Abstract
Background Because soil salinity is a major abiotic constraint affecting crop yield, much research has been conducted to develop plants with improved salinity tolerance. Salinity stress impacts many aspects of a plant’s physiology, making it difficult to study in toto. Instead, it is more tractable to dissect the plant’s response into traits that are hypothesized to be involved in the overall tolerance of the plant to salinity.
Scope and conclusions We discuss how to quantify the impact of salinity on different traits, such as relative growth rate, water relations, transpiration, transpiration use efficiency, ionic relations, photosynthesis, senescence, yield and yield components. We also suggest some guidelines to assist with the selection of appropriate experimental systems, imposition of salinity stress, and obtaining and analysing relevant physiological data using appropriate indices. We illustrate how these indices can be used to identify relationships amongst the proposed traits to identify which traits are the most important contributors to salinity tolerance. Salinity tolerance is complex and involves many genes, but progress has been made in studying the mechanisms underlying a plant’s response to salinity. Nevertheless, several previous studies on salinity tolerance could have benefited from improved experimental design. We hope that this paper will provide pertinent information to researchers on performing proficient assays and interpreting results from salinity tolerance experiments.
Keywords: Assessing salinity tolerance, salt-imposition systems, quantifying physiological traits, analysing salinity data, tolerance indices, salt stress phenotyping, osmotic stress
INTRODUCTION
Soil salinity is a global problem that affects approx. 20 % of irrigated land and reduces crop yields significantly (Qadir et al., 2014). The physiological responses of a plant to salinity are often complex and multi-faceted, which makes experiments difficult to design and interpret. Current plant physiology has advanced, given the development of so-called ‘omics-driven’ research. Physiological measurements have been revolutionized by new technologies, such as high-throughput phenotyping, bioinformatics and novel analytical methods that have enabled fields such as metabolomics to emerge. At a basic level, the response of plants to salinity can be described in two main phases: the shoot ion-independent response occurs first, within minutes to days, and is thought to be related to Na+ sensing and signalling (Gilroy et al., 2014; Roy et al., 2014). In this first phase, effects of salinity on water relations can be important, causing stomatal closure and the inhibition of leaf expansion (Munns and Termaat, 1986). The second phase, the ion-dependent response to salinity, develops over a longer period (days to weeks) and involves the build-up of ions in the shoot to toxic concentrations, particularly in old leaves, causing premature senescence of leaves and ultimately reduced yield or even plant death (Munns and Tester, 2008).
Three main salinity tolerance mechanisms have been proposed by Munns and Tester (2008): ion exclusion – the net exclusion of toxic ions from the shoot; tissue tolerance – the compartmentalization of toxic ions into specific tissues, cells and subcellular organelles; and shoot ion-independent tolerance – the maintenance of growth and water uptake independent of the extent of Na+ accumulation in the shoot. Other physiological components are also likely to contribute to salinity tolerance, such as the maintenance of plant water status, transpiration (T) and transpiration use efficiency (TUE) (Harris et al., 2010; This et al., 2010; Barbieri et al., 2012); leaf area (Maggio et al., 2007); seed germination (Foolad and Lin, 1997); production of antioxidants (Ashraf, 2009); early seedling growth (Kingsbury and Epstein, 1984); and harvest index (HI) (Gholizadeh et al., 2014). Very little is known about these physiological components, so understanding the effects of salinity on these processes needs further investigation. In addition, numerous factors can influence plants’ responses to salinity due to the complex nature of salinity tolerance. For example, the beneficial effect of calcium application to plants exposed to high levels of Na+ was reported back in 1902 by Kearney and Cameron (as reviewed by Lahaye and Epstein, 1971). Since then, the interaction between Na+ and Ca2+ has been extensively studied (as reviewed by Cramer, 2002), and nowadays several salinity experiments use Ca2+ supplemented to the medium to maintain Ca2+ activity (for further details see Supporting Methodologies, section 1).
In this review, we describe techniques that measure the impact of salinity on several physiological traits, such as growth, water relations, ion homeostasis, photosynthesis, yield components and senescence. It often can be difficult to identify which traits are the most important ones contributing to salinity tolerance in the given plant system. To ease this difficulty, we suggest the generation of graphs that show correlations between the proposed traits (e.g. leaf Na content) and a measure of salinity tolerance (e.g. salt tolerance index). Such correlations help to establish whether the measured traits are associated with each other (noting the limitation that a correlation cannot give definitive information on cause-and-effect relationships); the correlation coefficient can give an indication of which traits are the most important contributors to salinity tolerance (for the analysed plant in the analysed environment). Numerous studies have used two contrasting genotypes to characterize their salinity response at the transcriptional (e.g. Ouyang et al., 2007; Beritognolo et al., 2011), proteomic (e.g. Ma et al., 2012; Cui et al., 2015) or metabolomic (e.g. Widodo et al., 2009; Zhao et al., 2014) levels, but limited arguments advocate the reasoning of selecting such genotypes. When a selection of few contrasting genotypes is necessary, one should take into account the potential variability of the trait under study within the population and, if available, also consider genotypic information. Although the use of contrasting genotypes in such analyses is valid, we consider that this narrow selection is not representative of a species’ performance under salinity stress. Thus, we strongly encourage broadening these types of analyses by using several genotypes before speculating about a species’ performance.
We also suggest some guidelines for designing experiments and analysing data related to salinity tolerance, with the aim of facilitating the gathering and interpretation of accurate and useful physiological data. We have included Supporting Methodologies Section 1 in an attempt to facilitate the use of good quality experimental procedures that are crucial to the success of salinity studies. Key aspects include the experimental system (e.g. agar plates, hydroponics, soil-filled pots or the field), the extent of the salinity stress (levels of salt stress, timing of salt application and duration of treatment) and the biological system (species and genotype). In Supporting Methodologies Section 2, we provide further details on the indices explained in this review and how they can be derived from physiological measurements. Readers are also referred to excellent online resources, such as Prometheus Wiki (e.g. http://prometheuswiki.publish.csiro.au/tiki-index.php?page=Salinity) and the PlantStress website (http://www.plantstress.com/methods/index.asp).
The aim of providing extensive details in the Supporting Methodologies is to help new researchers as they begin to work in this field. That said, we also believe the extensive details are necessary because many papers are still being published with insufficient attention to important aspects of the experiments. Previously, Flowers (2004) examined in detail several papers that claimed significant effects of transgenic events on salinity tolerance. However, these papers were found to have provided insufficient evidence for the claims made for a range of reasons. Moreover, Claeys et al. (2014) also concluded that most of the published studies use very high levels of NaCl, and that the recorded phenotypes, which include expression data, are associated with severe stress responses. We hope that researchers new to this field can draw on some of the technical points raised in this paper and that new and experienced researchers alike will base their work on rigorous experimental methodologies such as those described in the Supporting Methodologies.
Quantifying the effects of salinity on plant growth: destructive and non-destructive approaches
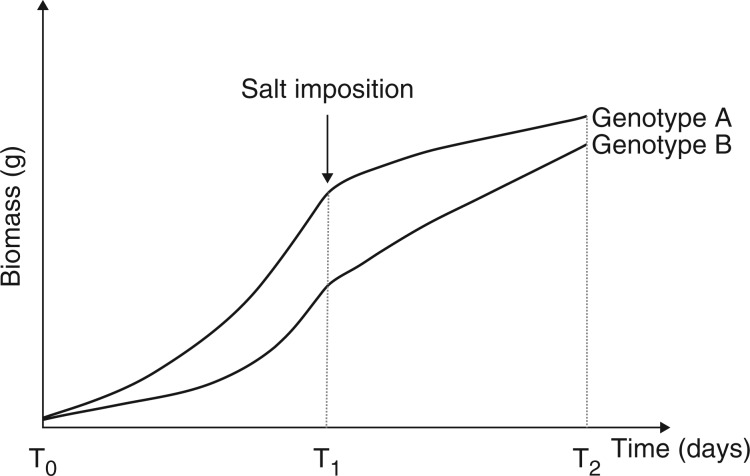
In an experimental setting, one of the first observable responses after salinity imposition is a reduction in shoot growth (Fig. 1, Supplementary data Movie S1). To describe this reduction in plant growth, two distinct approaches can be used: a destructive harvest, or a non-destructive approach using, for example, digital imaging.
Fig. 1.
Salinity tolerance should be calculated by measuring the effects of salinity on plant growth during the time of stress imposition and not during the lifetime of the plant. Growth of two hypothetical genotypes is shown, before (T0 to T1) and after (T1 to T2) imposition of salinity stress. Genotype A grows faster than Genotype B under control conditions, but its growth is inhibited more by salinity. If growth were measured by biomass increase from T0 to T2, Genotype A would appear to be more salt tolerant. However, if growth were measured only from T1 to T2, then Genotype B would appear to be more salt tolerant.
A destructive harvest involves separation of plants into parts, such as shoot from root, or into more parts, such as blade, petiole (or sheath), stem and root. Fresh mass and dry mass are then recorded; and other measurements can also be made, such as root length, plant height and leaf area. For detailed information about the key aspects to consider when designing such an experiment (e.g. selection of appropriate controls, stress imposition and the experimental system), see Supporting Methodologies Section 1. Although this destructive approach requires no specialized, expensive equipment, it often involves a substantial amount of manual handling, which, when combined with available space for growing plants, limits the number of time points for sampling. The relative decrease in plant biomass (RDPB) indicates the reduction in growth by comparing the total biomass of stressed and control plants at the end of the experiment [Supporting Methodologies Section 2, eqn (a)]. At a minimum, plants should be harvested at the time of the start of the salt stress and at the end of the period of interest, allowing the reduction in growth by salinity to be observed across the stress interval and not as an integral of the growth both before and after the salt stress. This is particularly important when comparing lines that have different rates of growth under control conditions and thus are likely to be different sizes at the beginning of the stress period. The stress interval can be defined as starting at the time of stress imposition (T1) until the endpoint of the experiment (T2, see Fig. 1). Measurements of the difference in growth reduction between control and stress treatments will be more accurate and usually greater when biomass increases are analysed only during the stress interval (T1 to T2).
It is important to mention that for plants that accumulate extremely high levels of salt in the shoot (e.g. halophytes such as Salicornia, Suaeda), the mass of the salt can become a significant fraction of the total mass. Hence, in these specific cases, we recommend the use of ash-free dry mass or ethanol-insoluble dry mass to quantify the reduction in growth – or at least to subtract the mass of NaCl from the total dry mass, given the amount of NaCl accumulated in the relevant plant part is known.
Another approach that should be considered is the use of dose–response curves, which can be applied to an individual or to several genotypes. In Arabidopsis, the evaluation of dose–response curves in salinity has shown that low concentrations of salt have little effect on growth, but at higher concentrations the relative growth rate quickly decreases as a quadratic function of the NaCl concentration (Claeys et al., 2014). A meta-analysis study using response curves showed that leaf-related parameters (i.e. leaf-specific area and leaf dry mass per unit area) were significantly affected by salinity (Poorter et al., 2009, 2010). Although time consuming to perform and analyse, dose–response curves may provide valuable insights into the genotypic and phenotypic differences in response to salinity. In the future, the use of big-data analysis and statistical methods may enable dose–response curves to be considered in associated studies.
The second approach for assessing plant growth in response to salinity uses image acquisition technology. In this non-destructive approach, images of plants are taken at defined time intervals and the biomass is deduced from pixel counts (Berger et al., 2012). With automated, high-throughput phenotyping facilities, such as The Plant Accelerator in Adelaide, Australia, and in facilities hosted by other partners of the International Plant Phenotyping Network (http://www.plant-phenotyping.org/), it is possible to obtain a daily estimate of plant biomass from the start of an experiment, before salt imposition, through to the end. These systems are powerful because they provide high time and spatial resolution. Because of this high resolution, it is possible to develop more detailed models of growth (Ward et al., 2015) and to estimate relative growth rates (RGRs) (Berger et al., 2012), as has recently been achieved for rice (Hairmansis et al., 2014; Campbell et al., 2015). Importantly, these measurements also allow assessment of the shoot’s ion-independent component of salt toxicity, which involves the inhibition of shoot growth from the moment of salt imposition (Berger et al., 2012), before salt has had time to accumulate in the shoot and significantly affect the shoot’s function [Supporting Methodologies Section 2, eqn (b)].
Image acquisition technologies are developing rapidly and a number of software tools are now freely available (http://www.plant-image-analysis.org/), enabling the efficient analysis of imaging data. To date, a plethora of imaging systems focus on shoot parameters (as recently reviewed by Fahlgren et al., 2015). These systems appear to be particularly robust and reliable to quantify shoot growth under controlled environments. Few systems to date have evaluated mature plants, as this is preferentially done in the field, such as reported by Weber et al. (2012) and reviewed by Araus and Cairns (2014). When evaluating mature plants, studies rarely focus on growth, but rather focus on predicting or measuring yield and yield-related parameters, which are a major objective for crop breeding (Weber et al., 2012; Araus and Cairns, 2014).
Root imaging is inherently difficult in the field (Reynolds et al., 2012; Wasson et al., 2012). ‘Shovelomics’ and similar methods, in which the roots are excavated and analysed, have been proposed as an approach to characterize root system architecture under field conditions, but these methods are time consuming and involve destructive analysis (Trachsel et al., 2011; Bucksch et al., 2014). Few studies have reported root imaging under natural conditions and without destructive harvesting, such as imaging of roots using the Growth and Luminescence Observatory for Roots (GLO-Roots) system (Rellán-Álvarez et al., 2015) or in a more artificial system using transparent growth media (such as gel or glass beads) (Courtois et al., 2013; Topp et al., 2013). Improvements to non-destructive analyses involve not only automated data gathering and data analyses, but also the development of new technologies that allow determination of root parameters as well as the development of models to recover the structures of plants using 3D models (Ward et al., 2015). We believe that these technologies will provide a step change in salinity research, especially when time resolution is incorporated to provide insights into the dynamic responses of plants to salinity. In the future, this will allow the design of new types of experiments that should enable, for example, the monitoring of changes in root architecture in response to salinity treatment.
Whether the destructive or non-destructive approach is chosen will depend on the biological question asked and on access to technologies. Destructive harvests are generally suitable for long-term salt stress experiments (many days/weeks), when the differences in growth parameters, such as biomass, are readily apparent. Non-destructive analyses, such as imaging, allow the monitoring of the same plant over multiple time points, such as before and after stress application, enabling the detection of small and dynamic differences in growth parameters. By following the growth of each plant throughout the experiment, it is possible to separate the effects of salt on the inhibition of the production of new leaves from the acceleration of senescence and death of old leaves.
A number of indices can be derived from biomass measurements (Supporting Methodologies Section 2). For instance, a commonly used index to compare different accessions and even species is the salt tolerance index (ST), calculated as the percentage of biomass production over a defined period under saline compared with non-saline conditions (Munns et al., 2002) [Supporting Methodologies Section 2, eqns (c) and (d)]. The tolerance index (TOL) measures the difference in biomass production between salt-treated and control conditions (Rosielle and Hamblin, 1981) [Supporting Methodologies Section 2, eqn (e)]. The stress tolerance index (STI) takes into account both the overall biomass production of the population under control conditions and the ability to maintain yield (or other growth parameters) under stress conditions, favouring the selection of genotypes that perform well under both control and stressed conditions (Fernandez, 1992) [Supporting Methodologies Section 2, eqn (f)].
A recognizable indication of salinity stress is a reduction in shoot growth, which, in turn, can change the allocation of biomass between roots and shoots. This change can be described using the root mass ratio (RMR). A lower relative RMR indicates that a plant reduces allocation of biomass to the roots upon salt stress to a greater extent than under control conditions [Supporting Methodologies Section 2, eqns (g)–(i)].
Salinity stress also affects cell expansion in young leaves, generally causing a decrease in leaf area (Munns and Tester, 2008). The leaf area ratio (LAR) is the ratio of leaf area to the leaf’s dry mass. Relative LAR (RLAR; the LAR under saline compared with control conditions) provides a measure of the effect of salinity on what is effectively leaf thickness. A reduction in RLAR under salinity stress may be adaptive, given the leaf’s thicker cell walls or perhaps greater volume into which salts could be sequestered [Supporting Methodologies Section 2, eqns (j)–(l)].
Quantifying the effects of salinity on plant water relations, transpiration and transpiration use efficiency
The effects of salinity stress on a plant’s water relations have been described previously in the classical literature (Munns and Passioura, 1984; Nobel, 1991). Two components of a plant’s water relations are water potential and hydraulic conductivity. Water potential refers to the potential energy of water relative to pure water, and therefore determines the direction of water movement, where water moves from a location with a higher water potential to a location with a lower water potential. Hydraulic conductivity refers to the ease with which water can flow from one location to another and therefore affects the rate of water movement. In the face of high salinity, a plant’s ability to control these two components is essential. Two additional components, in combination with other components, are the outputs of water potential and hydraulic conductivity, namely the maintenance of water levels in tissue (the primary determinant of cellular growth and function) and the maintenance of transpiration [T; along with transpiration use efficiency (TUE), a related component]. Both enable a plant to continue to grow. In this review, we briefly introduce water potential and hydraulic conductivity. We then focus on the maintenance of water levels and transpiration because of their myriad physiological consequences and because their high-throughput measurement is now possible.
To learn about the many methods used to quantify the energy levels of water in plants, the reader may consult an extensive classical literature on this subject, including many papers by Munns and Passioura (e.g. Munns and Passioura, 1984; Passioura and Munns, 2000; Nobel, 1991) and books by Nobel (2005) and Meidner and Sheriff (1976). General undergraduate textbooks such as that by Taiz et al. (2015) also treat this topic well.
It has been argued that salt-tolerant plants decrease the hydraulic conductance of their roots, thereby reducing the delivery of (salty) water to the shoot (Vysotskaya et al., 2010) and resulting in reduced water potential in their leaves (Gama et al., 2009). While it can be informative to assess the effects of salinity on hydraulic conductance in the roots using a high-pressure flow meter (following, for instance, the method of Tyree et al., 1995) or on water potential in the leaves using a pressure chamber (following, for instance, the method of Scholander et al., 1965), these are specialized measurements that are probably best made in laboratories skilled in such technologies.
The water fraction (WF) of a tissue can be assessed simply. WF is the water content of the shoot (under controlled conditions) as a fraction of the fresh mass of the shoot. In the context of this review, a plant with a higher relative water fraction (RWF, the WF under stress conditions relative to control conditions) is better able to maintain its water content in the shoot upon salt stress [Supporting Methodologies Section 2, eqns (m)–(o)].
Another related trait important to plant function is the ability to maintain water content in tissues at optimal levels in the face of environmental stress. Plants under stress often lose some water from their tissues, which can have rapid and large effects on cell expansion, cell division, stomatal opening, abscisic acid (ABA) accumulation, etc. (Hsiao and Xu, 2000). Most of these effects become evident with no change in turgor pressure, although water potential can become more negative due to osmotic potential becoming more negative (‘osmotic adjustment’ – the ability to change the osmotic potential by alteration of the concentration of salts and/or neutral solutes, thus reducing changes in pressure potential). Most of these effects can also become evident with very small reductions (<10 %) in tissue water content. Here we are describing relative water content (RWC), which has also been extensively used in the classical literature to determine the water status of a shoot relative to its fully hydrated state. Under saline conditions, plants usually adjust their osmotic potential to maintain turgor pressure and this can exacerbate difficulties with classically used methods to measure RWC (Boyer et al., 2008). As such, quantifying the effects of salinity on RWC is important and physiologically relevant. Methods to do this are included in Supporting Methodologies Section 2, eqn (p).
It has been proposed that the ability of plants to maintain normal rates of transpiration under saline conditions is an important indicator of salt tolerance, particularly because transpiration is related to normal rates of CO2 uptake for photosynthesis (Harris et al., 2010). However, assessment of a plant’s transpiration rate using porometers (Meidner and Sheriff, 1976) and infra-red gas analysers (Nobel, 1991) can be difficult due to rapid changes in stomatal conductance that can occur in both space and time (Munns et al., 2006). Thus, measures of T and TUE depend on the leaf, time of day and the particular part of the leaf from which the measurements are taken. In this review, we discuss T and TUE at the whole-plant level. Methods to measure T and TUE are described in Supporting Methodologies Section 2, eqn (q).
TUE is dependent on both the genotype and the environment (Vadez et al., 2014). The genes involved in TUE remain largely unknown (Masle et al., 2005), and the effects of salinity on TUE remain, to our knowledge, largely unstudied. Carbon isotope discrimination has been used for analysis of TUE (Farquhar et al., 1982), and it has been used successfully to improve the water use efficiency of wheat (Farquhar et al., 1982; Condon et al., 2004). We note that the tissue sampled for measurements of carbon isotope discrimination must be carefully chosen to obtain fair and relevant measures of TUE and the effect of salinity on TUE.
Quantifying the effects of salinity on ion relations
Maintaining ion homeostasis can be particularly challenging for plants under saline conditions, as the accumulation of toxic ions (i.e. Na+) can perturb the plant’s ability to control accumulation of other ions. In most species, Na+ appears to accumulate to toxic levels before Cl− does; thus, we focus here on Na+, because reducing Na+ in the shoot, while maintaining K+ homeostasis, is a key component of salinity tolerance in many cereals and other crops. However, in some perennials, Cl− accumulates in the shoot and inhibits photosynthesis whereas Na+ appears to be preferentially retained in the woody roots and stems (Flowers and Yeo, 1988).
Most research on salinity estimates the amount of elemental Na and K at the whole-tissue level (leaves or roots) using techniques such as atomic absorption spectroscopy (AAS), flame photometry (FP) or inductively coupled plasma mass spectrometry (ICP-MS); at the sub-cellular level, techniques such as X-ray microanalysis or secondary ion mass spectrometry (SIMS) can be employed (for a complete review of methods, see Conn and Gilliham, 2010). It should be clarified that the abbreviation Na, rather than Na+, is used deliberately here because these techniques generally quantify the element and not the ion.
It should also be noted that the Na or K concentration is the amount of the element per unit of volume (e.g. mm) whereas content refers to the amount of the element per unit of mass (e.g. μmol g−1 dry mass). There is confusion in the plant science literature over use of the term ‘content’, and we need to standardize this as a community to be in line with international commonly accepted usage in all other fields of science. The use of the term ‘content’ in other fields is clear – it is reserved for the amount per unit mass (Tolhurst et al., 2005; Dybkaer, 2007; Fuentes-Arderiu, 2013). Crucially, the Bureau International des Poids et Mésures (BIPM), the international organization of metrology, use the term ‘content’ in this way, such as can be seen at http://goo.gl/pEqQHW. Content can be calculated as the amount per unit mass (amount-of-substance content, with SI units of moles per gram) or as the mass per unit mass (which can be termed the ‘mass fraction’). The confusion in the plant literature arises from the use of content for the total amount of a substance in a particular organ – as opposed to the concentration. However, we suggest that the community use the term amount for this (with units of moles or kg), and express it as amount per organ.
We recommend calculating Na and K concentrations (i.e. in mm), rather than Na and K content (i.e. μmol g−1 dry mass) because the latter does not account for differences in the water status of tissues (particularly if one is comparing different accessions or species), and thus differences in concentration in the aqueous phase, which is the primary factor directly affecting transport and biochemical processes, might be missed. Of course, this does not apply in tissue that has visible significant signs of senescence and desiccation.
It is also beneficial to measure the amount of Na and K in the roots of the plant, as this will indicate how much Na+ is retained in the roots. Hydroponically grown plants are particularly suited for ion analyses as soil particles do not interfere with the collection of root material and ion analysis. Care must be taken to quantify Na and K in plant roots. Roots need to be rinsed for a short time with a Ca2+ solution to remove apoplastic Na+ (e.g. as in Davenport and Tester, 2000) to prevent damage to cells. It is also good practice to adjust the osmotic potential of the rinse solution to equal that of the growth solution, to prevent cell damage and remove the need for tissues to osmotically adjust. More details on choosing and processing samples for ion analysis are provided in Supporting Methodologies Section 1.
It has been proposed that the salinity tolerance of a plant is determined not only on the basis of the leaf Na+ concentration, but also on the ability of the plant to maintain high cellular K+ levels (Shabala and Cuin, 2008). It is therefore common to present the Na/K ratio for both roots and shoots; however, this ratio is affected mainly by changes in the Na+ concentration, which are commonly proportionally much greater than changes in the K+ concentration.
Ions that accumulate during salt stress can be compartmentalized into different types of cells in a particular organ. For example, X-ray microanalysis (a semi-quantitative method that identifies relative ion locations) of salt-stressed barley leaves showed that the ion composition in vacuoles of mesophyll cells differed from that of vacuoles of epidermal cells (Leigh and Storey, 1993).
The classical method for analysing ion fluxes within plants is the pulse-chase experiment, which involves exposing plants to radioisotopes such as 22Na+, 36Cl− or 42K+ (it is risky to use 86Rb+ as tracer for the flux of K+ because its behaviour can often differ markedly from that of K+, especially when making comparisons with Na+ flux). Although technically challenging, pulse-chase techniques are powerful for studying the unidirectional fluxes of Na+ and K+ into roots and the movements of ions in plants subjected to salinity stress (Davenport et al., 2007). It is important to note that the unidirectional flux of Na+ into roots appears to be quite high, requiring measurement over very short times (on the order of 5 min); otherwise, the efflux of the radioactive tracer begins to become significant, reducing the apparent rate of influx (reviewed by Tester and Davenport, 2003). Net fluxes can also be calculated by measuring increases in tissue ion concentration using sequential harvests. In addition, the use of extracellular vibrating ion-sensitive electrodes can be very useful for measuring net fluxes of ions (Shabala et al., 2013), as can changes in bulk concentrations in either external solutions or plant tissues upon a step change in external concentrations. These approaches, however, cannot be used for measuring unidirectional fluxes, and also have some limitations because of issues with selectivity of resins used in electrodes (e.g. of Na+-selective ionophores) and also the ability to detect depletion of ions when external ion concentrations are high (such as in saline solutions).
Accumulation of compatible solutes, such as glycine betaine, proline and polyols, in the cytoplasm is required to balance the decrease in water potential occurring in the vacuole due to ion accumulation in that compartment (dos Reis et al., 2012). It is well established that compatible organic solutes increase with salt stress; however, whether a greater increase in compatible solutes correlates with increased salinity tolerance in plants remains to be shown. For barley, at least, it appears that the more salt-tolerant varieties accumulate less compatible solutes than do the more sensitive varieties (Chen et al., 2007). Concentrations of glycine betaine, proline, polyols and sugars can be quantified using techniques such as high-performance liquid chromatography (HPLC) with UV detection (Naidu, 1998; Abraham et al., 2010) or gas-liquid chromatography (GLC) methods (Holligan, 1971). In some cases, as for proline, simpler methods have been established, such as a ninhydrin-based colorimetric assay (Abraham et al., 2010). A simple method to quantify sugars is a colorimetric assay using anthrone (Yemm and Willis, 1954).
Quantifying the effects of salinity on photosynthesis
Upon salinity stress, a substantial decrease in a plant’s stomatal aperture can be observed, but the rates of photosynthesis per unit leaf area sometimes remain unchanged (Munns and Tester, 2008). Previous work using two contrasting durum wheat genotypes showed that salinity stress caused a large decrease in stomatal conductance (gs) of both genotypes (James et al., 2002). Interestingly, the efficiency of photosystem II (PSII) in the tolerant wheat accession was unaffected, while there was a decline in the quantum yield of PSII photochemistry, coinciding with leaf ageing, higher Na+ and Cl– concentrations in the leaf, and chlorophyll degradation, in the sensitive genotype (James et al., 2002). Following stomatal closure, the internal reduction of CO2 decreases the activity of several enzymes including RuBisCo (Chaves et al., 2009), thus limiting carboxylation and reducing the net photosynthetic rate. The intercellular CO2 concentration (Ci) is another parameter that has been used to estimate the effects of salinity on photosynthesis (Seemann and Critchley, 1985; Redondo-Gomez et al., 2007; Stepien and Johnson, 2009). Under salinity, the CO2 assimilation rate (as a function of Ci) was shown to be better maintained by a salt-tolerant species, Eutrema salsugineum, compared with a sensitive-species, Arabidopsis (Stepien and Johnson, 2009). However, it is difficult to differentiate cause–effect relationships between photosynthesis (source) and growth reduction (sink); also, the effects of salinity on photosynthesis can be caused by alterations in the photosynthetic metabolism, or else by secondary effects caused by oxidative stress (Chaves et al., 2009).
To understand the impact of salinity on photosynthetic responses, many studies quantify the amount of chlorophyll in the leaf (expressed, for instance, as μg Chl g−1 tissue or μg Chl cm−2 tissue) (Arnon, 1949; Hiscox and Israelstam, 1979). However, under salinity stress, leaf expansion, associated with changes in leaf anatomy (smaller and thicker leaves), is reduced, resulting in higher chloroplast density per unit leaf area, which can lead to a reduction in photosynthesis as measured on a unit chlorophyll basis (Munns and Tester, 2008). Non-invasive methods that capture photosynthetic responses include measurements by infrared gas analysers (IRGAs) and pulse amplitude-modulated (PAM) chlorophyll fluorometers. In addition, the use of soil and plant analyser development (SPAD) meters to determine the chlorophyll content can also provide an estimate of leaf damage under stress. The chlorophyll content can be estimated using the SPAD index, which is the ratio between leaf thickness (as determined by the transmission of light in the IR range) and leaf greenness (as determined by the transmission of light in the red light range). The SPAD index has been shown to decrease under salinity compared to control conditions. The extent of this decrease has been shown to vary between barley accessions, suggesting a genetic control of this effect of salinity on the SPAD index (Adem et al., 2014). It should be noted that the interpretation of SPAD meter measurements is not straightforward because salinity stress can increase leaf thickness (Longstreth and Nobel, 1979) and thus influence SPAD meter readings in a way that is independent of effects of salinity on chlorophyll content (Li et al., 2009). Therefore, careful calibration of the system, as well as the use of species-specific calibration equations, are necessary to determine chlorophyll content (for further details the reader is referred to Richardson et al., 2002). Also, SPAD meter measurements should be carried out at the same time of day to avoid variation due to diurnal changes, and they should be performed on the same leaf and the same location on the leaves of every plant to reduce effects of spatial variation. Several studies have shown the existence of genotypic differences in photosynthetic responses due to salinity (James et al., 2002, 2008; El-Hendawy et al., 2005). To study the effects of salinity on the regulation of photosynthesis, consistency in the measurements is essential. Such measurements are dependent of the time of day, which leaf is measured and the position on the leaf where the measurements are taken. Extensive replication is required to obtain a representative measurement for a particular genotype. This, in turn, reduces capacity for comparative measurements.
Thermal imaging using IR thermography has also been used as an indication of stomatal regulation in response to abiotic stress (Jones, 1999). In barley plants subjected to salinity stress, there is a strong relationship between direct measurements of stomatal conductance and leaf temperature and these differences are dependent on genotype (Sirault et al., 2009). IR thermography measurements may be most profitably used to assess the early response to salinity stress (osmotic phase), before other plant processes confound the measurements, such as the build-up of salt in the leaves, causing changes in leaf morphology, and before age-associated decreases in stomatal conductance occur (James and Sirault, 2012). IR thermography should therefore be completed on young seedlings (leaf 2–3 stage for cereals), shortly after the final desired concentration of salinity is attained (at 3–5 d) (James and Sirault, 2012).
Quantifying the effects of salinity on plant senescence
Once the plant has accumulated Na+ in the shoot and suffers from the toxic effects of Na+, the most visible symptom is a yellowing, then browning, of leaves, due to leaf senescence and death. This effect is most visible in older leaves that have had a longer time to accumulate Na+ and suffer from the effects of that accumulation. However, it is notable that the leaves of some plants are better able than others to maintain greenness and photosynthetic function for longer in the presence of high levels of Na+ in tissues. The classical way to determine leaf senescence is by using a visual scoring method, which can be used to compare different plant genotypes affected by salinity. Such scoring methods can also be used in combination with growth analyses. An example of the scoring of growth and leaf damage in rice seedlings is presented in Table 1.
Table 1.
Standard evaluation system of visible salt damage in rice at the seedling stage (Gregorio et al., 1997)
| Score | Observation |
|---|---|
| 1 | Normal growth, no leaf symptoms |
| 3 | Nearly normal growth with some leaves and tips whitish and rolled |
| 5 | Growth severely retarded with most leaves rolled and only a few elongated |
| 7 | Complete growth arrest with most of the leaves dried and some plants dead |
| 9 | Almost all plants dead or dying |
Senescence can also be estimated in an automated set-up using, for instance, high-throughput fluorescence imaging. Plants are imaged after they have been exposed to salinity for an extended period when clear symptoms of Na+ toxicity are visible. This imaging analysis allows calculation of the area affected by salt-induced senescence (SIS) (Rajendran et al., 2009; Berger et al., 2012) [Supporting Methodologies Section 2, eqn (r)].
Tissue tolerance, at the shoot level, refers to the ability of plants to maintain tissue function in the face of high accumulation of Na in older leaves, where leaf senescence is observed first (rather than in younger leaves). SIS, however, is estimated at the whole shoot level; thus, SIS is not an ideal indicator for tissue tolerance. Consequently, to estimate tissue tolerance, measurements of senescence need to be made specifically in older leaves. One way to do this is to use image analysis models to separate individual leaves and to therefore enable measurements of parameters of each single leaf, rather than just those of the whole shoot (Ward et al., 2015). In effect, a full life history of each leaf could be developed, and the effects of salinity and genetic composition on that life history could be quantified. Such a procedure would allow the progression of senescence to be observed in a specific leaf (e.g. leaf 3 in cereals) and thus provide a more accurate estimation of tissue tolerance.
Quantifying the effects of salinity on yield-related parameters
The ultimate goal of salinity tolerance research is to increase salinity tolerance in crops for them to maintain yield under adverse conditions. Given that research conducted using pots/greenhouse conditions does not provide a reliable estimation of yield responses, fieldwork needs to be undertaken to quantify yield and yield-related parameters (yield components). Supporting Methodologies Section 1 provides further details about the choice of an appropriate experimental system. Soil salinity in the field is not only determined by the concentration of Na+ and Cl−, but also other ions such as Mg2+, Ca2+ and . In the field, soil salinity is often reported as electrical conductivity (ECa), which can be determined using instruments such as electromagnetic (EM38) soil mapping devices (Corwin and Lesch, 2013). Field trials require control (low salinity) and saline plots, with a level of stress depending on the species and the available irrigation. The use of check plots with a known genotype adapted to the region is a prerequisite, as is some degree of replication and accounting for spatial variation. Moreover, field trials should be replicated over at least two years to account for heterogeneity in the field and other environmental factors. Heterogeneity in field salinity is a significant issue in field trials in dryland environments; using irrigated fields can reduce spatial heterogeneity significantly, and irrigation with fresh and brackish water can be effective, at least on sandy soils.
Although plants’ sensitivity to salinity is higher during early seedling stage and reproductive stage, crops need to maintain functions at all stages of their life cycle to increase their ability to maintain yield under high salinity. For instance, yield can also be reduced during the vegetative stage by affecting parameters such as tiller number per plant in cereals such as rice (Zeng and Shannon, 2000) or wheat (Maas et al., 1994). The harvest index (HI) has been shown to be affected by salinity (Gholizadeh et al., 2014). A plant capable of maintaining HI under stress conditions will often have a higher yield [Supporting Methodologies Section 2, eqn (s)]. The reason for the maintenance of HI under salinity is not fully understood. Plausible reasons for changes in HI may include a lower shoot biomass reduction, maintenance of tiller number (Zeng and Shannon, 2000) or earlier flowering (Saade et al., 2016). Besides HI, other parameters, such as yield and yield components including seed/fruit mass, spikelets per spike (for cereals), spike length, fertility rates in the spikes and 1000-grain mass, have also been shown to be affected by salinity stress (Gholizadeh et al., 2014; Mishra et al., 2014). Measurements of yield for crops whose harvestable parts are below ground (such as tubers or modified roots) provide their own challenges, but experts in these crops have well-developed methodologies to address this. The principle that such work should be field-based remains.
Interpreting the physiological results
After all data collection and analyses are completed, the key question then is how the data should be interpreted to address the question, ‘What are the plants telling us?’ In this review, a couple of methods of data presentation and interpretation that lead to improved data analysis are described.
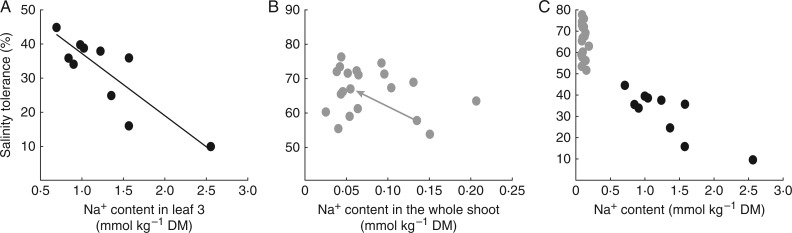
In the first example (Fig. 2), the salt tolerance index (ST) of different genotypes was plotted in relation to the Na+ content in the third leaf or whole shoot (Munns and James, 2003; Genc et al., 2007). A strong correlation between the Na+ content of leaves and ST is observed in the genotypes analysed in Fig. 2A, indicating that the Na+ content in the third leaf may be associated with salinity tolerance in these genotypes. On the other hand, no correlation is found between ST and the content of Na+ in shoots in the genotypes analysed in Fig. 2B. It is noteworthy that the lack of correlation in Fig. 2B does not necessarily mean that a particular trait (in this case the content of Na+ in shoots) does not play a part in salinity tolerance; this example illustrates that in some genotypes, the Na+ content in the shoot can contribute to salinity tolerance (Fig. 2A), while in others salinity tolerance appears to be due more to other factors (Fig. 2B). That Na+ exclusion can be important in some genotypes is shown by direct manipulations of Na+ accumulation in shoots, such as by simple genetic variation (Munns et al., 2012), or by addition of Ca2+, which reduces Na+ accumulation in shoots (Lahaye and Epstein, 1969). Although there is no obvious correlation, a decrease in Na+ content in shoots may still cause an increase in salinity tolerance, as indicated by the arrow in Fig. 2B.
Fig. 2.
Correlating the salt tolerance index (ST) with Na+ content. (A) A strong correlation is observed when plotting ST in relation to Na+ content in a number of genotypes of tetraploid wheat, indicating that the more tolerant genotypes accumulate less shoot Na+ (modified from Munns and James, 2003). (B) No correlation exists between ST and shoot Na+ content in 20 moderately stressed bread wheat varieties (modified from Genc et al., 2007). Although there is no obvious correlation, a decrease in Na+ content in shoots may still cause an increase in salinity tolerance, as indicated by the arrow. Note the different y-axes, because plants are different species of Triticum and were treated with different NaCl concentrations. Note also that values on the x-axes were obtained using different, although related, tissues. Genc et al. (2007) report a similar lack of correlation when using Na content of just the blade of leaf 3. (C) Data from A and B are plotted on the same axis. Figures used with permission of the publishers.
As shown in Fig. 2, any trait that is hypothesized to contribute to salinity tolerance (e.g. TUE, RMR or HI) may be plotted on the x-axis in relation to ST – where salinity tolerance is best measured by the ability to maintain yield in saline conditions relative to control conditions. A correlation between the trait of interest and ST in the analysed genotypes will be an indicator that the trait contributes to salinity tolerance in the plants being tested.
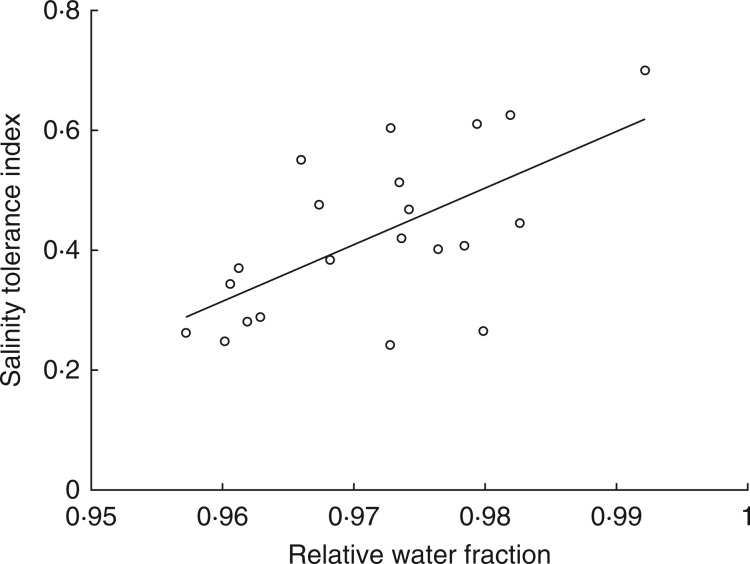
Another example is the positive correlation between ST and RWF as shown in Fig. 3. In this example, different Arabidopsis thaliana genotypes were exposed to 125 mm NaCl for 7 d (D. E. Jarvis, KAUST, unpubl. res.). Figure 3 shows the correlation between ST and RWF, indicating that this trait is associated, at least in part, with salinity tolerance. A possible explanation could be that plants that are better able to maintain their water status are more salt tolerant. Of course, these data could also be interpreted from the opposite perspective, i.e. that plants that are more salt tolerant are better able to maintain their water status. Cause and effect can often be difficult to disentangle.
Fig. 3.
Correlation between salt tolerance index (ST) and the relative water fraction (RWF) in Arabidopsis thaliana ecotypes. Plants were grown in hydroponics for 4 weeks according to Conn et al. (2013), and subjected to 7 d of salt stress after increasing salinity to 125 mm NaCl over three increments separated by 12 h each. CaCl2 was added to the medium to maintain constant Ca2+ activity. Unpublished data of Dr David E. Jarvis.
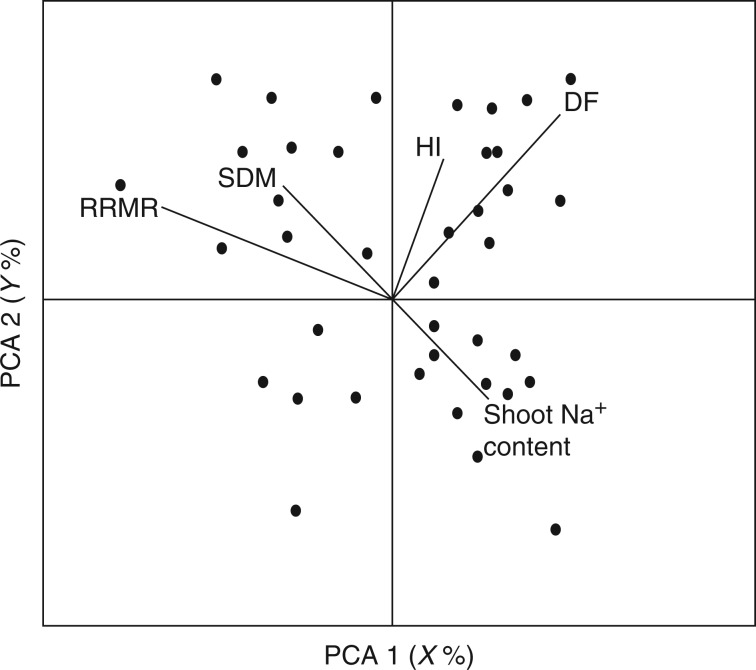
Simple correlation analyses (e.g. using pairwise regression) are often preferred, as their calculation is straightforward. However, complex and important traits might be difficult to dissect and a simple pairwise analysis may not yield satisfying results. In this case, data can also be analysed and presented using principal component analysis (PCA) or non-linear PCA. The use of PCA can provide an indication of the most important traits contributing to salinity tolerance in the materials and conditions under study. In the hypothetical example provided in Fig. 4, the distribution of the genotypes shows that PCA1 and PCA2 account for (X+Y) % of the total variability in the set of variables (traits) analysed in each genotype. In this example, PCA1 accounts for X % of the variability and it is strongly negatively correlated with the relative root mass ratio (RRMR) and, to a lesser extent, with the shoot dry mass (SDM). This is in contrast to days to flowering (DF), which is positively correlated with PCA1. DF is therefore a trait that is positively associated with salinity tolerance in this example. The figure indicates that HI, DF and shoot Na+ content are the best discriminating parameters for PCA2, explaining Y % of the variability. Moreover, traits such as DF and SDM, as well as DF and shoot Na+ content, are independent variables, whereas HI and DF are strongly correlated with each other. Under these hypothetical conditions, the shoot Na+ content and SDM are negatively correlated, suggesting that plants with lower shoot Na+ content have more shoot biomass under saline conditions.
Fig. 4.
Example of the use of principal component analysis (PCA) to assess the importance of traits contributing to salinity tolerance. The example traits used here are relative root mass ratio (RRMR), shoot dry mass (SDM), harvest index (HI), days to flowering (DF) and shoot Na+ content.
CONCLUSIONS
A proposed experimental route from design to data analysis should include classical screening of germplasm for breeding purposes, characterization of genes, and discovery of new quantitative trait loci (QTL) and, ultimately, genes using forward genetics.
Yeo et al. (1990) have suggested to study salinity tolerance not based on overall performance, but rather to look at traits that contribute to salinity tolerance (such as shoot Na+ content or plant vigour). It has been long considered that it is highly unlikely that one gene alone determines plant salinity tolerance, so a useful strategy to obtain salt-tolerant varieties is to pyramid several genes contributing to salinity tolerance (Yeo and Flowers, 1986; Yeo et al., 1990). The effects of salinity stress on plants are complex and results can be difficult to interpret if experiments are not designed carefully and if appropriate measurements are not made. To facilitate the interpretation of results from tests investigating effects of salinity on plants, we propose analyses of salinity responses not at the level of the whole plant (e.g. simply total plant biomass), but rather at the component (or trait) levels that are hypothesized to contribute to salinity tolerance. In the future, the relevance of such traits on maintenance of yield (and quality) under saline conditions can be tested in the field. The assessment of seedling survival or shoot Na+ content may not be meaningful as a predictor of salinity tolerance without other information, such as the effect of salinity on various growth parameters. In this review, we have aimed to describe methods used to measure some of the traits that may contribute to salinity tolerance. To allow useful measurements to be performed, we recommend systems and time scales that are appropriate for addressing particular biological questions.
New technologies and methods will improve quantitative comparisons within the same species by including different genotypes, and also of different species, such as Eutrema salsugineum with Arabidopsis thaliana, domesticated tomato with wild relatives such as Solanum cheesmaniae, and domesticated rice with wild relatives. Furthermore, if experimental conditions are accurately documented, comparisons of results obtained from different laboratories should be possible. Constant advances are being made to identify traits that are associated with salinity tolerance, such as measurements of HI and WUE, allowing us to get a better understanding of the complex network of traits that contribute to salinity tolerance. Undoubtedly, the rapid development of bioinformatic tools and high-throughput ‘omics’ platforms will boost the acquisition of physiological data, with consequent benefits to research on salinity tolerance in plants.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: sensitivity of rice to salt varies over the course of the life cycle [image from Singh et al. (2008), with permission of the authors and the publisher]. Table S1: range of salt concentrations advised for use with five species in hydroponic, soil-filled pots and field experiments. Movie S1: growth of wheat leaves decreases during salinity stress. Wheat plants were grown over a 13-d period. On day 9, the plant on the right side was treated with NaCl; the control plant is on the left side. Within 2 d, a clear inhibition of growth is visible in the wheat plant exposed to NaCl.
ACKNOWLEDGMENTS
The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). We thank Dr Christina Morris and Virginia Unkefer for editing the manuscript and Dr David E. Jarvis for providing us with unpublished data.
Footnotes
These authors contributed equally to this work.
LITERATURE CITED
- Abraham E, Hourton-Cabassa C, Erdei L, Szabados L. 2010. Methods for determination of proline in plants. Methods in Molecular Biology 639: 317–331. [DOI] [PubMed] [Google Scholar]
- Adem GD, Roy SJ, Zhou M, Bowman JP, Shabala S. 2014. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biology 14: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Cairns JE. 2014. Field high-throughput phenotyping: the new crop breeding frontier. Trends in Plant Science 19: 52–61. [DOI] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M. 2009. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnology Advances 27: 84–93. [DOI] [PubMed] [Google Scholar]
- Barbieri G, Vallone S, Orsini F, Paradiso R, et al. 2012. Stomatal density and metabolic determinants mediate salt stress adaptation and water use efficiency in basil (Ocimum basilicum L.). Journal of Plant Physiology 169: 1737–1746. [DOI] [PubMed] [Google Scholar]
- Berger B, de Regt B, Tester M. 2012. Trait dissection of salinity tolerance with plant phenomics. Methods in Molecular Biology (Clifton, N.J.) 913: 399–413. [DOI] [PubMed] [Google Scholar]
- Beritognolo I, Harfouche A, Brilli F, et al. 2011. Comparative study of transcriptional and physiological responses to salinity stress in two contrasting Populus alba L. genotypes. Tree Physiology 31: 1335–1355. [DOI] [PubMed] [Google Scholar]
- Boyer JS, James RA, Munns R, Condon TAG, Passioura JB. 2008. Osmotic adjustment leads to anomalously low estimates of relative water content in wheat and barley. Functional Plant Biology 35: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Bucksch A, Burridge J, York LM, et al. 2014. Image-based high-throughput field phenotyping of crop roots. Plant Physiology 166: 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MT, Knecht AC, Berger B, Brien CJ, Wang D, Walia H. 2015. Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiology 168: 1476–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. 2009. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany 103: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP, Shabala S. 2007. Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. Journal of Experimental Botany 58: 4245–4255. [DOI] [PubMed] [Google Scholar]
- Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D. 2014. What is stress? Dose–response effects in commonly used in vitro stress assays. Plant Physiology 165: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon AG, Richards RA, Rebetzke GJ, Farquhar GD. 2004. Breeding for high water-use efficiency. Journal of Experimental Botany 55: 2447–2460. [DOI] [PubMed] [Google Scholar]
- Conn S, Gilliham M. 2010. Comparative physiology of elemental distributions in plants. Annals of Botany 105: 1081–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, et al. 2013. Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin DL, Lesch SM. 2013. Protocols and guidelines for field-scale measurement of soil Salinity distribution with ECa-directed soil sampling. Journal of Environmental and Engineering Geophysics 18: 1–25. [Google Scholar]
- Courtois B, Audebert A, Dardou A, et al. 2013. Genome-wide association mapping of root traits in a japonica rice panel. PLoS One 8: e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR. 2002. Sodium-calcium interactions under salinity stress In: Läuchli A, Lüttge U, eds. Salinity: environment - plants - molecules. Dordrecht: Springer, 205–227. [Google Scholar]
- Cui DZ, Wu DD, Liu J, et al. 2015. Proteomic analysis of seedling roots of two maize inbred lines that differ significantly in the salt stress response. PLoS One 10: e0116697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M. 2000. A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiology 122: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus ANA, Tester M. 2007. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant, Cell & Environment 30: 497–507. [DOI] [PubMed] [Google Scholar]
- dos Reis SP, Lima AM, de Souza CRB. 2012. Recent molecular advances on downstream plant responses to abiotic stress. International Journal of Molecular Sciences 13: 8628–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybkaer R. 2007. The meaning of ‘concentration’. Accreditation and Quality Assurance 12: 661–663. [Google Scholar]
- El-Hendawy SE, Hu YC, Schmidhalter U. 2005. Growth, ion content, gas exchange, and water relations of wheat genotypes differing in salt tolerances. Australian Journal of Agricultural Research 56: 123–134. [Google Scholar]
- Fahlgren N, Gehan MA, Baxter I. 2015. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Current Opinion in Plant Biology 24: 93–99. [DOI] [PubMed] [Google Scholar]
- Farquhar G, O’Leary M, Berry J. 1982. On the relationship between carbon isotope discrimination and the intercellular carbon dioxide concentration in leaves. Functional Plant Biology 9: 121–137. [Google Scholar]
- Fernandez GCJ. 1992. Effective selection criteria for assessing plant stress tolerance. In: Proceedings of the international symposium on adaptation of vegetable and other food crops in temperature and water stress Taiwan, 257–270.
- Flowers TJ. 2004. Improving crop salt tolerance. Journal of Experimental Botany 55: 307–319. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Yeo AR. 1988. Ion relations of salt tolerance In: Baker D, Halls J, eds. Solute transport in plant cells and tissues. Harlow: Longman, 392–413. [Google Scholar]
- Foolad MR, Lin GY. 1997. Genetic potential for salt tolerance during germination in Lycopersicon species. Hortscience 32: 296–300. [Google Scholar]
- Fuentes-Arderiu X. 2013. Concentration and content. Biochemia Medica 23: 141–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama PBS, Tanaka K, Eneji AE, Eltayeb AE, El Siddig K. 2009. Salt-induced stress effects on biomass, photosynthetic rate, and reactive oxygen species-scavenging enzyme accumulation in common bean. Journal of Plant Nutrition 32: 837–854. [Google Scholar]
- Genc Y, McDonald GK, Tester M. 2007. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell & Environment 30: 1486–1498. [DOI] [PubMed] [Google Scholar]
- Gholizadeh A, Dehghania H, Dvorakb J. 2014. Determination of the most effective traits on wheat yield under saline stress. Agricultural Advances 3: 103–110. [Google Scholar]
- Gilroy S, Suzuki N, Miller G, et al. 2014. A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends in Plant Science 19: 623–630. [DOI] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD. 1997. Screening rice for salinity tolerance. IRRI discussion papers No. 22. Manila (Philippines): International Rice Research Institute.
- Hairmansis A, Berger B, Tester M, Roy SJ. 2014. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice (N Y) 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BN, Sadras VO, Tester M. 2010. A water-centred framework to assess the effects of salinity on the growth and yield of wheat and barley. Plant and Soil 336: 377–389. [Google Scholar]
- Hiscox JD, Israelstam GF. 1979. A method for the extraction of chlorophyll from leaf tissue without maceration. Canadian Journal of Botany 57: 1332–1334. [Google Scholar]
- Holligan PM. 1971. Routine analysis by gas-liquid chromatography of soluble carbohydrates in extracts of plant tissues 11. Review of techniques used for separation, identification and estimation of carbohydrates by gas-liquid chromatography. New Phytologist 70: 239. [Google Scholar]
- Hsiao TC, Xu LK. 2000. Sensitivity of growth of roots versus leaves to water stress: biophysical analysis and relation to water transport. Journal of Experimental Botany 51: 1595–616. [DOI] [PubMed] [Google Scholar]
- James RA, Sirault XR. 2012. Infrared thermography in plant phenotyping for salinity tolerance. Methods in Molecular Biology 913: 173–189. [DOI] [PubMed] [Google Scholar]
- James RA, Rivelli AR, Munns R, Caemmerer Sv. 2002. Factors affecting CO2 assimilation, leaf injury and growth in salt-stressed durum wheat. Functional Plant Biology 29: 1393–1403. [DOI] [PubMed] [Google Scholar]
- James RA, von Caemmerer S, Condon AGT, Zwart AB, Munns R. 2008. Genetic variation in tolerance to the osmotic stress component of salinity stress in durum wheat. Functional Plant Biology 35: 111–123. [DOI] [PubMed] [Google Scholar]
- Jones HG. 1999. Use of thermography for quantitative studies of spatial and temporal variation of stomatal conductance over leaf surfaces. Plant, Cell & Environment 22: 1043–1055. [Google Scholar]
- Kingsbury RW, Epstein E. 1984. Selection for salt-resistant spring wheat. Crop Science 24: 310–315. [Google Scholar]
- Lahaye PA, Epstein E. 1969. Salt toleration by plants: enhancement with calcium. Science 166: 395–396. [DOI] [PubMed] [Google Scholar]
- Lahaye PA, Epstein E. 1971. Calcium and salt toleration by bean plants. Physiologia Plantarum 25: 213–218. [Google Scholar]
- Leigh RA, Storey R. 1993. Intercellular compartmentation of ions in barley leaves in relation to potassium nutrition and salinity. Journal of Experimental Botany 44: 755–762. [Google Scholar]
- Li JW, Yang JP, Fei PP, et al. 2009. Responses of rice leaf thickness, SPAD readings and chlorophyll a/b ratios to different nitrogen supply rates in paddy field. Field Crops Research 114: 426–432. [Google Scholar]
- Longstreth DJ, Nobel PS. 1979. Salinity effects on leaf anatomy: consequences for photosynthesis. Plant Physiology 63: 700–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HY, Song LR, Shu YJ, et al. 2012. Comparative proteomic analysis of seedling leaves of different salt tolerant soybean genotypes. Journal of Proteomics 75: 1529–1546. [DOI] [PubMed] [Google Scholar]
- Maas EV, Lesch SM, Francois LE, Grieve CM. 1994. Tiller development in salt-stressed wheat. Crop Science 34: 1594–1603. [Google Scholar]
- Maggio A, Raimondi G, Martino A, De Pascale S. 2007. Salt stress response in tomato beyond the salinity tolerance threshold. Environmental and Experimental Botany 59: 276–282. [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD. 2005. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436: 866–70. [DOI] [PubMed] [Google Scholar]
- Meidner H, Sheriff DW. 1976. Water and plants. New York: Wiley. [Google Scholar]
- Mishra YK, Dwivedi DK, Pramila P. 2014. Consequence of salinity on biological yield, grain yield and harvest index in rice (Oryza sativa L.) cultivars. Environment and Ecology 32: 964–968. [Google Scholar]
- Munns R, James RA. 2003. Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant and Soil 253: 201–218. [Google Scholar]
- Munns R, Passioura JB. 1984. Hydraulic resistance of plants. III. Effects of NaCl in barley and lupin Australian Journal of Plant Physiology 11: 351–359. [Google Scholar]
- Munns R, Termaat A. 1986. Whole-plant responses to salinity. Australian Journal of Plant Physiology 13: 143–160. [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Munns R, Husain S, Rivelli AR, et al. 2002. Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. Plant and Soil 247: 93–105. [Google Scholar]
- Munns R, James RA, Lauchli A. 2006. Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany 57: 1025–1043. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, et al. 2012. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30: 360–364. [DOI] [PubMed] [Google Scholar]
- Naidu BP. 1998. Separation of sugars, polyols, proline analogues, and betaines in stressed plant extracts by high performance liquid chromatography and quantification by ultra violet detection. Functional Plant Biology 25: 793–800. [Google Scholar]
- Nobel PS. 1991. Physicochemical and environmental plant physiology. London: Academic Press. [Google Scholar]
- Nobel PS. 2005. Physicochemical and environmental plant physiology. Amsterdam: Elsevier Academic Press. [Google Scholar]
- Ouyang B, Yang T, Li HX, et al. 2007. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. Journal of Experimental Botany 58: 507–520. [DOI] [PubMed] [Google Scholar]
- Passioura JB, Munns R. 2000. Rapid environmental changes that affect leaf water status induce transient surges or pauses in leaf expansion rate. Australian Journal of Plant Physiology 27: 941–948. [Google Scholar]
- Poorter H, Niinemets U, Poorter L, Wright IJ, Villar R. 2009. Causes and consequences of variation in leaf mass per area (LMA): a meta-analysis. New Phytologist 182: 565–588. [DOI] [PubMed] [Google Scholar]
- Poorter H, Niinemets U, Walter A, Fiorani F, Schurr U. 2010. A method to construct dose–response curves for a wide range of environmental factors and plant traits by means of a meta-analysis of phenotypic data. Journal of Experimental Botany 61: 2043–2055. [DOI] [PubMed] [Google Scholar]
- Qadir M, Quillerou E, Nangia V, et al. 2014. Economics of salt-induced land degradation and restoration. Natural Resources Forum 38: 282–295. [Google Scholar]
- Rajendran K, Tester M, Roy SJ. 2009. Quantifying the three main components of salinity tolerance in cereals. Plant Cell & Environment 32: 237–249. [DOI] [PubMed] [Google Scholar]
- Redondo-Gomez S, Mateos-Naranjo E, Davy AJ, et al. 2007. Growth and photosynthetic responses to salinity of the salt-marsh shrub Atriplex portulacoides. Annals of Botany 100: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Álvarez R, Lobet G, Lindner H, et al. 2015. GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. eLife 4: e07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds MP, Pask AJD, Mullan DM. 2012. Physiological breeding I: interdisciplinary approaches to improve crop adaptation. Mexico, DF (Mexico: ): CIMMYT. [Google Scholar]
- Richardson AD, Duigan SP, Berlyn GP. 2002. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytologist 153: 185–194. [Google Scholar]
- Rosielle AA, Hamblin J. 1981. Theoretical aspects of selection for yield in stress and non-stress environments. Crop Science 21: 943–946. [Google Scholar]
- Roy SJ, Negrão S, Tester M. 2014. Salt resistant crop plants. Current Opinion in Biotechnology 26: 115–124. [DOI] [PubMed] [Google Scholar]
- Saade S, Maurer A, Shahid M, et al. 2016. Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Scientific Reports 6: 32586. [DOI] [PMC free article] [PubMed]
- Scholander PF, Bradstreet ED, Hemmingsen EA, Hammel HT. 1965. Sap pressure in vascular plants. Science 148: 339–346. [DOI] [PubMed] [Google Scholar]
- Seemann JR, Critchley C. 1985. Effects of salt stress on the growth, ion content, stomatal behaviour and photosynthetic capacity of a salt-sensitive species, Phaseolus vulgaris L. Planta 164: 151–162. [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA. 2008. Potassium transport and plant salt tolerance. Physiologia Plantarum 133: 651–669. [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala L, Bose J, Cuin T, Newman I. 2013. Ion flux measurements using the MIFE technique. Methods in Molecular Biology 953: 171–183. [DOI] [PubMed] [Google Scholar]
- Singh RK, Gregorio GB, Ismail AM. 2008. Breeding rice varieties with tolerance to salt stress. Journal of Indian Society of Coastal Agricultural Research 26: 16–21.
- Sirault XRR, James RA, Furbank RT. 2009. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Functional Plant Biology 36: 970–977. [DOI] [PubMed] [Google Scholar]
- Stepien P, Johnson GN. 2009. Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiology 149: 1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E, Møller IM, Murphy A. 2015. Plant physiology and development. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tester M, Davenport R. 2003. Na+ tolerance and Na+ transport in higher plants. Annals of Botany 91: 503–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This D, Comstock J, Courtois B, et al. 2010. Genetic analysis of water use efficiency in rice (Oryza sativa L.) at the leaf level. Rice 3: 72–86. [Google Scholar]
- Tolhurst TJ, Underwood AJ, Perkins RG, Chapman MG. 2005. Content versus concentration: Effects of units on measuring the biogeochemical properties of soft sediments. Estuarine, Coastal and Shelf Science 63: 665–673. [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, et al. 2013. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proceedings of the National Academy of Sciences of the United States of America 110: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. 2011. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant and Soil 341: 75–87. [Google Scholar]
- Tyree MT, Patiño S, Bennink J, Alexander J. 1995. Dynamic measurements of root hydraulic conductance using a high-pressure flowmeter in the laboratory and field. Journal of Experimental Botany 46: 83–94. [Google Scholar]
- Vadez V, Kholova J, Medina S, Kakkera A, Anderberg H. 2014. Transpiration efficiency: new insights into an old story. Journal of Experimental Botany 65: 6141–53. [DOI] [PubMed] [Google Scholar]
- Vysotskaya L, Hedley PE, Sharipova G, et al. 2010. Effect of salinity on water relations of wild barley plants differing in salt tolerance. Annals of Botany Plants 2010: plq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B, Bastian J, van den Hengel A, et al. 2015. A model-based approach to recovering the structure of a plant from images Computer Vision - ECCV 2014 Workshops. Berlin: Springer International. [Google Scholar]
- Wasson AP, Richards RA, Chatrath R, et al. 2012. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. Journal of Experimental Botany 63: 3485–3498. [DOI] [PubMed] [Google Scholar]
- Weber VS, Araus JL, Cairns JE, Sanchez C, Melchinger AE, Orsini E. 2012. Prediction of grain yield using reflectance spectra of canopy and leaves in maize plants grown under different water regimes. Field Crops Research 128: 82–90. [Google Scholar]
- Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. 2009. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. Journal of Experimental Botany 60: 4089–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. 1954. The estimation of carbohydrates in plant extracts by anthrone. Biochemical Journal 57: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AR, Flowers TJ. 1986. Salinity resistance in rice (Oryza sativa L) and a pyramiding approach to breeding varieties for saline soils. Australian Journal of Plant Physiology 13: 161–173. [Google Scholar]
- Yeo AR, Yeo ME, Flowers SA, Flowers TJ. 1990. Screening of rice (Oryza sativa L) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theoretical and Applied Genetics 79: 377–384. [DOI] [PubMed] [Google Scholar]
- Zeng L, Shannon MC. 2000. Salinity effects on seedling growth and yield components of rice. Crop Science 40: 996–1003. [Google Scholar]
- Zhao XQ, Wang WS, Zhang F, Deng JL, Li ZK, Fu BY. 2014. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS One 9: e108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.