Abstract
Proteins associated with the murine cytomegalovirus (MCMV) viral particle were identified by a combined approach of proteomic and genomic methods. Purified MCMV virions were dissociated by complete denaturation and subjected to either separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and in-gel digestion or treated directly by in-solution tryptic digestion. Peptides were separated by nanoflow liquid chromatography and analyzed by tandem mass spectrometry (LC-MS/MS). The MS/MS spectra obtained were searched against a database of MCMV open reading frames (ORFs) predicted to be protein coding by an MCMV-specific version of the gene prediction algorithm GeneMarkS. We identified 38 proteins from the capsid, tegument, glycoprotein, replication, and immunomodulatory protein families, as well as 20 genes of unknown function. Observed irregularities in coding potential suggested possible sequence errors in the 3′-proximal ends of m20 and M31. These errors were experimentally confirmed by sequencing analysis. The MS data further indicated the presence of peptides derived from the unannotated ORFs ORFc225441-226898 (m166.5) and ORF105932-106072. Immunoblot experiments confirmed expression of m166.5 during viral infection.
Murine cytomegalovirus (MCMV), a member of the betaherpesvirus family, shares 45.2% sequence identity with human CMV (HCMV) and is currently the most commonly used animal model for the study of CMV-induced disease. The MCMV genome has a size of 230 kbp and was originally estimated to encode 170 proteins (45). The MCMV virion consists of double-stranded viral DNA surrounded by an icosahedral capsid, a complex proteinaceous tegument, and a lipid membrane (34). Although the composition of HCMV particles has been addressed (3, 10), the protein composition of the MCMV virion has not been examined in detail.
The majority of MCMV genes have been annotated based on their homologues in HCMV (45) and, consequently, many open reading frames (ORFs), including genes encoding some structural proteins, lack experimental confirmation. Analysis of regions unique to MCMV has been even more limited. There is, therefore, a need for characterization of the composition of MCMV particles, as well as the confirmation of putative structural and functional homologues of HCMV gene products.
The traditional method of analyzing the protein composition of a virus preparation consists of denaturation in sodium dodecyl sulfate (SDS), separation of the constituent proteins by electrophoresis, and identification either by immunological methods or by sequencing of visible bands via Edman degradation or mass spectrometry (MS). This approach has been used to analyze other herpesvirus family members, such as HCMV (3), murine gammaherpesvirus 68 (9), Epstein-Barr virus (18), and Kaposi's sarcoma-associated herpesvirus (36). Direct digestion of viral particles and “shotgun” sequencing by liquid chromatography-tandem MS (LC-MS/MS) was first explored with the much simpler adenovirus type 5 proteome (13). Although adenovirus particles consist of only 11 well-documented viral proteins, this analysis illustrated the sensitivity of LC-MS/MS and its applicability to the study of viral proteomes.
The approach presented here consists of two separate yet complementary strategies. In addition to resolution of MCMV viral proteins by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by tryptic digestion and MS/MS analysis, we subjected purified MCMV particles to direct in-solution digestion. Digestion of virus particles in situ produces a complex mixture of tryptic fragments which can be separated by nanoflow LC followed by MS/MS. Omission of a gel separation step enhances recovery of peptides derived from proteins prone to aggregation, hydrophobic peptides, or proteins of low abundance. The drawback of this approach is that information on the molecular weight of the polypeptide source of the tryptic fragment in question is lost.
Analysis on the protein level must be supported by an accurate analysis on the genomic level. Statistical gene prediction programs have been used previously for viral genome annotation. The GeneMark program (7a), for example, has been used to identify genes in the genomes of bovine herpesvirus 4 (65) as well as other viruses (19, 32, 51, 53). More recent studies have used information from newly sequenced, closely related CMV genomes (17), as well as tools which derive amino acid positional patterns from protein databases to predict protein coding regions in viruses (2, 35).
A particular feature of MCMV is the heterogeneity of the nucleotide composition of its genome. This heterogeneity makes the statistical models for gene prediction, trained from the genomic sequence as a whole, not sufficiently accurate in the regions in which the composition deviates from the average. To eliminate these effects, a Bayesian segmentation method (43) was used to segment the genome into regions of uniform nucleotide composition. These regions were then analyzed independently using a virus-specific version of the self-training gene-finding program GeneMarkS (5).
In order to analyze the proteomic data, GeneMark was used to generate a database of putative MCMV ORFs. This database contains viral ORFs previously annotated (45), as well as MCMV genes newly predicted by GeneMark. Searching this bioinformatic database with the obtained MS proteomic data confirmed a number of annotated MCMV-derived gene products and led to the detection of peptides attributed to novel MCMV genes. In addition, it allowed the detection of sequencing errors in two MCMV virion-associated proteins.
MATERIALS AND METHODS
Cells and viruses.
Mouse embryonic fibroblasts (MEFs) of C57BL/6 mice were grown in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum, penicillin, streptomycin, and 2 mM glutamine. NIH 3T3 cells (ATCC CRL1658) and 10.1 cells (22) were cultured in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum, penicillin, streptomycin, and 2 mM glutamine. The MCMV wild-type viruses used in this study were either MCMV strain Smith or the bacterial artificial chromosome (BAC)-derived wild-type recombinant MW97.01 (56).
Purification and isolation of MCMV.
Virus was produced in 10.1 cells or low-passage primary C57BL/6 MEFs infected at a low multiplicity of infection with MCMV strain Smith (ATCC VR-1399). Cell supernatants were harvested 24 h after 100% cytopathic effect and cleared by centrifugation in a Sorval GSA rotor (3,500 rpm, 30 min, 4°C). Extracellular virus was then pelleted at 11,500 rpm for 2 h at 4°C. Pelleted material was resuspended in 2 ml of phosphate-buffered saline (PBS) and homogenized four times using a tight-fitting 1-ml Dounce homogenizer. Residual debris was removed by microcentrifugation in an Eppendorf centrifuge (2,000 rpm, 10 min, 4°C). Supernatants containing viral particles were loaded onto a 5-to-30% continuous dextran gradient in SW28 centrifugation tubes and subjected to ultracentrifugation (76,000 × g, 1 h, 4°C). Purified virus was visible as an opalescent white band located at a concentration of 15 to 20% dextran. The virus-containing band was removed using a 21-gauge needle, diluted 1:1 in PBS, layered onto a 10-to-30% dextran gradient, and centrifuged at 76,000 × g for 1 h at 4°C. Banded material was isolated, diluted in 10 ml of PBS, and pelleted in an SW55Ti rotor at 31,000 rpm for 1.5 h at 4°C. Pelleted material was resuspended in a minimal amount of PBS and snap-frozen at −80°C. Purity was confirmed by electron microscopy of negatively stained preparations.
Protein gel electrophoresis.
Viral preparations were denatured, boiled in reducing sample buffer, and then separated on a 10% acrylamide SDS-PAGE gel. Proteins were visualized by Coomassie blue staining.
Tryptic digestion of gel fragments and analysis by electrospray MS/MS (LC-MS/MS).
In-gel tryptic digestion was performed essentially as described previously (26). The samples were applied to a nanoflow LC system (Waters CapLC, Medford, Mass.) equipped with a Picofrit column (75-μm inner diameter, 9.8 cm; NewObjective, Woburn, Mass.) at a flow rate of approximately 150 nl/min using a Nanotee (Waters) 16/1 split (initial flow rate, 5.5 μl/min). The LC system was directly coupled to a QTOF micro tandem mass spectrometer (Waters). Analysis was performed in survey scan mode, and parent ions with intensities greater than 7 were sequenced in MS/MS mode using the MassLynx 4.0 software (Micromass).
Proteomic analysis of MCMV-derived peptides and database generation.
Individual MS/MS spectra were submitted to MASCOT version 1.8 (Matrixscience) (41) and searched against the NCBI nonredundant database (NCBInr) or against a custom-made database consisting of MCMV gene products predicted using a virus-specific version of GeneMarkS. In addition, two new databases were generated. One contained protein translations of all possible ORFs greater than 20 amino acids. The other included protein translations of all sequences between two consecutive stop codons. Spectra obtained from the MS analysis were searched against all three databases. These databases allowed for the identification of novel MCMV genes by an exhaustive search among all possible virus-derived protein sequences. Proteins were scored using a probability-based MOWSE algorithm, and the scores were reported in the form of −10 · log(P), where P is the probability that the observed match is a random event (41). Matches with scores indicating a less-than-5% probability of being a random match were judged as significant. In all searches, oxidation of methionine, carbamidomethylation of cysteine residues, acetylation of N termini, and sodiated glutamic acid and aspartic acid were considered possible modifications. Individual peptide interpretations were ranked as significant if they had a MASCOT MOWSE score of greater than 20 and a minimum of four consecutive b or y ions present in MS/MS spectra. At least two peptides were found for each identified ORF except for ORF105932-106072.
MCMV DNA analysis.
The sequences of the Smith strain of MCMV and Maastricht strain of rat CMV (RCMV) were obtained from RefSeq (NC_004065 and NC_002512). The analysis of the MCMV genome was performed essentially as described elsewhere (33), with the following modifications. The MCMV genome was divided into several regions by using the Bayesian segmentation method (43), modified to keep the number and the minimal size of segments suitable for subsequent model training. The borders between segments were then adjusted to not interrupt any ORF larger than 300 bp.
The individual segments were analyzed with the GeneMarkS program, using either heuristic models defined in the first iteration (for segments shorter than 17 kbp) or models defined at a point of reaching convergence in the self-training process. Since the GeneMark program (7a), developed prior to GeneMarkS (5), has the ability to identify profiles of coding potential that visualize the predicted genes in more detail than GeneMarkS, we also built models for the GeneMark program. These models were built for each genomic segment by using protein-coding and noncoding regions identified by GeneMarkS. GeneMark models were constructed for each genomic segment by using the final noncoding and protein-coding parse, and these models were used to generate the GeneMark graphical outputs. The GeneMark outputs were used to detect irregularities in the protein-coding potential in both previously annotated and newly predicted genes. Putative frameshifts identified in this analysis were further studied through similarity searches in protein databases.
Sequencing of m20 and M31.
To sequence the regions of m20 and M31, PCR fragments were generated using the plasmid pSLFRTKn as template DNA with the primers 5′-m20 (5′-CTGATCCTGACCGCACGCCCG) and 3′-m20 (5′-TCGTGCACGGATGTTCGGAATCG) for m20 and with the primers 5′-M31 (5′-TACGATCTCTGCTTCAGAGAGG) and 3′-M31 (5′-AATTACACTACAGGCATATACACAGC) for M31. PCR fragments were sequenced in both directions with the same primers by the Harvard Medical School Microbiology DNA Sequencing Facility.
BAC mutagenesis and construction of recombinant MCMVs.
Recombinant MCMV was generated from the MCMV BAC genome pSM3fr (56) by mutagenesis in Escherichia coli as described previously (55). For construction of an MCMV genome with a 3′-terminal addition of the hemagglutinin (HA) tag sequence to the putative m166.5 ORF, a PCR fragment was generated using the plasmid pSLFRTKn (1) as template DNA and the contiguous primers 5′-m166.5 (5′-AGGGCGGCGGTGGTGGTGGTTTTCTTCGGTTTGCTACAGTACAAGGACGACGACGACAAGTAA) and 3′-m166.5-HA (5′TCTGGTGTCTCGCGTGGCTCGTCAGCCGCCCCGCGCTCGTACCCCTACGACGTCCCCGACTACGCCTGACGTCGTGGAATGCCTTCGAATTC) (the HA sequence is underlined). For construction of an MCMV genome with a 3′-terminal addition of the HA tag sequence to the m166 ORF, a PCR fragment was generated using the plasmid pSLFRTKn-SBP-HA (unpublished data) as template DNA and the contiguous primers 5′-m166 (5′-CAACGGTTTAATAATAGAAAAGGAAATCGGGTCACAGTCCACAAGGACGACGACGACAAGTAA) and 3′-m166-HA (5′-GGCGGACCGGCCTATGAGATACTCGTGAACGAGGAGACGGCGTCCTCCGACGAGAAGACCACCGGCTGGAG). The PCR fragments were inserted into pSM3fr by homologous recombination in E. coli, which led to deletion of the native m166.5 and m166 stop codons, respectively, and the in-frame insertion of the HA tag sequence. Correct mutagenesis was confirmed by restriction pattern analysis and sequencing of the 3′-terminal part of the tagged genes. The recombinant viruses MCMV-m166.5-HA and MCMV-m166-HA were generated from the respective BACs by transfection of MEFs.
Immunoblot analysis.
For detection of HA-tagged proteins m166 and m166.5, NIH 3T3 cells were infected at a multiplicity of infection of 10, and total cell lysates were harvested 5 and 24 h postinfection. Immunoblot analysis was performed with anti-human influenza virus HA epitope-specific rat monoclonal antibody 3F10 directly coupled to peroxidase (Roche). Proteins were visualized using the ECL Plus chemiluminescence system (Amersham Biosciences).
RESULTS
MCMV was purified from the supernatant of infected primary MEFs or 10.1 cells 24 h after 100% cytopathic effect. The preparation was homogenized and passed over two consecutive dextran sedimentation velocity gradients. Purified MCMV was examined by negative stain electron microscopy and found to contain predominantly single-capsid virion particles with the typical morphology described for MCMV (14) (Fig. 1A). Virus preparations attained maximum purity after passage through two gradients. Additional gradient purifications resulted in a uniform decrease in the total yield of viral material (data not shown). To compare commonly used methods of herpesvirus purification, positive density-negative viscosity glycerol tartrate gradient centrifugation and sucrose gradient centrifugation were carried out in parallel. As determined by SDS-PAGE analysis and electron microscopy, the best purification was obtained using dextran sedimentation velocity gradient centrifugation.
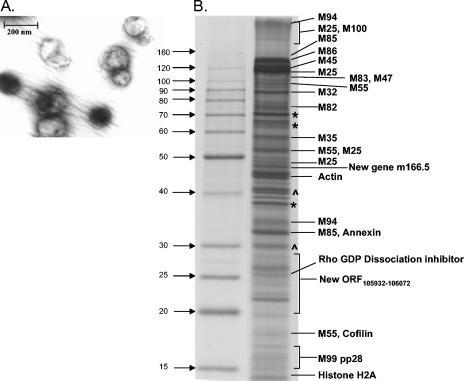
FIG. 1.
Characterization of isolated MCMV virions. (A) Electron microscopy picture of purified MCMV particles. (B) Analysis of virus particles isolated from supernatant of infected cells. Isolated protein was separated by SDS-10% PAGE, and individual polypeptides were excised from the gel, digested with trypsin, and analyzed by MS/MS. MCMV-encoded polypeptides and host cell proteins are indicated. Contaminating serum proteins (e.g., serum albumin) and unidentified polypeptides are marked with an asterisk and a caret, respectively.
Identification of MCMV virion-associated proteins.
The polypeptide composition of MCMV preparations was analyzed by SDS-PAGE. After Coomassie blue staining, individual bands were excised and remaining regions of the gel were divided into sections of 1 cm in height. These gel slices were then subjected to in-gel digestion with trypsin, followed by high-performance LC and MS/MS as described previously (26).
Due to the incompleteness of current MCMV annotations in commonly used protein databases, it was necessary to construct a unique database of predicted MCMV gene translations against which to search the obtained peptide MS/MS spectra. This new database was created based on the GeneMarkS program predictions. In an attempt to prevent search bias due to the small database size, searches were also conducted against a merged database of SwissProt and the newly assembled MCMV entries. The large size of the latter database, however, greatly increased the MASCOT MOWSE cutoff score and decreased the sensitivity of this approach. As no prominent MCMV peptide also matched proteins in the SwissProt database, this step was considered unnecessary for subsequent analysis. Using these newly constructed databases, we identified 19 viral proteins by in-gel digestion analysis (Fig. 1B).
In order to identify virion-associated components of lower abundance, direct in-solution digestion of isolated virions was carried out in parallel. This method eliminates peptide purification steps and minimizes the loss of proteolytic fragments from virion proteins present in low copy numbers. Furthermore, virion-associated proteins with molecular masses of 10 kDa or less are more likely to be resolved using this assay because of the difficulties inherent in resolving low-molecular-weight proteins by gel matrices. This approach revealed a total of 58 virus-derived gene products associated with the virions (Table 1).
TABLE 1.
MCMV ORFs associated with MCMV virions identified by MS/MS
| ORF | Strand | Position
|
HCMVe | Comment (reference)a | Mass (kDa)
|
% Cov- eragef | No. of spectrag | ||
|---|---|---|---|---|---|---|---|---|---|
| From | To | Predicted | Observed | ||||||
| m02 | 999 | 1979 | Glycoprotein family m02 (36) | 36.6 | 39 | 10 | |||
| m18 | C | 17071 | 20193 | Highly antigenic early gene (23) | 108.5 | 14 | 6 | ||
| m20b | C | 20581 | 23044 | 89.4 | 6 | 2 | |||
| M25 | 26015 | 28813 | UL25 | Tegument protein (60) | 130, 105, 95 | 48 | 54 | 91 | |
| m25.2 | C | 30245 | 31657 | US22 | US22 homologue (30) | 51.8 | 44 | 9 | |
| M28 | C | 34486 | 35778 | UL28 | 47.3 | 15 | 4 | ||
| M31c | 37278 | 39068 | UL31 | 66.1 | 18 | 5 | |||
| M32 | C | 39280 | 41436 | UL32 | (HCMV: pp150) (25) | 78.6 | 90 | 37 | 20 |
| M35 | 47465 | 45909 | UL35 | UL25 family member, virulence factor (52) | 58.1 | 58 | 41 | 8 | |
| m39 | C | 52484 | 53200 | 25.6 | 59 | 4 | |||
| M43 | C | 55351 | 57144 | UL43 | Immunoregulatory gene (49) | 67.0 | 55 | 11 | |
| M44 | C | 57885 | 59120 | UL44 | DNA binding phosphoprotein (27) | 44.6 | 22 | 3 | |
| M45 | C | 59515 | 63038 | UL45 | Ribonucleotide reductase homolog, anti- apoptotic (11) | 120 | 120 | 34 | 12 |
| M46 | C | 63040 | 63924 | UL46 | (HCMV: minor capsid binding protein) (20) | 33.2 | 36 | 8 | |
| M47 | 63923 | 67045 | UL47 | (HCMV: tegument protein) (4) | 118.1 | 115 | 9 | 3 | |
| M48 | 67042 | 73491 | UL48 | Large tegument protein (45) | 238.5 | 22 | 18 | ||
| m48.2 | C | 73571 | 73867 | UL48.5 | Smallest capsid protein (8) | 9.8 | 87 | 9 | |
| M51 | C | 76515 | 77216 | UL51 | 25.1 | 27 | 2 | ||
| M54 | C | 79701 | 82994 | UL54 | DNA polymerase (45) | 123.8 | 17 | 8 | |
| M55 | C | 83003 | 85816 | UL55 | Glycoprotein B (44) | 130, 105, 52 | 99, 52, 18 | 41 | 20 |
| M56 | C | 85716 | 88112 | UL56 | (HCMV: terminase subunit, tegument protein) (7) | 89.0 | 25 | 8 | |
| M57 | C | 88319 | 91894 | UL57 | (HCMV: Single-stranded DNA binding protein) (39) | 131.4 | 10 | 6 | |
| M69 | C | 96193 | 98721 | UL69 | (HCMV: tegument protein) (58) | 93.0 | 28 | 11 | |
| M70 | C | 99010 | 101904 | UL70 | (HCMV: helicase-primase subunit) (39) | 109.8 | 11 | 5 | |
| M71 | 101903 | 102802 | UL71 | 32.9 | 65 | 6 | |||
| M72 | C | 103031 | 104236 | UL72 | dUTPase (31) | 45.0 | 13 | 2 | |
| M74 | C | 104496 | 105812 | UL74 | 49.1 | 27 | 6 | ||
| M75 | C | 106110 | 108287 | UL75 | Glycoprotein H (61) | 81.3 | 32 | 8 | |
| M77 | 108931 | 110817 | UL77 | (HCMV: pyruvoyl decarboxylase) (63) | 68.6 | 5 | 2 | ||
| M80 | 113414 | 115507 | UL80 | Assembly protein-protease (28) | 74 | 7 | 4 | ||
| M82 | C | 115711 | 117507 | UL82 | Upper matrix phosphoprotein, pp71 (16) | 67.4 | 75 | 32 | 11 |
| M83 | C | 117614 | 120043 | UL83 | Lower matrix phosphoprotein, pp65 (16) | 90.9 | 110 | 20 | 10 |
| M85 | C | 122192 | 123124 | UL85 | Minor capsid protein (only found in gel) (3) | 34.6 | 34, 140 | 52 | 15 |
| M86 | C | 123199 | 127260 | UL86 | (HCMV: major capsid protein) (12) | 151.4 | 150 | 42 | 41 |
| M87 | 127383 | 130163 | UL87 | 102.4 | 21 | 6 | |||
| M88 | 130243 | 131523 | UL88 | (HCMV: virion protein) (3) | 47.3 | 21 | 4 | ||
| m90 | C | 132920 | 133876 | 35.8 | 17 | 4 | |||
| M94 | 136234 | 137271 | UL94 | (HCMV: virion-associated protein) (57) | 37.7 | >160, 35 | 55 | 23 | |
| M95 | 138282 | 139535 | UL95 | 45.8 | 20 | 5 | |||
| M97 | 140141 | 142072 | UL97 | Phosphotransferase (46) | 71.1 | 47 | 10 | ||
| M98 | 142101 | 143769 | UL98 | (HCMV: alkaline nuclease) (48) | 62.0 | 23 | 4 | ||
| M99 | 143723 | 144061 | UL99 | Virion-associated phosphoprotein (15) | 11.9 | 15-17 | 67 | 8 | |
| M100 | C | 144296 | 145411 | UL100 | Glycoprotein M (47) | 42.3 | >160 | 50 | 15 |
| M102 | 145596 | 148034 | UL102 | (HCMV: helicase-primase subunit) (39) | 91.0 | 24 | 8 | ||
| M104 | C | 149113 | 151227 | UL104 | Structural protein (50) | 80.6 | 31 | 12 | |
| M105 | 151028 | 153874 | UL105 | (HCMV: helicase-primase subunit) (39) | 106.4 | 40 | 11 | ||
| m107 | 161983 | 162678 | 24.6 | 29 | 4 | ||||
| M116 | C | 167205 | 169142 | UL116 | 66.1 | 6 | 2 | ||
| m117.1 | 169541 | 170956 | 45.3 | 7 | 2 | ||||
| M121 | C | 175679 | 177775 | UL121 | 73.2 | 28 | 6 | ||
| m147 | C | 206862 | 207299 | 16.9 | 32 | 6 | |||
| m150 | C | 208789 | 207623 | Member of MGPh family m145 (45) | 42.8 | 28 | 5 | ||
| m151 | C | 208814 | 209983 | Member of MGP family m145 (45) | 42.4 | 28 | 3 | ||
| m163 | C | 221875 | 222645 | 19.1 | 19 | 3 | |||
| m165 | C | 223280 | 224278 | 35.8 | 10 | 2 | |||
| m166.5d | C | 225441 | 226898 | New gene | 48 | 42, 48 | 31 | 10 | |
Comments in parentheses preceded by “HCMV:” indicate a gene function annotated solely from the homologue in HCMV.
Different 5′ extension based on a frameshift sequencing error at position 20958 in the original sequence.
Extended C terminus based on insertion of a G at position 38803.
New gene located between m166 and m167.
HCMV homologue.
Percent coverage of protein by polypeptides detected by MS.
Number of polypeptides detected from this ORF by MS.
MGP, potential membrane glycoprotein.
Analysis of MCMV virion-associated proteins. (i) Capsid proteins.
All four annotated MCMV capsid proteins, m48.2, M85, M86, and M46, were found associated with MCMV virions by the in-solution digestion approach. In contrast, only two capsid proteins, M85 and M86, were recovered from in-gel digestion. M46 is the homologue of UL46, the minor capsid protein of HCMV. UL46 aggregates upon heating and is unable to enter the resolving gel during SDS-PAGE (20). This characteristic of the protein appears to apply to M46 as well. m48.2 has a predicted molecular mass of only 9.8 kDa. Therefore, it was not resolved by the 10% acrylamide matrix used in this study.
(ii) Glycoproteins.
The MCMV glycoproteins gB, gM, and gH (M55, M100, and M75, respectively) were identified by the in-solution digestion approach. In addition, m74, the positional homologue of HCMV gO, was also identified. In-gel analysis identified only M55 and M100. gL, previously identified as associated with purified virions (62), was not found by either in-gel or in-solution analysis.
(iii) Tegument proteins.
In-solution digestion identified nine proteins homologous to HCMV tegument proteins: lower and upper matrix phosphoproteins (M83 and M82), large tegument protein (M48), pp150 (M32), M25, M47, M94, M99, and M51 (Table 1). In-gel analysis detected seven of these proteins (Fig. 1B). Although M78 has been reported to be an MCMV tegument protein (38), it was not detected by either in-solution digestion analysis or in-gel digestion analysis.
(iv) Replication proteins.
Eleven loci encoding trans-acting factors have been identified as being required for transient complementation of HCMV oriLyt-dependent DNA replication (39). Six of these proteins comprise the structure required to initiate and perform DNA synthesis. Five of these homologues in MCMV, DNA polymerase (M54), polymerase accessory protein (M44), major DNA binding protein (M57), and all proposed subunits of the helicase-primase complex (M70, M102, and M105) were found with high degrees of confidence by in-solution digestion of the virion preparations (Table 1).
(v) Immunomodulatory proteins.
The virion preparation contained a number of proteins suggested to interact with the immune system, including M43, a protein thought to influence T helper cell responses in vivo (49), and M45, a protein thought to have multiple functions during viral infection (11, 40). In-gel digestion analysis identified one of these proteins.
(vi) Additional viral proteins.
In addition to known tegument and capsid proteins, a number of other viral proteins were identified in association with purified virions. The assembly protein M80 and the DNA packaging protein M56 were both detected. In addition M98, the viral exonuclease, and M97, the viral protein kinase, were present. Lastly, M72, the dUTPase, was found albeit with a slightly lower degree of confidence. This suggests it may be present in low copy numbers per virion.
(vii) Annotated proteins with unknown function.
Several proteins of unknown function, not previously described to be present in the virion, were identified by in-solution digestions. These included m18, m25.2, M28, M31, M35, m39, M71, M87, M88, m90, M95, m107, M121, m150, m151, m163, and m165 (Table 1). These proteins were not detected by analysis of in-gel fragments, indicating that they are probably present at low copy numbers. Identification of these proteins as virion associated may provide some direction to further in vivo and in vitro characterizations of their function.
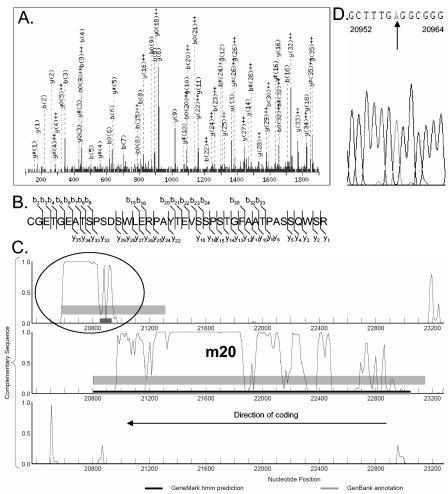
Evidence for a C-terminal m20 gene extension.
MS analysis identified two peptides with significant MOWSE-based probability scores originating from ORFc20579-21313 (Fig. 2A to C). The peptides appeared to cluster in the beginning of the reading frame and terminate where m20 begins in reading frame 2. GeneMarkS predicted a high coding potential for this region (Fig. 2C). This suggested that the peptides identified were an extension of m20, shifted to a different reading frame by a sequencing error in the reported Smith sequence. Sequence analysis of region 20600 to 21200 confirmed that the G at position 20958 in the current annotation is incorrect (Fig. 2D). This changes the ORF of the C-terminal end of m20, extending the sequence from position 23045 to 20579 instead of the originally predicted 20805.
FIG. 2.
Evidence for a 3′ extension of the MCMV m20 gene. (A) MS/MS spectrum of the 1,449.75 (M+H)3+ precursor ion of the tryptic peptide fragment CGETGEATSPSDSWLERPAYTEVSSPSTGFAATPASSQWSR as interpreted by MASCOT with a score of 45. (B) The matching b and y ions of the peptide found by MS/MS analysis. (C) Putative coding region at position 20579 to 21313 overlapping the 5′ region of the m20 gene. The three reading frames of the complementary sequence are displayed. The location of the tryptic peptide identified from this ORF, corresponding to the MS/MS spectrum in panel A, is indicated as a black box. Thin grey bars indicate genes annotated by Rawlinson et al. (45). Black bars represent ORFs predicted as protein coding by GeneMarkS, and grey areas indicate regions of interest with moderate coding potential generated by GeneMark. (D) DNA sequencing analysis revealed an incorrect insertion (G) at position 20958 of the MCMV genome (Smith strain NCBI 004065). Removal of this G extended the C terminus of m20 from position 20805 to position 20579.
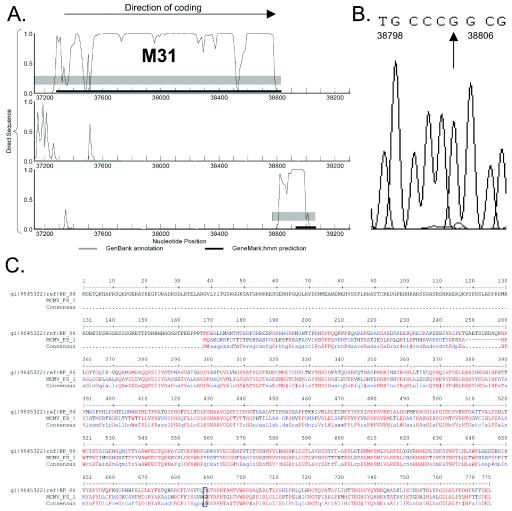
Frameshift identified in the M31 gene.
Similar to the case for m20, a small ORF located C terminally to the M31 gene, but in a different reading frame, was predicted to have high coding potential (Fig. 3A). Resequencing of the M31 gene region revealed an extra G at position 38803 (Fig. 3B). This additional nucleotide shifts the reading frame of the second half of M31 to encompass this newly identified coding region and also restores full-length homology of M31 to the R31 protein of RCMV (Fig. 3C).
FIG. 3.
Identification of a frameshift in the MCMV M31 gene. (A) A region with high coding potential was detected at the C-terminal end of the M31 gene in reading frame 3. The three reading frames of the direct sequence are displayed. (B) DNA sequencing analysis determined a missing G at position 38803. (C) Insertion of G38803 restored full-length homology of M31 to RCMV protein R31. The box indicates the amino acid which corresponds to the corrected codon from the insertion.
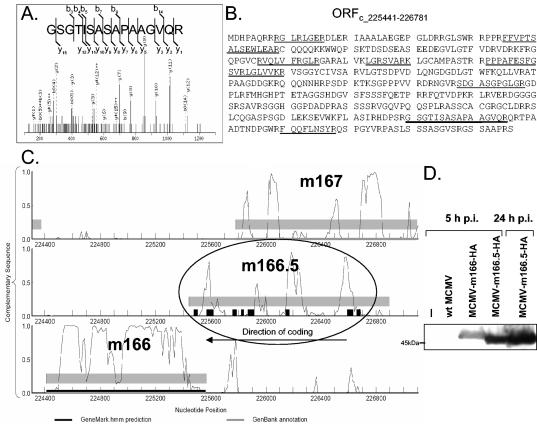
Novel MCMV genes.
MASCOT analysis of our MS data using a database of translations of all possible ORFs or all possible regions between stop codons of MCMV led to the identification of peptides from two previously unannotated ORFs. In-solution digestion analysis and SDS-PAGE analysis of virus material identified peptides from MCMV ORFc225441-226898. This ORF is located between m166 and m167 on the complementary strand of MCMV (Fig. 4) and specifies an ORF of 446 amino acids, hereafter referred to as m166.5 (Fig. 4B). It overlaps significantly with m167 and was marginally predicted by GeneMark (Fig. 4C). Analysis of virus material by SDS-PAGE revealed a polypeptide migrating at 48 kDa that matched m166.5. In-solution digestion and SDS-PAGE analysis identified 10 peptides from this region, resulting in 31% sequence coverage (Fig. 4B and C). Peptides from m166.5 were detected in two out of three in-solution virus preparations.
FIG. 4.
Expression of m166.5 confirmed by MS and Western blot analysis. (A) MS/MS spectrum of precursor ion 715.26 (uppercase 2+) was interpreted as peptide GSGTISASAPAAGVQR by MASCOT, with a score of 44. Detected b and y ions are indicated. (B) Sequence of m166.5. Tryptic peptides detected by MS are underlined. (C) GeneMark probability plot of the m166 MCMV gene region. The three reading frames of the complementary sequence are displayed. m166.5 partially overlaps with m167, and similar probabilities of coding potential are seen for both reading frames. Peptides found by MS/MS analysis are indicated as black boxes. Thin grey bars represent genes annotated by Rawlinson et al. (45), black bars are genes predicted by GeneMarkS, and grey areas indicate regions of interest with moderate coding potential. (D) Western blot analysis of NIH 3T3 endothelial cells infected with MCMV containing an HA-tagged m166.5 gene. Infected cells were harvested after the indicated times, lysed, and analyzed by SDS-PAGE and immunoblotting with an anti-HA antibody.
To confirm expression of the newly predicted m166.5 gene, NIH 3T3 cells were infected with recombinant MCMV-m166.5-HA possessing a C-terminal fusion of the HA tag to the m166.5 ORF. As control, the recombinant virus MCMV-m166-HA with an HA fusion to the m166 ORF was used. Total cell lysates were harvested 5 and 24 h postinfection, and immunoblot analysis was performed. As shown in Fig. 4D, a specific band for the HA-tagged m166.5 ORF with a size slightly larger than 45 kDa could be detected at 5 and 24 h postinfection. These data confirmed that a protein is expressed from ORF m166.5 during MCMV infection.
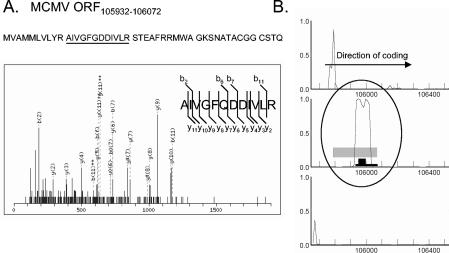
A peptide with a significant MOWSE score was also identified from the region of ORF105932-106072 by in-solution digest analysis. This ORF codes for a possible protein of 44 amino acids in an unannotated region of the MCMV genome predicted to have high coding potential by GeneMark (Fig. 5). The virus-derived peptide was also isolated from the 20- to 29-kDa range of our SDS-PAGE gel during in-gel analysis (Fig. 1B).
FIG. 5.
Evidence for expression of the predicted ORF105932-10672. (A) The MS/MS spectrum of the precursor ion 673.21 (uppercase 2+) corresponds to the tryptic fragment AIVGFQDDIVLR with a score of 48 by MASCOT. Matching b and y ions are indicated. This virus-derived peptide was detected in the virion preparation analyzed by SDS-PAGE and isolated from the 20- to 29-kDa range. The tryptic peptide fragment detected within the sequence of ORF105932-106072 by MS is underlined. (B) Gene predicted by GeneMarkS, with high coding potential. The three reading frames of the direct sequence are displayed. A black box indicates the tryptic fragment identified by MS/MS. The predicted gene and regions with moderate coding potential are indicated as black and gray bars, respectively.
Cellular proteins.
Identification of peptides from a number of cellular proteins was accomplished by conducting MASCOT searches using the NCBInr database. The results from in-gel analysis mirrored those obtained from in-solution digestion, with the predominant cellular protein being actin. In addition, annexin, cofilin, histone H2A, elongation factor 1α, glyceraldehyde-3-phosphate dehydrogenase, and rho GDP dissociation inhibitor were present and detectable by in-solution and in-gel digestion (Fig. 1B and Table 2).
TABLE 2.
Cellular proteins associated with MCMV particles
| Cellular protein | SwissProt ID | Other matchesa | Mass (kDa)
|
Comment (reference) | MS spectrab | % Cov- eragec | |
|---|---|---|---|---|---|---|---|
| Predicted | Observed | ||||||
| γ-Actin, cytoplasmic | gi:6752954 | β-actin, α-actin 1 | 42 | 45 | Associated with HCMV preparations (3) | 14 | 42 |
| Annexin I (lipocortin I) | gi:1351942 | Annexin V | 35.7 | 33 | Associated with HCMV preparations (59) | 3 | 11 |
| Cofilin | gi:116849 | 18.5 | 18 | Associated with HIV particles (38a) | 2 | 16 | |
| EF-1 α (EF-Tu) | gi:1169475 | 50 | 5 | 22 | |||
| Glyceraldehyde-3-phosphate dehydrogenase | gi:120702 | 35.7 | 1 | 6 | |||
| Histone H2A | gi:121961 | Histone H2B | 14 | 14 | 6 | 20 | |
| Rho GDP dissociation inhibitor | gi:21759130 | 23.3 | 26 | 2 | 7 | ||
Other cellular proteins matching the same set of peptides.
Number of polypeptides detected from this protein by MS.
Percent coverage of protein by polypeptides detected by MS.
DISCUSSION
Traditional analysis of the composition of herpesvirus virions consists of denaturation of a virus preparation in SDS, separation of the constituent proteins by electrophoresis, and identification of polypeptides by sequencing visible bands with Edman degradation or MS. This approach has been used to analyze HCMV (3), murine gammaherpesvirus 68 (9), Epstein-Barr virus (18), and Kaposi's sarcoma-associated herpesvirus (36). This method, however, is unlikely to be sufficiently sensitive to detect low-abundance virion proteins. Recovery of highly hydrophobic peptides or low-abundance components excised from acrylamide gels is often poor. In addition, at least one of the viral glycoproteins in the HCMV particle cannot be identified in this manner due to its tendency to aggregate upon heating and its subsequent inability to enter the resolving gel (20).
Here, both in-gel analysis and in-solution digestion are used as complementary approaches to examine the protein content of MCMV particles. The complex nature of the MCMV viral particle, and the relative scarcity of information regarding its composition, warranted this approach for confirmation and comparison purposes. Standard in-gel analysis allowed the resolution of various molecular weight forms of MCMV virion components, including novel proteolytic products of the M25 ORF. In-solution digestion of MCMV particles afforded the detection of proteins present in lower copy numbers, as well as the identification of two putative novel gene products.
As expected, we identified the majority of CMV virion constituents previously reported in the literature. Analysis of our in-solution digestion confirmed the presence of all four capsid proteins, M85, M86, m48.2, and M46. In addition, all but one of the reported MCMV tegument proteins were unambiguously detected by this method. M78, a G-protein-coupled, seven-transmembrane receptor, has previously been described as a virion component (38). We only detected a single peptide derived from M78 in one out of four virion preparations. It did not, therefore, meet our threshold criteria to be considered significant. The reason for this discrepancy is not clear, but the transmembrane structure of this protein may account for its inaccessibility to tryptic digestion.
In our viral preparations, the relative abundance of tegument protein M25 was striking. We identified 91 individual peptides derived from M25. In contrast, the second-most-abundantly detected protein, major capsid protein M86, yielded only 41 peptides. As a result of our in-gel analysis, it is also apparent that M25 exists in a number of proteolytic forms within the virion. Previous reports have identified 130-, 105-, and 95-kDa forms of this protein in infected cell lysates and 130-kDa forms of this protein in supernatant virions (60). Our analysis identified not only the 130-kDa form but also several other forms at 200, 52, and 48 kDa, as assessed by their position of migration on SDS-PAGE. Mutagenesis of the M25 region by transposon insertion results in no loss of titer in NIH 3T3 cells or organs of infected BALB/c mice (64) and suggests that this protein is dispensable for viral replication. The function of this abundant, yet apparently nonessential, virion protein warrants further investigation.
Contrary to expectation M115, a homologue of an HCMV glycoprotein, was not detected. In HCMV, glycoprotein L forms a trimer with glycoproteins M and O (24). This trimer is an essential structure for HCMV entry into cells. Although previous work by Xu and colleagues suggested that an MCMV glycoprotein L homologue exists and is expressed during infection (62), it was not detected in this study by either in-solution digestion or in-gel digestion analysis. The small size and heavy glycosylation of this protein may result in poor recovery of tryptic fragments. In addition, glycoprotein L may be differentially expressed during infection.
Similar to previous reports on HCMV (3), actin is also associated with MCMV viral particles. Annexin II is known to bind HCMV glycoprotein B (42). We found peptides derived from annexin I and annexin V in our preparations, suggesting that MCMV may interact with these proteins. Peptides were also recovered from cofilin, histone H2A, elongation factor 1α, glyceraldehyde-3-phosphate dehydrogenase, and cadherin EGF LAG seven-pass G-type receptor 2 (Table 2). Bovine immunoglobulin G and serum albumin were detected by both in-gel digestion and in-solution digestion. This was presumably due to the abundance of these proteins in tissue culture media.
The method of purification used in this paper does not necessarily remove secreted viral proteins released into the medium that could adsorb onto the surface of extracellular virions. Furthermore, none of the currently used purification methods can fully resolve virion particles from abundant serum and host cell proteins, such as bovine serum albumin, bovine immunoglobulin G, or actin. As seen by SDS-PAGE, the fraction of nonspecific cellular protein contamination was modest in relation to actual viral constituents (Fig. 1B), but further biochemical analysis is warranted in investigating the role of the individual gene products identified in this study.
The in-solution digestion approach has revealed the presence of MCMV replication proteins associated with the virion. Although the presence of these proteins might be attributed to nonspecific contamination of virus preparations, there is evidence to suggest the contrary. Shenk and colleagues have demonstrated that HCMV virions contain mRNA specific for immediate-early genes (10). Given the importance for rapid DNA replication, transcription, and translation during the viral life cycle, it would not be surprising to find that particles enter cells ready to copy their DNA as well as begin translation of immediate-early gene products. Mar and colleagues reported DNA polymerase activity associated with purified HCMV virions (29), supporting the notion that the replication complex may also be incorporated into MCMV virions. These proteins may be present in only a few copies per virion, possibly explaining why they escape detection with traditional, less-sensitive methods.
A common approach to herpesvirus genome annotation is the identification as a protein coding region any ORF longer than 300 bp with less than 60% overlap with an adjacent ORF (45, 54). These rules can cause substantial overannotation, especially in genomes that have high G+C content, such as MCMV. The prediction algorithm used in this study, an MCMV-modified version of GeneMarkS, takes into account particular characteristics of viruses, including their small size, overlapping genes, repeated regions, circular genomes, and translation start context.
Computer analysis of the MCMV genome using GeneMarkS, as described above, led to the discovery of 12 putative novel genes (data not shown). Our complementary proteomic analysis suggested the existence of two of these genes as being present in the virion. Expression of one of these genes, m166.5, during viral infection was confirmed by generating recombinant MCMV expressing an HA-tagged form of this protein. This recombinant virus will be useful to further explore the biological function of this viral gene.
The second ORF identified, ORF105932-106072, codes for a possible translation product of 44 amino acids in an unannotated region of the MCMV genome. BLAST search using the NCBI server and the SwissProt database revealed sequence identity to a fragment of a putative spliced M73 transcript described as an unpublished finding by Dallas et al. (gi:762817). This suggests that this fragment may be part of a larger, spliced gene. Consistent with this hypothesis, the peptide derived from this region was detected by SDS-PAGE analysis in the range of 20 to 29 kDa. The full sequence of this gene remains unknown, as peptides were not detected that corresponded to the rest of the M73 putative spliced sequence.
Generation of graphical output based on GeneMark predictions allowed visual examination of irregularities in the protein-coding potential of currently annotated genes. This examination led to the identification of sequencing errors in the m20 and M31 genes. Restoration of proper coding sequence led to full-length homology of the M31 gene to its R31 counterpart in RCMV (Fig. 3C).
The inability to detect our other newly predicted genes may be due to the fact that, even if these ORFs are expressed, the products encoded may not be structural proteins, or they may be of too low abundance to be detected. Further experiments are required to address these possibilities.
Our combined analysis represents an improved strategy for determining the protein composition of virus particles. The standard method of virion analysis by SDS-PAGE was compared to in-solution digestion and MS/MS analysis of viral proteins. In-solution digestion followed by MS/MS greatly improved sensitivity, increasing the number of identified proteins from 19 (in-gel analysis) to 58 (in-solution analysis). Although the functional relevance of these newly identified proteins during the viral life cycle is unclear, the data indicate that the particle composition of MCMV is more complex than previously thought.
Acknowledgments
We thank Alexei Paskhin for help in performing computations for the genomic segmentations at the preprocessing step for gene finding.
M.B., A.L., V.M., and R.M. have been supported in part by NIH grants HG00783 and TW005899. H.L.P. is supported by NIH grant AI33456-05, and B.M.K is supported by a Multiple Myeloma Research Foundation Senior Research Award. M.W. is supported by a Human Frontier Science Program Organization long-term fellowship.
REFERENCES
- 1.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahr, U., and G. Darai. 2004. Re-evaluation and in silico annotation of the Tupaia herpesvirus proteins. Virus Genes 28:99-120. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogner, E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12:115-127. [DOI] [PubMed] [Google Scholar]
- 7.Bogner, E., M. Reschke, B. Reis, T. Mockenhaupt, and K. Radsak. 1993. Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology 196:290-293. [DOI] [PubMed] [Google Scholar]
- 7a.Borodovsky, M., and J. McInich. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17:123-133. [Google Scholar]
- 8.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortz, E., J. P. Whitelegge, Q. Jia, Z. H. Zhou, J. P. Stewart, T. T. Wu, and R. Sun. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77:13425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 11.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 12.Chee, M., S. A. Rudolph, B. Plachter, B. Barrell, and G. Jahn. 1989. Identification of the major capsid protein gene of human cytomegalovirus. J. Virol. 63:1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chelius, D., A. F. Huhmer, C. H. Shieh, E. Lehmberg, J. A. Traina, T. K. Slattery, and E. Pungor, Jr. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J. Proteome Res. 1:501-513. [DOI] [PubMed] [Google Scholar]
- 14.Chong, K. T., and C. A. Mims. 1981. Murine cytomegalovirus particle types in relation to sources of virus and pathogenicity. J. Gen. Virol. 57:415-419. [DOI] [PubMed] [Google Scholar]
- 15.Cranmer, L. D., C. Clark, and D. H. Spector. 1994. Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99). Virology 205:417-429. [DOI] [PubMed] [Google Scholar]
- 16.Cranmer, L. D., C. L. Clark, C. S. Morello, H. E. Farrell, W. D. Rawlinson, and D. H. Spector. 1996. Identification, analysis, and evolutionary relationships of the putative murine cytomegalovirus homologs of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J. Virol. 70:7929-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 18.Dolyniuk, M., R. Pritchett, and E. Kieff. 1976. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J. Virol. 17:935-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson, W., K. S. Clopper, W. J. Britt, and M. K. Baxter. 1996. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J. Virol. 70:5680-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey, D. M., and A. J. Levine. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 5:2375-2385. [DOI] [PubMed] [Google Scholar]
- 23.Holtappels, R., N. K. Grzimek, D. Thomas, and M. J. Reddehase. 2002. Early gene m18, a novel player in the immune response to murine cytomegalovirus. J. Gen. Virol. 83:311-316. [DOI] [PubMed] [Google Scholar]
- 24.Huber, M. T., and T. Compton. 1999. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J. Virol. 73:3886-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahn, G., B. C. Scholl, B. Traupe, and B. Fleckenstein. 1987. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J. Gen. Virol. 68:1327-1337. [DOI] [PubMed] [Google Scholar]
- 26.Kinter, M., and N. E. Sherman. 2000. Protein sequencing and identification using tandem mass spectrometry. Wiley & Sons, New York, N.Y.
- 27.Loh, L. C., W. J. Britt, C. Raggo, and S. Laferte. 1994. Sequence analysis and expression of the murine cytomegalovirus phosphoprotein pp50, a homolog of the human cytomegalovirus UL44 gene product. Virology 200:413-427. [DOI] [PubMed] [Google Scholar]
- 28.Loutsch, J. M., N. J. Galvin, M. L. Bryant, and B. C. Holwerda. 1994. Cloning and sequence analysis of murine cytomegalovirus protease and capsid assembly protein genes. Biochem. Biophys. Res. Commun. 203:472-478. [DOI] [PubMed] [Google Scholar]
- 29.Mar, E. C., P. C. Patel, and E. S. Huang. 1981. Human cytomegalovirus-associated DNA polymerase and protein kinase activities. J. Gen. Virol. 57:149-156. [DOI] [PubMed] [Google Scholar]
- 30.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerle, M., M. Rapp, P. Lucin, and U. H. Koszinowski. 1995. Characterization of a conserved gene block in the murine cytomegalovirus genome. Virus Genes 10:73-80. [DOI] [PubMed] [Google Scholar]
- 32.Mesyanzhinov, V. V., J. Robben, B. Grymonprez, V. A. Kostyuchenko, M. V. Bourkaltseva, N. N. Sykilinda, V. N. Krylov, and G. Volckaert. 2002. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317:1-19. [DOI] [PubMed] [Google Scholar]
- 33.Mills, R., M. Rozanov, A. Lomsadze, T. Tatusova, and M. Borodovsky. 2003. Improving gene annotation of complete viral genomes. Nucleic Acids Res. 31:7041-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 35.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, S. A., S.-H. Park, P. Lee, A. Bendelac, and T. E. Shenk. 2002. Murine cytomegalovirus m02 gene family protects against natural killer cell-mediated immune surveillance. J. Virol. 76:885-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 98:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder II, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patrone, M., E. Percivalle, M. Secchi, L. Fiorina, G. Pedrali-Noy, M. Zoppe, F. Baldanti, G. Hahn, U. H. Koszinowski, G. Milanesi, and A. Gallina. 2003. The human cytomegalovirus UL45 gene product is a late, virion-associated protein and influences virus growth at low multiplicities of infection. J. Gen. Virol. 84:3359-3370. [DOI] [PubMed] [Google Scholar]
- 41.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 42.Pietropaolo, R. L., and T. Compton. 1997. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J. Virol. 71:9803-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramensky, V. E., V. Makeev, M. A. Roytberg, and V. G. Tumanyan. 2000. DNA segmentation through the Bayesian approach. J. Comput. Biol. 7:215-231. [DOI] [PubMed] [Google Scholar]
- 44.Rapp, M., M. Messerle, B. Buhler, M. Tannheimer, G. M. Keil, and U. H. Koszinowski. 1992. Identification of the murine cytomegalovirus glycoprotein B gene and its expression by recombinant vaccinia virus. J. Virol. 66:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawlinson, W. D., F. Zeng, H. E. Farrell, A. L. Cunningham, A. A. Scalzo, T. W. Booth, and G. M. Scott. 1997. The murine cytomegalovirus (MCMV) homolog of the HCMV phosphotransferase (UL97pk) gene. Virology 233:358-363. [DOI] [PubMed] [Google Scholar]
- 47.Scalzo, A. A., C. A. Forbes, N. J. Davis-Poynter, H. E. Farrell, and P. A. Lyons. 1995. DNA sequence and transcriptional analysis of the glycoprotein M gene of murine cytomegalovirus. J. Gen. Virol. 76:2895-2901. [DOI] [PubMed] [Google Scholar]
- 48.Sheaffer, A. K., S. P. Weinheimer, and D. J. Tenney. 1997. The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J. Gen. Virol. 78:2953-2961. [DOI] [PubMed] [Google Scholar]
- 49.Singh, R., E. Haghjoo, and F. Liu. 2003. Cytomegalovirus M43 gene modulates T helper cell response. Immunol. Lett. 88:31-35. [DOI] [PubMed] [Google Scholar]
- 50.Spaete, R. R., R. C. Gehrz, and M. P. Landini. 1994. Human cytomegalovirus structural proteins. J. Gen. Virol. 75:3287-3308. [DOI] [PubMed] [Google Scholar]
- 51.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam, A., J. Zhu, R. Hai, E. Haghjoo, T. Tong, X. Zhan, S. Lu, and F. Liu. 2003. Murine cytomegalovirus with a transposon insertional mutation at open reading frame M35 is defective in growth in vivo. J. Virol. 77:7746-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tu, A. H., L. L. Voelker, X. Shen, and K. Dybvig. 2001. Complete nucleotide sequence of the mycoplasma virus P1 genome. Plasmid 45:122-126. [DOI] [PubMed] [Google Scholar]
- 54.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wing, B. A., G. C. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright, J. F., A. Kurosky, E. L. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, C. A., M. E. Carlson, S. C. Henry, and J. D. Shanley. 1999. The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology 262:265-276. [DOI] [PubMed] [Google Scholar]
- 61.Xu, J., P. B. Dallas, P. A. Lyons, G. R. Shellam, and A. A. Scalzo. 1992. Identification of the glycoprotein H gene of murine cytomegalovirus. J. Gen. Virol. 73:1849-1854. [DOI] [PubMed] [Google Scholar]
- 62.Xu, J., A. A. Scalzo, P. A. Lyons, H. E. Farrell, W. D. Rawlinson, and G. R. Shellam. 1994. Identification, sequencing and expression of the glycoprotein L gene of murine cytomegalovirus. J. Gen. Virol. 75:3235-3240. [DOI] [PubMed] [Google Scholar]
- 63.Yoakum, G. H. 1993. Mapping a putative pyruvoyl decarboxylase active site to human cytomegalovirus open reading frame UL77. Biochem. Biophys. Res. Commun. 194:1207-1215. [DOI] [PubMed] [Google Scholar]
- 64.Zhan, X., M. Lee, G. Abenes, I. Von Reis, C. Kittinunvorakoon, P. Ross-Macdonald, M. Snyder, and F. Liu. 2000. Mutagenesis of murine cytomegalovirus using a Tn3-based transposon. Virology 266:264-274. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann, W., H. Broll, B. Ehlers, H. J. Buhk, A. Rosenthal, and M. Goltz. 2001. Genome sequence of bovine herpesvirus 4, a bovine rhadinovirus, and identification of an origin of DNA replication. J. Virol. 75:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]