Abstract
ND10 structures are disrupted during herpes simplex virus type 1 (HSV-1) infection by viral regulatory protein ICP0. The significance of this effect remains controversial, partly because of a report that high-level expression of the major ND10 promyelocytic leukemia (PML) protein precludes ND10 disruption yet does not inhibit HSV-1 infection. Here we demonstrate dramatic reorganization of ND10 during HSV-1 infection by live-cell microscopy, even in the presence of overexpressed PML.
ND10 (also known as promyelocytic leukemia [PML] nuclear bodies) are discrete nuclear foci that contain several cellular proteins involved in diverse processes such as transcription, chromatin structure, DNA repair, and apoptosis (1, 7, 23, 24, 30). ND10 are dynamic structures that disassemble during mitosis and apoptosis and are structurally affected by many forms of stress, such as heat shock, heavy metal treatment, and a variety of drugs that alter cellular metabolism. ND10 are also intimately associated with the replication of many nuclear replicating DNA viruses in that parental viral genomes frequently associate with ND10, and it is at these locations that viral immediate-early (IE) gene transcription occurs and from which viral DNA replication compartments develop (5, 18, 20, 22, 29; reviewed in references 8 and 21). In the case of herpes simplex virus type 1 (HSV-1), recent evidence suggests that the association of viral genomes with ND10 may be functionally important, because genomes that are so associated in the initial stages of infection have an increased probability of developing into a replication compartment (14, 15, 28). Further evidence for a role of ND10 in HSV-1 replication comes from a strong implication that ND10-like structures assemble in association with and in response to incoming HSV-1 genomes in the absence of IE regulatory protein ICP0 (14). ICP0 is a ubiquitin E3 ligase that brings about the destruction of ND10 in wild-type HSV-1 infections through inducing the degradation of the principal core component of ND10, the PML protein. The ability of ICP0 to disrupt ND10 by this mechanism correlates very well with its role in stimulating HSV-1 lytic infection and reactivation from quiescence or latency (2, 3, 11, 16; for reviews see references 9, 17, 26, and 27). All the above evidence suggests but does not prove that ND10 structures have important roles in HSV-1 infection.
In contrast, it has been reported that high-level expression of PML by using a baculovirus engineered to express proteins in mammalian cells leads to the formation of large ND10 complexes that are not disrupted during HSV-1 infection (19). The presence of very high levels of PML in this situation did not impede HSV-1 infection (19), a conclusion that is consistent with that of a previous study (6) and with unpublished data from this laboratory. Therefore, there is consistent evidence that high levels of PML do not inhibit HSV-1 infection, at least in the situations so far examined. However, because large ND10-like structures remained during HSV-1 infection of cells expressing very high levels of PML, it was concluded that the disruption of ND10 has no functional role (19). If true, this is a highly important conclusion, because it places in doubt the significance of a substantial body of published work and interpretation that is relevant not only to HSV-1 but also to a spectrum of DNA viruses. On the other hand, treatments that inhibit the disruption of ND10 by ICP0 inhibit the formation of replication compartments, progression to efficient early gene expression, and production of viral progeny in low-multiplicity HSV-1 infections (4, 5, 13). In view of this apparently conflicting evidence, this study was initiated to test the hypothesis that high-level expression of PML resulted in ND10 structures that were resistant to disruption during HSV-1 infection. Instead of relying on extrapolations from fixed-cell images, the fate of ND10 and PML protein distribution was followed by time-lapse microscopy of live infected cells.
Baculovirus Ac.CMV.EYFP-PML contains the PML (isoform IV) cDNA with an N-terminal enhanced yellow fluorescent protein (EYFP) tag downstream of the human cytomegalovirus (HCMV) IE promoter/enhancer (15). This isoform of PML was chosen because it gives a pattern of SUMO-modified forms that bear a strong resemblance to those of the endogenous protein and because, like endogenous PML (11, 25), its sensitivity to the effects of ICP0 have been well characterized (2, 25). Analogous baculoviruses expressing Sp100 (isoform A) and hDaxx were constructed (Ac.CMV.EYFP-Sp100 and Ac.CMV.EYFP-Daxx). Infection of Vero cells with these viruses indicated that all three proteins were expressed at very high levels (estimated to be of the order of 100-fold in excess of the endogenous proteins), and in the case of both PML and Sp100, modified forms consistent with the normal conjugation of these proteins to SUMO-1 were evident (Fig. 1). Vero cells in a coverslip glass chamber unit were coinfected with Ac.CMV.ECFP-PML (28) and Ac.CMV.EYFP-Sp100, and the following day the unit was placed in a live-cell microscopy suite equipped with full environmental control and motorized accessories for detection of EYFP and enhanced cyan fluorescent protein (ECFP) in the same sample (15). Control experiments showed that in uninfected cells the fluorescent PML and Sp100 proteins extensively colocalized in ND10 structures that, in interphase cells, did not show extensive changes in appearance or composition over the time periods used for the following experiments (data not shown).
FIG. 1.
High-level expression of ND10 proteins PML, Sp100, and hDaxx by infection with baculoviruses containing HCMV promoter-driven expression cassettes. Vero cells were infected at an MOI of 100 insect cell PFU per cell with viruses Ac.CMV.EYFP-PML, Ac.CMV.EYFP-Sp100, and Ac.CMV.EYFP-Daxx, and total cell extracts were prepared 16 h later. Aliquots of infected and mock-infected samples were loaded onto gels in the indicated ratios and then were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using antibodies 5E10, SpGH, and r1866, respectively (described in reference 12), that recognize both exogenous and endogenous proteins. Under these conditions, expression of the former exceeds that of the latter by approximately two orders of magnitude.
Additional wells of Vero cells infected with Ac.CMV.ECFP-PML and Ac.CMV.EYFP-Sp100 were prepared, and the next day the cells were infected with HSV-1 strain 17 (multiplicity of infection [MOI], 10). Soon after virus adsorption the sample was placed in the microscope system, and then fields of view were chosen and images were captured at 10-min intervals for periods of 3 to 4 h. Selected images from a typical time sequence are shown in Fig. 2 and 3, and the complete movie is available in the supplementary data (Movie S1). Consistent with previous data (19), high-level expression of PML caused the assembly of large ND10-like foci, and most cells infected with both baculoviruses contained both PML and Sp100. As HSV-1 infection progressed, Sp100 was lost from the ND10-like foci and the proportion of Sp100 that was diffusely spread through the nucleus increased. While some punctate PML structures remained throughout, they clearly underwent a dramatic rearrangement during the first few hours of infection. Movie S1 shows that the foci became less distinct and even appeared to go out of focus before becoming more distinct again towards the end of the sequence. While movement of some of the foci out of the focal plane cannot be discounted, this type of behavior occurred in all infected cells examined by time-lapse microscopy, and it did not occur in uninfected cells. Visual inspection of the samples confirmed the reduction in numbers of PML foci during infection, illustrating that the results were not due to loss of camera focus. Rather, the PML foci became less distinct, disaggregated, and highly mobile, and then the more stable distinct structures that were formed towards the end of the sequence appeared to arise from the merging of PML material from ND10 remnants. Similar results were obtained with Vero cells coinfected with baculoviruses Ac.CMV.ECFP-PML and Ac.CMV.EYFP-Daxx (Fig. 2 and 3). We conclude that although large foci of PML can be retained after overexpression of PML in HSV-1-infected cells, the PML structures undergo substantial changes in both composition (loss of Sp100 and hDaxx) and morphology (disaggregation and then merging of remnant material). Although it was not possible to directly identify HSV-1-infected cells simultaneously in these experiments, controls using fixed cells and antibody staining confirmed that all the Vero cells became HSV-1 infected at this MOI (data not shown). The changes that we have observed here would not have been detectable in fixed-cell experiments, because fixation does not allow the fate of individual PML foci to be followed over time.
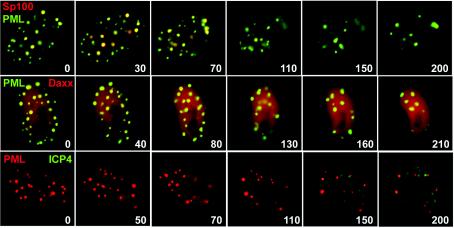
FIG. 2.
Visualization of the disruption of ND10 during HSV-1 infection. The upper two rows show Vero cells in a coverslip glass chamber unit that were infected with Ac.CMV.ECFP-PML and Ac.CFP.EYFP-Sp100 at MOI of 100 insect PFU per cell; the following day the cells were infected with HSV-1 strain 17 at an MOI of 10 PFU per cell. After adsorbing the virus for 30 min, the cells were washed and the medium was replaced with Dulbecco's modified Eagle's medium without phenol red and containing 1% fetal calf serum. The sample was then examined by live-cell microscopy as described previously (15). Images were taken every 10 min, starting 90 min after the initial addition of the virus. The top row of the uppermost pair of rows shows the ECFP image (PML), and the lower row shows EYFP (Sp100). The numbers at the bottom right of each panel indicate the time in minutes relative to this 90-min time point. The middle two rows show an analogous experiment following preinfection with baculoviruses Ac.CMV.ECFP-PML and Ac.CMV.EYFP-Daxx. The bottom two rows show HFFF-2 cells in a coverslip glass chamber unit that were infected with Ac.CMV.EYFP-PML at an MOI of 100 insect PFU per cell; the following day the cells were infected with vECFP-ICP4 at an MOI of 10 PFU per cell and were examined by live-cell microscopy following the same time protocol described for the uppermost panels. The EYFP signal (PML) is shown in the upper of the two rows, and the ECFP signal (ICP4) is shown in the lower row.
FIG. 3.
Merged color images corresponding to the rows of images in Fig. 2. The upper row shows the PML (green) and Sp100 (red) signals from the panels in the upper two rows of Fig. 2, the middle row shows the PML (green) and hDaxx (red) signals from the middle pair of rows in Fig. 2, and the bottom row shows the PML (red) and ICP4 (green) signals from the bottom set of rows in Fig. 2. The numbers refer to the time point in minutes from the start of the sequence, in each case starting 90 min after addition of the relevant HSV-1 virus.
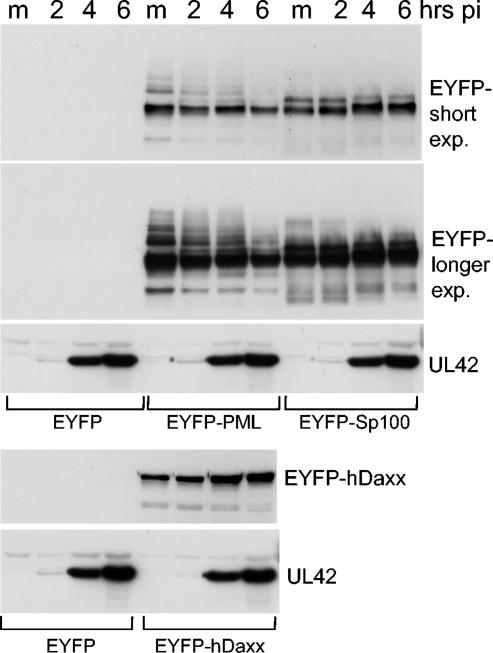
The fate of the overexpressed ND10 proteins during infection of Vero cells was investigated by Western blotting. Vero cells were infected with baculoviruses expressing either EYFP alone or the EYFP-PML, EYFP-Sp100, or EYFP-hDaxx fusion proteins. The MOIs used were the same as those in the live-cell studies. The following day replicate samples were infected with HSV-1 strain 17 (MOI, 10) and were harvested 2, 4, and 6 h later. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting, the filters were probed with an anti-EYFP antibody and an antibody to detect the viral DNA replication protein UL42, a marker for the progression of the infection. Figure 4 shows that infection with HSV-1 caused a slight reduction in the major PML band (presumed to be unmodified by SUMO-1), but there was a marked reduction in the slower migrating SUMO-modified forms. This observation is consistent with the study of Lopez et al. (19) and presumably reflects the disruption of the ND10 structures that we have detected in this study. In the case of Sp100, there were relatively minor changes in the comparatively less abundant lower mobility forms and, rather than any decrease in the level of the major Sp100 band, there was a modest increase during HSV-1 infection that is likely due to the HSV-1 transactivators enhancing expression from the HCMV promoter in the baculovirus genome nonspecifically. This behavior contrasts with the loss of endogenous Sp100 seen during HSV-1 infection, although even in this case the loss is preferentially of the presumed SUMO-modified forms (25). Other than a similar transactivation effect, we did not observe any major changes to the levels of baculovirus-expressed hDaxx during HSV-1 infection (Fig. 4), consistent with the behavior of endogenous hDaxx (data not shown). High-level expression of EYFP-PML or the other fusion proteins did not compromise the efficiency of UL42 expression by HSV-1 in high-multiplicity infections of Vero cells (Fig. 4), in agreement with the conclusions of Lopez et al. (19).
FIG. 4.
Effect of HSV-1 infection on ND10 proteins expressed at high levels. Sets of four wells of Vero cells were infected at 100 insect cell PFU per cell with either Ac.CMV.EYFP (a baculovirus similar to the others used in this study, except expressing EYFP alone), Ac.CMV.EYFP-PML, Ac.CMV.EYFP-Sp100, or Ac.CMV.EYFP-Daxx as indicated. The following day one well of each set was mock infected and the others were infected with HSV-1 strain 17 at an MOI of 10. The wells were harvested at 2, 4, or 6 h after virus adsorption, and total cell proteins were analyzed by Western blotting using either an anti-EYFP rabbit serum (Abcam) or anti-UL42 monoclonal antibody Z1F11. The EYFP protein is not visible in these panels, as it has a far greater gel mobility than the fusion proteins. m, mock control, not infected with HSV-1. pi, postinfection; exp., exposure.
To test whether this was a cell-type-specific phenomenon and to provide a direct control for virus infection, human fibroblast HFFF-2 cells were infected with Ac.CMV.EYFP-PML alone and the fate of the PML foci was followed after infection with HSV-1 virus vECFP-ICP4, which expresses viral IE protein ICP4 linked to ECFP (14, 15). ICP4 is a transcriptional regulator that associates with parental viral genomes and accumulates in viral replication centers (14 and references therein). Again, although PML foci remained throughout the infection period, there was a dramatic reorganization of the PML protein followed by its accumulation into a reduced number of foci (Fig. 2 and 3 and Movie S2). Although the remaining PML foci appear to arise from the merging of PML material from the original structures, the interpretation of these image sequences requires some caution because of the uncertainties introduced by the time intervals between the images and possible movement of foci from the optimal focal plane. With these limitations in mind, it nonetheless appears that after infection many of the PML foci become surrounded by increased diffuse fluorescence in the vicinity of the original structure, suggesting a marked efflux of PML protein. At later times this diffuse fluorescence is less marked and the PML foci become distinct and well-defined again, superficially similar to their appearance before infection. The reorganization of the PML foci began at an early stage of infection, before extensive synthesis of ICP4 was detectable by the image capture conditions used. Note, however, that detection of ECFP-ICP4 is not very sensitive in the live-cell microscopy system; staining of fixed cells by using antibodies readily detects ICP4 as early as 1 h after infection of Vero cells at an MOI of 10 (data not shown). The examples shown in Fig. 2 and 3 focus on cells that contain large but not exceptionally extensive PML aggregates. However, during the course of these experiments it was observed that even the largest PML aggregates were subject to disruption during HSV-1 infection in both Vero and HFFF-2 cells (data not shown).
These experiments clearly show that high-level expression of PML produces large aggregates of PML that nonetheless are subject to modification in terms of both content and morphology during HSV-1 infection. While overexpression of PML itself does not impede HSV-1 infection (6, 19), the conclusion that ND10 disruption is not important for productive HSV-1 infection (19) is not proven. It is possible that disruption of PML structures through the ubiquitin E3 ligase activity of ICP0 releases or changes the activity of activators and/or repressors that impinge on the efficiency of HSV-1 infection. Thus, the targeting of PML and ND10 by ICP0 may not be to relieve an inhibition mediated by PML itself, but more of a means to affect other factors that are ND10 associated. Accordingly, high-level expression of PML may not impede HSV-1 infection, because it does not abrogate ND10 disruption and the resultant downstream effects on these other factors. On the other hand, the observations reported here do not prove that ND10 disruption is an important or essential process during HSV-1 infection.
It will remain extremely difficult to design experiments that assess directly the role of ND10 disruption in the progression of HSV-1 infection. We note that in these experiments and in several previous studies (4, 5), and even in the absence of ICP0 (14), the commitment of cells into the HSV-1 lytic cycle and the development of viral replication compartments is accompanied by significant changes in the morphology of ND10. Whether the processes of viral genome association with ND10 and subsequent ND10 disruption underlie positive or negative influences on HSV-1 replication (or elements of both) or are merely effects without functional significance remains to be determined; these issues have been discussed in detail elsewhere (10, 14). Whatever the solution to these complicated questions, it is clear that at the descriptive level, modification or disruption of ND10 is intimately linked to the commitment of cells to lytic HSV-1 infection and the development of replication compartments.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council and by the award of a short-term fellowship to A.Z. by the European Molecular Biology Organization.
We are grateful for the support of Demetrios Spandidos (Crete Medical School) in enabling A.Z. to take up the EMBO fellowship in Glasgow. We thank Gerd Maul (Wistar Institute) for providing an hDaxx cDNA clone and Wei-Li Hsu and Anne Orr for unpublished work investigating whether high-level expression of ND10 proteins impedes HSV-1 infection.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutell, C., A. Orr, and R. D. Everett. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkham, J., D. M. Coen, C. B. Hwang, and S. K. Weller. 2001. Interactions of Herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 7.Eskiw, C. H., and D. P. Bazett-Jones. 2002. The promyelocytic leukemia nuclear body: sites of activity? Biochem. Cell Biol. 80:301-310. [DOI] [PubMed] [Google Scholar]
- 8.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 10.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of Herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, H., and B. Roizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 22.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 23.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 24.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 27.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 30.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.