Abstract
Unlike naive CD8+ T cells, antigen-experienced memory CD8+ T cells persist over time due to their unique ability to homeostatically proliferate. It was hypothesized that memory cells might differentially regulate the expression of genes that control the cell cycle to facilitate homeostatic proliferation. To test this, the expression levels of 96 different cell cycle regulatory genes were compared between transgenic naive and memory CD8+ T cells that specifically recognize the GP33-41 epitope of lymphocytic choriomeningitis virus (LCMV). It was discovered that relative to naive cells, memory cells overexpress several important genes that control the transition between G1 and S phase. Some of these genes include those encoding cyclins D3, D2, B1, C, and H, cyclin-dependent kinases (cdk's) 4 and 6, the cdk inhibitors p16, p15, and p18, and other genes involved in protein degradation and DNA replication. Importantly, these differences were observed both in total populations of LCMV-specific naive and memory CD8+ cells and in LCMV-specific CD8+ T-cell populations that were in the G1 phase of the cell cycle only. In addition, the expression differences between naive and memory cells were exaggerated following antigenic stimulation. The fact that memory cells are precharged with several of the major factors that are necessary for the G1- to-S-phase transition suggests they may require a lower threshold of stimulation to enter the cell cycle.
Historically, lymphocytic choriomeningitis virus (LCMV) infection has been proven to be an excellent model for studying CD8+-T-cell activation and development, because infection with LCMV provides a robust CD8+-T-cell response and viral clearance is primarily dependent on CD8+ T cells (6). Naive, antigen-specific CD8+ T cells respond to an LCMV infection by eliminating the target cell, which is facilitated by increased expression of several antiviral effector molecules, such as perforin, granzyme B, gamma interferon, and tumor necrosis factor alpha. In addition, the activated cells upregulate the interleukin 2 (IL-2) receptor and begin to synthesize IL-2, leading to rapid proliferation. The newly activated or “effector” T cells continue to proliferate until shortly after the virus is cleared, with the number of antigen-specific CD8+ cells peaking at day 8 postinfection. After antigen clearance, the pool of effector cells contracts until it ultimately constitutes approximately ∼5 to 10% of the total T-cell pool. Current evidence indicates that the antigen-specific “memory” cells remaining after the contraction phase progressively developed from the pool of effector cells by slowly altering their gene expression pattern to provide them with unique characteristics. For example, memory cells have the ability to more rapidly secrete antiviral cytokines and to kill infected cells than their naive precursors. In addition, one of the hallmark features of memory cells is their ability to homeostatically proliferate independently of contact with antigen. After antigen-specific memory cells are initially generated, their numbers are maintained by a gradual turnover of the population. Antigen-specific memory cells will divide approximately once per 1 to 2 months (∼1 to 5% of memory cells are in cycle at any given point in time), yet the total number of those cells does not change, suggesting there are equal rates of cell death and division in a given specific memory-cell population (8, 12, 14, 26).
Although many details about the molecular mechanisms that control memory-cell homeostasis are not yet clear, several critical components that govern this process are beginning to emerge. First, it has become apparent that the cytokine IL-15 is required for memory-cell homeostatic proliferation (2, 9, 17, 24, 25, 27). In mice lacking IL-15 or the IL-15 receptor alpha chain, memory CD8+ T cells fail to undergo homeostatic proliferation and self-renewal, thus leading to a gradual reduction in their number over time (2). Interestingly, some evidence suggests that IL-7 can substitute for IL-15 to support homeostasis in immunodeficient mice, as long as it is not made limiting by competition from other cell types (9, 17, 23, 24). Second, it has been shown that memory cells can survive in the absence of major histocompatibility complex (MHC) class I contact, unlike their naive counterparts (22). These two distinct properties of memory cells, the ability to homeostatically proliferate in an IL-15-dependent manner and the ability to survive without MHC class I contact, suggest that they have altered their gene expression pattern relative to their naive progenitors and are thus equipped to interact with and respond to their environment in different ways. In a comprehensive paper by Kaech et al. (13), memory cells were shown to have increased expression of genes encoding T-cell-effector molecules, such as gamma interferon, perforin, and granzyme B, in addition to a variety of genes involved in signal transduction, cell migration, apoptosis, and cell division pathways. It is reasonable to assume that some of these gene expression changes are fundamentally responsible for providing memory cells with the ability to undergo homeostatic division. Interestingly, the authors demonstrated that cyclins E1, E2, and B1 are overexpressed in memory cells by nearly twofold. From these observations, it was hypothesized that memory cells might differentially express additional genes that regulate the cell cycle, and a closer examination was warranted. In work described here, the expression of 96 different genes that regulate the cell cycle was compared in purified populations of LCMV antigen-specific naive and memory cells. It was discovered that relative to naive CD8+ T cells, memory CD8+ T cells express higher levels of mRNAs that correspond to a variety of cell cycle regulatory genes, particularly those that control the G1-S-phase transition. To exclude the possibility that these observed differences in gene expression resulted from inclusion of the small percentage of memory cells that were undergoing homeostatic proliferation, the analysis was performed with similar results on sorted populations of cells that were only in the G1 phase of the cell cycle, based on DNA content. In addition, it was discovered that the differences in expression of these genes between naive and memory cells was exaggerated following antigenic stimulation.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were purchased from the National Cancer Institute (Frederick, Md.). Thy1.1+ P14 mice bearing the Db-GP33-specific T-cell receptor were backcrossed to C57BL/6 and have been previously described (5). LCMV Armstrong virus was propagated, titers were determined, and virus was used as previously described (1). P14 chimeric immune mice were generated by adoptively transferring ∼106 total splenocytes from a naive P14 transgenic animal into naive B6 mice by intravenous injection. The chimeric animals were then infected with 2 × 105 PFU of LCMV Armstrong by intraperitoneal injection. All chimeric immune animals were sacrificed at >30 days postinfection. All experiments involving the use of mice were performed in compliance with all federal and institutional guidelines and policies.
Isolation of T-cell subsets.
Single-cell suspensions were prepared in RPMI containing 10% fetal calf serum by mechanical disruption of spleens. Red blood cells were lysed in 0.83% ammonium chloride. CD8+ cells were initially purified by magnetic bead selection using anti-CD8 beads (Miltenyi Biotec, Auburn, Calif.). Enriched CD8+ cells were stained with anti-CD8 and anti-Thy1.1 antibodies and further purified by fluorescence-activated cell sorting (FACS) to select antigen-specific, Thy1.1 CD8 double-positive cells. The purity for each population averaged ≥94% across several experiments.
Flow cytometry, Hoechst staining, and cell sorting.
Single-cell suspensions of spleen were prepared, and 106 cells were stained in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (FACS buffer) for 30 min at 4°C followed by three washes in FACS buffer. Cells were then fixed in 2% paraformaldehyde diluted in PBS. Samples were acquired on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.), and the data were analyzed by using CELLQuest software (Becton Dickinson Immunocytometry Systems). The anti-CD8 (clone 53-6.7) and anti-CD44 (clone IM7) antibodies were purchased from BD PharMingen. LCMV GP33-41 MHC class I/peptide tetramers were prepared and used as described previously (21).
Cell culture and peptide stimulation.
Total splenocytes were cultured for 24 h with or without 1 μg of GP33-41 peptide/ml in RPMI plus 10% fetal calf serum, 40 × 106 in each well of 6-well plate. Following stimulation, the cells were subjected to magnetic bead separation and cell sorting as described above.
RNA isolation and amplification.
RNA was extracted from sorted cells using TRIZOL reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The entire amount of mRNA isolated from approximately 5 × 105 to 1 × 106 cells per sample was linearly amplified two times as previously described (13).
Gene expression analysis.
Five micrograms of 2× amplified RNA was reverse transcribed into single-stranded cDNA and was then used as the substrate for probe synthesis according to the SuperArray (Frederick, Md.) [32P]dCTP radioactive labeling protocol, using the Mouse Q-series cell cycle array kit. Probe hybridization and blot washes were performed according to the manufacturer's instructions. Following hybridization, the arrays were subjected to phosphorimager analysis by using a Molecular Dynamics Typhoon imager and software (Amersham, Piscataway, N.J.). The relative expression level of each gene was quantified and normalized to the signal of the cyclophilin A and rpl13a control genes on the same membrane. The relative expression level of each gene was compared by using arbitrary units that were calculated with the following formula:
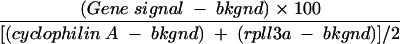 |
where bkgnd is background level.
RESULTS
To test the hypothesis that naive and memory CD8+ T cells may differentially regulate the expression of genes that control the cell cycle, mRNA from each cell type was comparatively analyzed using the Mouse Q-series cell cycle gene arrays produced by SuperArray, Inc. Ninety-six different genes that control the cell cycle are represented on each array, including cyclin-dependent kinases (cdk's), cyclins, cdk inhibitors, cdk phosphatases, and cdk kinases. In addition, genes essential for DNA damage and mitotic spindle checkpoints and genes in some ubiquitin-conjugation pathways are represented.
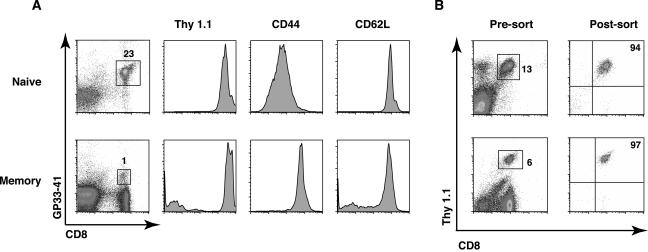
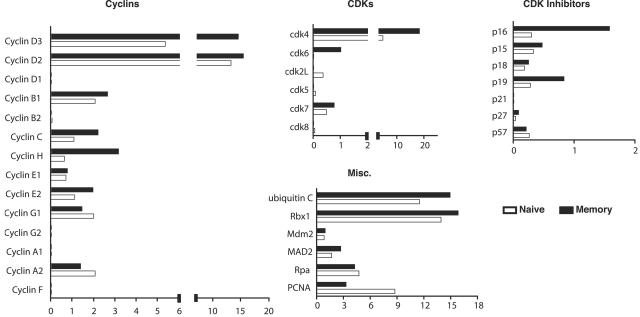
Naive CD8+ T cells were isolated directly from the spleens of uninfected P14 transgenic mice, which express T cell receptors that specifically recognize the GP33-41 epitope of LCMV in the context of MHC class I (5). Transgenic P14 memory CD8+ T cells were harvested >30 days postinfection from the spleens of mice that initially received 106 total P14 transgenic splenocytes by intravenous injection, followed by an acute infection with LCMV. Figure 1A shows a typical example of FACS analysis of cells harvested from naive and memory mice. The P14 transgenic CD8+ cells in both naive and memory mice stain positive for GP33-41 MHC class I tetramer, the congenic marker Thy1.1, and the surface marker CD62L. CD44 is expressed at low to intermediate levels on naíve cells but is present at high levels on memory cells. In multiple independent experiments, both naive and memory cells were initially purified using anti-CD8 magnetic beads and then were stained with anti-CD8 and anti-Thy1.1 antibodies and sorted to >94% purity. A typical example of the FACS analysis from one such stain and sort is shown in Fig. 1B. The mRNA from sorted cells of each type was then isolated, linearly amplified, radioactively labeled, and hybridized to the gene arrays. The data were analyzed and quantified by using a phosphorimager. To determine the relative amount of expression for each gene, the hybridization signal for each gene was calculated as a percentage of the combined average expression of two control genes, encoding cyclophilin A and rpl13A, following background correction. Gene expression data from three independent experiments were averaged and are summarized in Fig. 2. The results indicate that memory cells express higher levels of several types of genes that regulate the cell cycle than do naive cells. These genes include some cyclins, cdk's, cdk inhibitors, and factors involved in DNA replication, mitosis, and protein degradation.
FIG. 1.
Characterization of the naive and memory P14 transgenic CD8 T cells used for subsequent gene expression analysis. (A) Splenocytes from naive P14 transgenic or LCMV-immune P14 chimeric mice were isolated and stained with MHC class I tetramer specific for the LCMV GP33-41 epitope, anti-CD8, anti-Thy1.1, anti-CD44, and anti-CD62L antibodies and then analyzed by flow cytometry. Thy1.1, CD44, and CD62L expression is shown for tetramer and CD8 double-positive cells. (B) Thy 1.1 and CD8 double-positive transgenic T cells were sorted to >94% purity prior to RNA isolation. RNA was isolated from cells sorted as indicated here and was analyzed as described in the legend to Fig. 2.
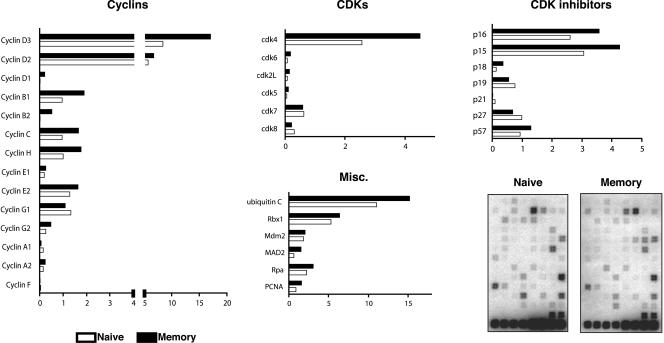
FIG. 2.
Relative expression of cell cycle control genes in naive and memory CD8+ T cells. P14 transgenic naive and memory CD8+ T cells were sorted as described in the legend to Fig. 1. Total RNA from these cells was harvested and subjected to two rounds of linear amplification, labeling with [32P]dCTP, and array hybridization. Two representative arrays are shown. The relative expression level of each gene was quantified by phosphorimager analysis, normalized to the signal of the cyclophilin A and rpl13a control genes on the same membrane, and expressed as arbitrary units. The formula used to calculate these units is described in Materials and Methods. The relative expression of cyclins, cyclin-dependent kinases, cdk inhibitors, and miscellaneous proteins involved in DNA replication and mitosis is indicated as an average for three independent experiments.
The comparison revealed that memory cells produce elevated levels of cyclins D3, D2, B1, C, and H. Notably, cyclin D3 was expressed in approximately twofold excess in memory cells. Cyclins D3 and D2 appeared to be the most abundantly expressed cyclins in both naive and memory cells, and they were among the most abundantly expressed of all genes examined, assuming that the hybridization efficiencies for each gene were similar. In addition, it was revealed that cdk4 and cdk6, the major G1-phase cyclin-dependent kinases, are overexpressed by approximately twofold in memory cells. The cdk inhibitors p16, p15, and p18 were also overexpressed in memory cells, while the inhibitor p27 was slightly elevated in naive cells. The levels of the remaining cdk inhibitors appeared similar.
The ubiquitin-mediated protein degradation pathway is important for controlling the concentration of cell cycle regulatory proteins. Ubiquitin C and Rbx1 are encoded by two genes involved in ubiquitination that were both slightly upregulated in memory cells. Ubiquitin C is one of several genes that encode a polyubiquitin protein that is thought to be posttranslationally processed into ubiquitin monomers (3). Kamura et al. (15) found that yeast Rbx1 is a subunit and activator of the SCF-Cdc4 complex that is required for ubiquitination of the cyclin-dependent kinase inhibitor Sic1 and for the G1-to-S cell cycle transition.
Other factors that were upregulated in memory cells include genes involved in DNA replication, such as proliferating cell nuclear antigen (PCNA), a subunit for DNA polymerase delta (16). Replication protein A (Rpa) (14 kDa), a single-stranded DNA binding protein that is involved in DNA damage repair (28), and mitotic arrest deficient protein 2 (Mad2) (7), which is required for execution of the mitotic checkpoint, were both also overexpressed in memory cells.
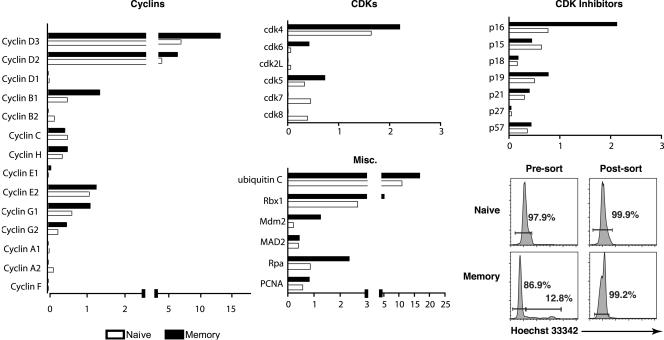
A primary concern was that these observed differences in the expression of cell cycle regulatory genes between naive and memory cells might be due to the fact that a small percentage of memory cells (1 to 3%) are in cycle at any given point in time, whereas naive cells essentially do not proliferate unless they are stimulated by antigen. To address this issue, naive and memory cells were stained with Hoechst 33342 prior to sorting (Fig. 3). The amount of fluorescence attributed to the Hoechst stain indicates DNA content, with the majority of cells having a diploid genome (i.e., residing in the G1 peak). Those cells that have progressed into S, G2, and M phases have a corresponding increase in fluorescence intensity. Only the cells that were clearly in the G1 peak in each sample were harvested by sorting and used for the array analysis. The data from two independent experiments were averaged and are shown in Fig. 3.
FIG. 3.
Expression of cell cycle control genes in transgenic, G1 sorted naive, and memory CD8+ T cells. Naive and memory P14 transgenic CD8+ T cells were stained with Hoechst 33342, and the cells with 2N DNA content (G1) were purified. RNA was isolated from the sorted cells and analyzed as described in the legend to Fig. 2 and in the Methods section. The relative expression of cyclins, cyclin-dependent kinases, cdk inhibitors, and miscellaneous proteins involved in DNA replication and mitosis is indicated as an average for two independent experiments.
The gene expression profiles observed in Hoechst-stained, G1 sorted cells were very similar to those observed in cells that were not stained with Hoechst, with a few notable differences (Fig. 3). Specifically, the differences in the expression of cyclins cyclins C and H were diminished. Interestingly, two previously unobserved differences appeared in the G1 sorted comparison. First, the cdk inhibitor p16 appeared to be overexpressed in the G1 sorted memory cells by two- to threefold. Second, mouse double minute 2 homolog (Mdm2), an inhibitor of the p53 oncogene, also appeared to be upregulated in G1 sorted memory cells. Finally, Mad2, which at first appeared to be overexpressed in memory cells, seemed to have equivalent expression between G1 sorted naive and memory samples.
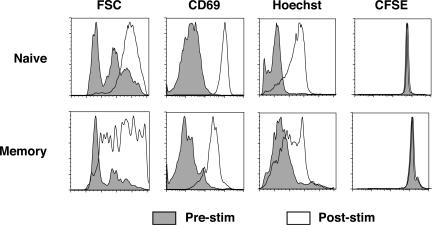
Given the observed differences in expression of these genes in resting naive and memory cells and a recent report that memory cells enter the cell cycle slightly sooner than naive cells following antigen stimulation (29), it was hypothesized that additional differences in cell cycle gene expression might be observed after antigen stimulation. To test this, splenocytes from transgenic naive and immune chimeric mice were isolated and stimulated for 24 h with 1 μg of GP33-41 peptide/ml. As shown in Fig. 4, the cells responded to stimulation by upregulating the surface expression of CD69. In addition, the stimulated cells displayed an increase in forward scatter and Hoechst 33342 staining, which indicates that they were enlarging and synthesizing DNA in preparation for division. However, the cells had not yet divided at 24 h poststimulation, as indicated by staining with CFSE (carboxy-fluorescein diacetate, succinimidyl ester). The antigen-specific cells were sorted and their RNA was harvested and analyzed as described above. The data from two independent experiments were averaged and are shown in Fig. 5. The results indicate that most of the genes that were examined were induced to a higher degree in memory cells than in naive cells. Following stimulation, memory cells expressed higher levels of every cyclin mRNA that was detectable with the exceptions of cyclins E1 and A2. Moreover, the differences between memory and naive cells were exaggerated compared to the unstimulated samples. Peptide stimulation dramatically induced cdk4 in memory cells at nearly six-fold higher levels than in naive cells. cdk6 expression remained higher in memory cells following stimulation but was not substantially increased relative to levels for unstimulated samples. Interestingly, antigen also induced the expression of most cdk inhibitors to a higher degree in memory cells than in than naive cells. Notably, p16 and p19 were upregulated by eight- and twofold, respectively. The expression of other factors, such as ubiquitin c, Rbx1, Mdm2, and Mad2, was elevated in both samples following stimulation, but the relative levels between memory and naive cells remained unchanged. In contrast, PCNA was elevated to higher levels in naive cells following antigen stimulation.
FIG. 4.
Activation state of transgenic cells following peptide stimulation. P14 transgenic T cells were labeled with CFSE, stimulated for 24 h with 1 μg of GP33-41 peptide/ml, stained with Hoechst 33342 and anti-CD8, anti-Thy1.1, and anti-CD69 antibodies, and then analyzed by flow cytometry. CD8 Thy1.1 double-positive cells are shown in each histogram (gates not shown).
FIG. 5.
Relative expression of cell cycle control genes in naive and memory CD8+ T cells following antigen stimulation. Transgenic splenocytes from naive and immune chimeric mice were harvested and stimulated for 24 h in vitro with 1 μg of GP33-41 peptide/ml. Following stimulation, the cells were stained with anti-CD8 and anti-Thy1.1 antibodies and sorted, and mRNA from each population was harvested and analyzed as described in the legend to Fig. 2. The data represent an average for two independent experiments.
DISCUSSION
Over the past few years, it has become increasingly clear that as naive CD8+ T cells develop into memory cells following antigen exposure, they obtain unique qualities that can be attributed to changes in their gene expression profiles. One of the hallmark characteristics of CD8+ memory T cells is their ability to homeostatically proliferate (12). It was thus hypothesized that memory cells may regulate the expression of genes that control the cell cycle differently than their naive precursors. To test this, mRNAs from naive and memory cells were linearly amplified, radioactively labeled, hybridized to a commercially available cell-cycle-gene-specific array, and comparatively analyzed. The results indicate that several genes, having a variety of different influences on the cell cycle, are expressed at higher levels in memory cells than in naive cells. This observation was made from a comparison of whole populations of antigen-specific naive and memory cells in addition to naive and memory cells that were sorted based on DNA content to isolate only those cells that had not progressed beyond the G1 phase of the cell cycle.
The experiments described here indicate that cyclins D3, D2, B1, C, and H are all expressed at higher levels in resting CD8+ memory cells than in naive cells. Of these, cyclins D3 and D2 appeared to be the most abundantly expressed in both naive and memory cells. Cyclin D1 was essentially not detectable in either population. This result is supported by previously published reports. First, a recent paper by Veiga-Fernandes et al. demonstrated that cyclins D3 and D2 are overexpressed in antigen-specific memory cells (29). Second, a recent report of global changes in the gene expression of purified antigen-specific memory cells has indicated that cyclin B1 is overexpressed relative to levels in naive cells (13). The work reported here confirms and extends both of these observations.
Cyclins D3 and D2 play important roles in the transition from the G phase (1) to the S phase of the cycle. In combination with cdk4 or cdk6, the D-type cyclins induce expression of cyclins E1 and E2, which are active at the peak of the G1-to-S-phase transition (18). Overexpression of either cyclin E1 or cyclin E2 in mammalian cells accelerates the G1 phase, indicating that these cyclins may be rate limiting for G1 progression (11). Interestingly, recent work has shown that ligation of the costimulatory molecule 4-1BB enhances cell cycle progression in CD8+ T cells via both IL-2-dependent and -independent pathways (18). It does this in part by inducing the expression of cyclins D2 and D3. It is plausible that increased expression of cyclins D3 and D2 in memory cells is the result of prior or increased signaling through 4-1BB during the process of antigen-induced activation or by continued signaling after antigen clearance. Corresponding to increased levels of the D-type cyclins, it was also observed that there were increases in the amounts of their binding partners, the major G1-phase cyclin-dependent kinases cdk4 and cdk6. cdk4 and cdk6 are the main G1 kinases that cause progression into S phase when bound to D-type cyclins. Cyclin B1, also upregulated in memory cells, is predominantly expressed during the G2-M-phase transition and complexes with cdk1 (cdc2) to form mitosis-promoting factor, which is responsible for causing the cell to enter mitosis following DNA replication (4). Interestingly, B1 can shuttle between the nucleus and the cytoplasm. A recent paper by Moore et al. demonstrated that the cdk1-cyclin B1 complex can also stimulate entry into S phase if the complex is relocated from the cytoplasm to the nucleus and stimulated with the cdc25 phosphatase (20). The fact that memory cells are precharged with several of the major factors that are required to signal entry into S phase suggests that they may require a lower threshold of stimulus to begin DNA synthesis.
The idea that memory cells might be a step closer to the initiation of DNA synthesis is supported by the observation that the expression of two genes involved in DNA synthesis and repair, the PCNA and Rpa genes, is also increased by approximately twofold in memory cells. First, PCNA is a cofactor for DNA polymerase delta, which is unique among the DNA polymerases in that it has 3′-5′ exonuclease proofreading activity (19). DNA polymerase delta is thus thought to be important for maintaining the fidelity of DNA replication. Coincidentally, Rpa (replication protein A; 14 kDa) is a single-stranded DNA binding protein that is involved in DNA damage repair and was found to be overexpressed as well.
Additional support for the hypothesis that memory cells might have an altered threshold for entering S phase comes from the observation that Mdm2, a major negative regulator of the p53 oncogene, was also upregulated in G1 sorted memory cells. The well-characterized p53 oncogene induces the expression of factors that inhibit cell division. Mdm2 binds p53 and targets it for ubiquitination and subsequent degradation, thus promoting survival and cell cycle progression (10). It has been shown that DNA damage induces phosphorylation of p53, leading to decreased interaction with Mdm2. In light of the fact that the concentrations of many proteins that control the cell cycle, including p53, are regulated by ubiquitination, it is not that surprising to see a corresponding upregulation in memory cells of some of the factors, such as Mdm2, ubiquitin c, and Rbx1, that are important in the ubiquitination pathway.
In summary, it appears that memory cells express elevated levels of several factors that control the G1-S-phase transition, including the major G1 cyclins and cyclin-dependent kinases. The reason that the levels of these transcripts are elevated is not yet known, but it could be due to a number of things, such as the following: (i) changes in chromatin structure at these loci, (ii) increased levels or activation of a common transcription factor that regulates these genes, (iii) an accumulation of transcript from previous cycles, (iv) enhanced or prior signaling through 4-1BB, (v) stimulation by cytokines, such as IL-15 or IL-7, or (vi) increased sensitivity to cytokine signaling. Regardless, this preloading of memory cells with factors that are required for entry into S phase could have the effect of lowering the stimulus threshold that is required to induce proliferation and may contribute to the ability of memory cells to homeostatically divide.
Acknowledgments
This work was generously supported by grant RO1-AI30048 from the National Institutes of Health and by a postdoctoral fellowship from The Cancer Research Institute.
REFERENCES
- 1.Ahmed, R., A. Salmi, L. D. Butler, J. M. Chiller, and M. B. Oldstone. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 160:521-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, T. C., E. J. Wherry, D. Boone, K. Murali-Krishna, R. Antia, A. Ma, and R. Ahmed. 2002. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 195:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Board, P. G., M. Coggan, R. T. Baker, J. Vuust, and G. C. Webb. 1992. Localization of the human UBC polyubiquitin gene to chromosome band 12q24.3. Genomics 12:639-642. [DOI] [PubMed] [Google Scholar]
- 4.Brandeis, M., I. Rosewell, M. Carrington, T. Crompton, M. A. Jacobs, J. Kirk, J. Gannon, and T. Hunt. 1998. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc. Natl. Acad. Sci. USA 95:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandle, D., K. Brduscha-Riem, A. C. Hayday, M. J. Owen, H. Hengartner, and H. Pircher. 1995. T cell development and repertoire of mice expressing a single T cell receptor alpha chain. Eur. J. Immunol. 25:2650-2655. [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier, M., and A. Zajac. 1999. Lymphocytic choriomeningitis virus, p. 575-605. In R. Ahmed and I. Chen (ed.), Persistent viral infections. John Wiley & Sons, Ltd., London, United Kingdom.
- 7.Dobles, M., V. Liberal, M. L. Scott, R. Benezra, and P. K. Sorger. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101:635-645. [DOI] [PubMed] [Google Scholar]
- 8.Dutton, R. W., L. M. Bradley, and S. L. Swain. 1998. T cell memory. Annu. Rev. Immunol. 16:201-223. [DOI] [PubMed] [Google Scholar]
- 9.Goldrath, A. W., P. V. Sivakumar, M. Glaccum, M. K. Kennedy, M. J. Bevan, C. Benoist, D. Mathis, and E. A. Butz. 2002. Cytokine requirements for acute and basal homeostatic proliferation of naive and memory CD8+ T cells. J. Exp. Med. 195:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 11.Gudas, J. M., M. Payton, S. Thukral, E. Chen, M. Bass, M. O. Robinson, and S. Coats. 1999. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol. Cell. Biol. 19:612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jameson, S. C. 2002. Maintaining the norm: T-cell homeostasis. Nat. Rev. Immunol. 2:547-556. [DOI] [PubMed] [Google Scholar]
- 13.Kaech, S. M., S. Hemby, E. Kersh, and R. Ahmed. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111:837-851. [DOI] [PubMed] [Google Scholar]
- 14.Kaech, S. M., E. J. Wherry, and R. Ahmed. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2:251-262. [DOI] [PubMed] [Google Scholar]
- 15.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 16.Kelman, Z. 1997. PCNA: structure, functions and interactions. Oncogene 14:629-640. [DOI] [PubMed] [Google Scholar]
- 17.Kieper, W. C., J. T. Tan, B. Bondi-Boyd, L. Gapin, J. Sprent, R. Ceredig, and C. D. Surh. 2002. Overexpression of interleukin (IL)-7 leads to IL-15-independent generation of memory phenotype CD8+ T cells. J. Exp. Med. 195:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, H. W., K. O. Nam, S. J. Park, and B. S. Kwon. 2003. 4-1BB enhances CD8+ T cell expansion by regulating cell cycle progression through changes in expression of cyclins D and E and cyclin-dependent kinase inhibitor p27kip1. Eur. J. Immunol. 33:2133-2141. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. Y., and N. L. Toomey. 1987. Human placental DNA polymerase delta: identification of a 170-kilodalton polypeptide by activity staining and immunoblotting. Biochemistry 26:1076-1085. [DOI] [PubMed] [Google Scholar]
- 20.Moore, J. D., J. A. Kirk, and T. Hunt. 2003. Unmasking the S-phase-promoting potential of cyclin B1. Science 300:987-990. [DOI] [PubMed] [Google Scholar]
- 21.Murali-Krishna, K., J. D. Altman, M. Suresh, D. J. Sourdive, A. J. Zajac, J. D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177-187. [DOI] [PubMed] [Google Scholar]
- 22.Murali-Krishna, K., L. L. Lau, S. Sambhara, F. Lemonnier, J. Altman, and R. Ahmed. 1999. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science 286:1377-1381. [DOI] [PubMed] [Google Scholar]
- 23.Prlic, M., and S. C. Jameson. 2002. Homeostatic expansion versus antigen-driven proliferation: common ends by different means? Microbes Infect. 4:531-537. [DOI] [PubMed] [Google Scholar]
- 24.Prlic, M., L. Lefrancois, and S. C. Jameson. 2002. Multiple choices: regulation of memory CD8 T cell generation and homeostasis by interleukin (IL)-7 and IL-15. J. Exp. Med. 195:F49-F52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schluns, K. S., K. Williams, A. Ma, X. X. Zheng, and L. Lefrancois. 2002. Cutting edge: requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J. Immunol. 168:4827-4831. [DOI] [PubMed] [Google Scholar]
- 26.Sprent, J., and C. D. Surh. 2001. Generation and maintenance of memory T cells. Curr. Opin. Immunol. 13:248-254. [DOI] [PubMed] [Google Scholar]
- 27.Tan, J. T., B. Ernst, W. C. Kieper, E. LeRoy, J. Sprent, and C. D. Surh. 2002. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J. Exp. Med. 195:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umbricht, C. B., L. F. Erdile, E. W. Jabs, and T. J. Kelly. 1993. Cloning, overexpression, and genomic mapping of the 14-kDa subunit of human replication protein A. J. Biol. Chem. 268:6131-6138. [PubMed] [Google Scholar]
- 29.Veiga-Fernandes, H., and B. Rocha. 2004. High expression of active CDK6 in the cytoplasm of CD8 memory cells favors rapid division. Nat. Immunol. 5:31-37. [DOI] [PubMed] [Google Scholar]