγAA analog, followed by immunoprecipitation. As shown in Fig. 3B, although NS4B can bind ATP, this appears to be significantly less efficient than GTP binding (Fig. 3B, top, lane 4 versus 5). Again, no labeling of the control GFP with either ATP or GTP was detected (Fig. 3B, top, lanes 6 and 7). These results are in good agreement with the observation that NS4B, unlike poliovirus 2C, contains additional conserved amino acids implicated in GTP binding by the members of the G protein family (31) (Fig. 1C).
Mutation of NS4B's NBM impairs GTP binding.
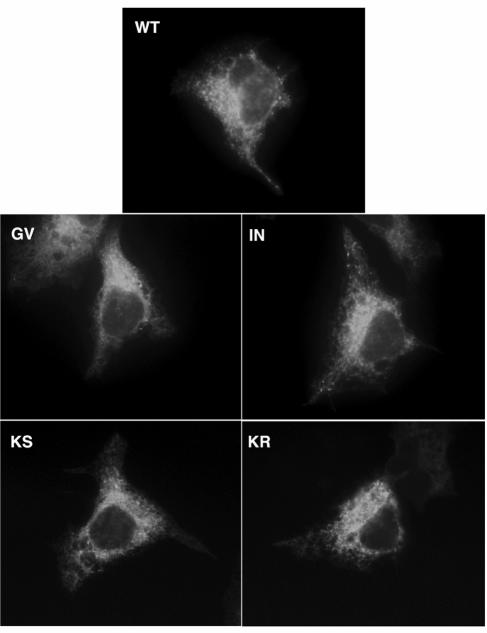
Genetic mutation of the NBM in a variety of nucleotide-binding proteins can both impair nucleotide binding and disrupt NBM-mediated functions. We therefore sought to test the hypothesis that mutations within the NS4B NBM can similarly impair GTP binding. For this, we designed four mutant proteins, each harboring a single amino acid mutation within the NS4B NBM (Fig. 1C). Ile131Asn is a single amino acid change at the X2 position of the NS4B NBM, a position that has been shown to be critical for NBM function in other proteins (31, 29). Lys135Ser and Lys135Arg are single amino acid changes at a position previously reported to confer an intermediate phenotype when mutated in the poliovirus system (31). Gly129Val is a single amino acid mutation at the highly conserved first position of the NBM A motif consensus sequence. These mutations were introduced into NS4B-GFP, and GTP binding assays were performed. As shown in Fig. 3C, top, the G129V and I131N NBM mutant proteins exhibited a two- to threefold reduction in GTP binding on average compared to the wild-type NS4B-GFP protein (lanes 10 and 9 versus 8). In contrast, the K135S and K135R NBM mutant proteins reduced GTP binding activity to a lesser degree (lanes 11 and 12 versus 8). This was not simply the result of an obvious gross effect of the mutations on folding, as the apparent intracellular expression levels and distribution patterns of mutant and wild-type proteins appeared identical by fluorescence microscopy (Fig. 4). Moreover, Western analysis with an anti-GFP antibody of the immunoprecipitates again revealed comparable levels of expression of these proteins (Fig. 3C).
FIG. 4.
Mutations within NS4B's NBM are not associated with obvious changes in protein expression level or intracellular distribution pattern. Huh-7 cells plated on coverslips were transfected with plasmids encoding wild-type NS4B-GFP (WT) or NS4B-GFP with one of the following NBM mutations: Gly129Val (GV), Ile131Asn (IN), Lys135Ser (KS), or Lys135Arg (KR). Eighteen hours posttransfection the cells were fixed and imaged by a fluorescence microscope. Note that all of these proteins display the same reticular membrane localization pattern with distinct foci located in the cytoplasm that is characteristic of wild-type NS4B.
NS4B has GTPase activity which is mediated by the NBM.
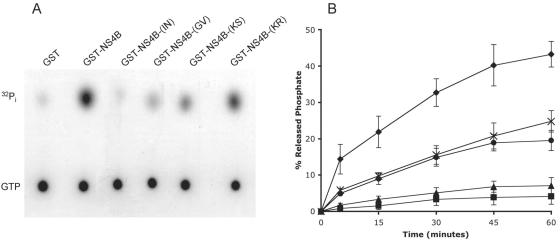
We next sought to test the hypothesis that the NS4B NBM mediates, in addition to GTP binding, GTP hydrolysis. For this, wild-type NS4B and the four NBM mutant proteins were fused in frame with N-terminal GST tags. The resultant fusion proteins, termed GST-NS4B, GST-NS4B-(IN), GST-NS4B-(GV), GST-NS4B-(KS), and GST-NS4B-(KR), were expressed in E. coli BL21 and purified with glutathione beads, as described previously (30). The purified proteins were then tested for their ability to hydrolyze GTP by a standard GTPase assay wherein release of phosphate from [γ-32P]GTP was monitored by quantitative TLC, essentially as described previously (25). Not only does NS4B have GTPase activity, but also the GTPase activity is sensitive to disruption of the NBM (Fig. 5). Indeed, the targeted mutations could either partially (K135S and K135R) or nearly completely (G129V and I131N) abolish GTPase activity. Mutations that mildly affect nucleoside triphosphate binding in other proteins have been reported to affect nucleotide hydrolysis more dramatically (21). It appears that a similar situation exists for NS4B.
FIG. 5.
NS4B has GTPase activity, which is mediated by the NBM. Equal amounts of purified GST, GST-NS4B, and the NBM mutant proteins GST-NS4B(GV), GST-NS4B(IN), GST-NS4B(KS), and GST-NS4B(KR) were incubated with [γ-32P]GTP. Aliquots were collected every 15 min and subjected to TLC to allow separation of hydrolyzed 32Pi from GTP, followed by autoradiography and phosphorimager analysis. (A) Representative TLC plate. Locations of GTP and 32Pi standards are indicated on the left. (B) GTPase activity of wild-type NS4B (⧫) and G129V (▴), I131N (▪), K135S (•), and K135R (×) mutant proteins is plotted as a function of time. When present, any detectable hydrolysis of GTP in the GST control was used for background subtraction purposes. Each data point represents the average of at least four independent determinations. The error bars represent standard deviations.
Disrupting NS4B's NBM inhibits HCV RNA replication.
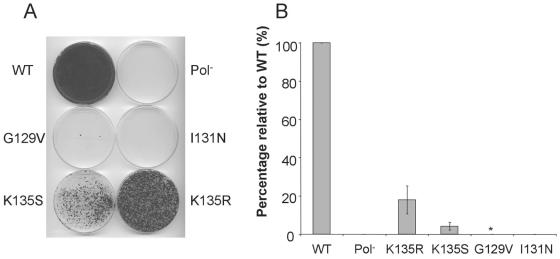
To test the hypothesis that the NS4B NBM is important for HCV replication, we introduced the above series of point mutations within the NBM into high-efficiency HCV replicons (Fig. 6B). The latter were then assayed in standard replicon colony formation assays, as previously described (3, 6). A replicon carrying a lethal mutation in the active site of the viral polymerase (GDD→AAG), NS5B, was used as a negative control (16). The tested mutations had a variety of significant effects on replication (Fig. 7). While mutating the A motif Lys to either Ser (K135S) or Arg (K135R) was associated with intermediate levels of replication (2 and 18% respectively), mutating the first Gly of the A motif to Val (G129V) dramatically inhibited replication with only rare colony formation. No colonies could be isolated when the A motif Ile was changed to Asn (I131N). The appearance of colonies on the G129V plates provided us with the opportunity to perform reversion analyses. RNA was therefore extracted from the rare colonies isolated from such experiments, amplified by RT-PCR, and sequenced. While it is possible that secondary-site revertants may appear, our sequence analysis to date has revealed the presence of only primary-site revertants. The latter do, however, provide additional evidence for the requirement of maintaining a functional NBM.
FIG. 6.
Engineered mutations in NS4B's NBM. (A) Consensus sequences for all NBM-containing proteins, all HCV isolates, and HCV genotype 1b. X, any amino acid. (B) Single-amino-acid point mutations (gray) were engineered into the NBM of NS4B at the indicated positions. (C) Effect on poliovirus replication of mutations engineered at the same positions in the poliovirus 2C protein's NBM (22, 31). N.D., not done.
FIG. 7.
Genetic disruption of NS4B's NBM impairs HCV RNA replication. Replication of HCV replicons harboring the mutations depicted in Fig. 5B were assayed by colony formation assays. (A) Wild-type and mutant replicons were electroporated into Huh-7 cells, and G418-resistant colonies were selected and stained with crystal violet. These replicons contain the gene for neomycin phosphotransferase (6). Each dot represents a colony of Huh-7 cells that was able to grow in the presence of G418 due to the presence of efficiently replicating intracellular replicons. WT, Bart79I (wild type) (3, 6); Pol−, Bart79I with a lethal mutation in NS5B (16); KR, Bart79I with a Lys135Arg point mutation; KS, Bart79I with a Lys135Ser point mutation; GV, Bart79I with a Gly129Val point mutation; IN, Bart79I with an Ile131Asn point mutation. A representative plate is shown. (B) Percentages of colonies relative to wild-type control. Note that rare colonies were obtained with the G129V mutant protein (*) and that none were obtained with the Pol− or I131N mutant proteins.
Our results suggest that the NBM within NS4B is essential for mediating NS4B's role in HCV replication in vitro. The requirement of a NBM for productive viral infection in vivo is further suggested by the conservation of this motif across natural HCV isolates of all genotypes (Fig. 1C; data not shown). The dramatic effect on replication would not appear to be simply the result of altered expression, misfolding, or mislocalization of the mutant proteins. Indeed Western blot analyses of proteins expressed in transfected cells and fluorescence analysis of the wild-type protein and various NBM mutant NS4B proteins fused to GFP revealed no obvious differences in protein levels (Fig. 3C, middle) and intracellular distribution (Fig. 4), respectively. In addition, the mutations in the NBM do not seem to disrupt targeting of the protein to the endoplasmic reticulum (ER) (14, 19), as the colocalization of mutant NS4Bs with the ER marker PDI is similar to that of wild-type protein (data not shown). Finally, examination of HCV sequences from clinical isolates published in public databases reveals that wobble mutations occur at the codon sites that we mutated, strongly suggesting that it is unlikely that our mutations disrupt a critical RNA structure or sequence. Rather, these subtle mutations appear to affect a key regulatory site in the NS4B amino acid sequence. Interestingly, when analogous mutations were introduced into the A motif of poliovirus 2C, similar quantitative and qualitative effects on viral replication were observed (Fig. 6C) (22, 31). Although speculative, the decreased replication potential of the K135R mutant replicon compared to that of the parental genotype 1b replicon is also intriguing. Arginine at this position of the NS4B NBM is found in HCV genotype 3, a genotype that clinically is much more responsive to interferon therapy than genotype 1b (15).
Current structure prediction models do not offer a conclusive assignment of the secondary structure of NS4B in the NBM region. This likely reflects the relative paucity of solved crystal structures of membrane proteins such as NS4B within the databases used to construct the prediction program algorithms. Nevertheless, it may well be that the NBM is located within a globular cytoplasmic domain. Certainly, recent empirical studies using glycosylation markers suggest that this domain is within the cytoplasm as opposed to the ER lumen (19). Alternatively, the NBM may reside close to the interface between the cytoplasm and ER membrane in a “cytoplasmic pocket,” perhaps created by NS4B oligomerization, within which the NBM is exposed to nucleotides.
Our results suggest that efficient binding and hydrolysis of nucleotides by NS4B are required for viral replication. These results do not rule out the possibility that the NS4B NBM mediates binding of nucleotides not only as single molecules but also as part of a polynucleotide structure (such as RNA). In the latter scenario, by simultaneously binding cellular membranes and RNA, NS4B might serve in channeling the replication complex through the vesicular membranous structure or contribute to the structural integrity of the replication complex by anchoring it to membranes.
GTPases typically are associated with a variety of regulatory factors (7). Based on our characterization of NS4B we would now expect it to also have such partners of cellular and/or viral origin. GTPases, such as Dynamin, can also hydrolyze other nucleotides (20). We recently became aware that an ATPase function has been proposed for NS4B (WO 99/01582), and this is consistent with our ATP binding data. That different types of nucleotides can bind NS4B suggests that nucleoside analogs traditionally contemplated for use against the NS5B polymerase activity may now be considered for targeting the GTPase function of NS4B as well.
Finally, these results reveal that the NBM within NS4B represents an attractive new potential target for anti-HCV therapy in its own right. Because the amino acid sequence immediately adjacent to either side of the NBM region that we mutated is highly conserved across HCV isolates, yet very different from that contained in known host cell GTP-binding proteins, highly selective inhibitors can be readily envisaged. Such compounds represent an exciting potential addition to current anti-HCV combination therapy regimens.
Acknowledgments
We thank Benjamin Aroeti, Allen Cooper, Harry Greenberg, Karla Kirkegaard, and Charles Rice for helpful discussions and critical reading of the manuscript.
S.E. is a recipient of the Stanford Dean's fellowship award. M.E. is a recipient of an American Liver Foundation Award. This work was also supported by RO1DK066793 and a Burroughs Welcome Career Award (to J.S.G.).
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1980. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 100:390-399. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K.J., and A. Kolykhalov, and C. M. Rice. 2001. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Chu, P. W., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. [DOI] [PubMed] [Google Scholar]
- 5.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice and J. S. Glenn. 2003. The amphipathic helix-dependent localization of NS5A mediates HCV RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 8.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glenn, J. S., J. C. Marsters, and H. B. Greenberg. 1998. Use of a prenylation inhibitor as a novel antiviral agent. J. Virol. 72:9303-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 12.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudi, S. R. P., B. C. Craig, and J. A. Frangos. 1996. Fluid flow rapidly activates G proteins in human endothelial cells—involvement of G proteins in mechanochemical signal transduction. Circ. Res. 79:834-839. [DOI] [PubMed] [Google Scholar]
- 14.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 15.Knolle, P. A., S. Kremp, T. Hohler, F. Krummenauer, P. Schirmacher, and G. Gerken. 1998. Viral and host factors in the prediction of response to interferon-alpha therapy in chronic hepatitis C after long-term follow-up. J. Viral Hepat. 5:399-406. [DOI] [PubMed] [Google Scholar]
- 16.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarus, L. H., and R. Barzilai. 1974. Association of foot-and-mouth disease virus replicase with RNA template and cytoplasmic membranes. J. Gen. Virol. 23:213-218. [DOI] [PubMed] [Google Scholar]
- 18.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 19.Lundin, M., M. Monne, A. Widell, G. von Heijne, and M. A. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda, K., T. Nakata, Y. Noda, R. Sato-Yoshitake, and N. Hirokawa. 1992. Interaction of Dynamin with microtubules: its structure and GTPase activity investigated by using highly purified Dynamin. Mol. Biol. Cell 3:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin, M. S., R. Casais, J. M. Alonso, and F. Parra. 2000. ATP binding and ATPase activities associated with recombinant rabbit hemorrhagic disease virus 2C-like polypeptide. J. Virol. 74:10846-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirzayan, C., and E. Wimmer. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547-555. [DOI] [PubMed] [Google Scholar]
- 23.Neves, S. R., P. T. Ram, and R. Iyengar. 2002. G protein pathways. Science 296:1636-1639. [DOI] [PubMed] [Google Scholar]
- 24.Pfeffer, S. 2003. Membrane domains in the secretory and endocytic pathways. Cell 112:507-517. [DOI] [PubMed] [Google Scholar]
- 25.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 26.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 27.Rice, C.M. 1996. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, p. 931-959. Lippincott-Raven Publications, Philadelphia, Pa.
- 28.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 29.Seeburg, P. H., W. W. Colby, D. J. Capon, D. V. Goeddel, and A. D. Levinson. 1984. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature 312:71-75. [DOI] [PubMed] [Google Scholar]
- 30.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 31.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73:1977-1986. [DOI] [PubMed] [Google Scholar]
- 32.Valencia, A., P. Chardin, A. Wittinghofer, and C. Sander. 1991. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 30:4637-4648. [DOI] [PubMed] [Google Scholar]