Abstract
The integrated form of human immunodeficiency virus type 1 (HIV-1) DNA is classically considered to be the sole template for viral gene expression. However, several studies have suggested that unintegrated viral DNA species could also support transcription. To determine the contribution of the different species of HIV-1 DNA to viral expression, we first monitored intracellular levels of various HIV-1 DNA and RNA species in a single-round infection assay. We observed that, in comparison to the precocity of HIV-1 DNA synthesis, viral expression was delayed, suggesting that only the HIV-1 DNA species that persist for a sufficient period of time would be transcribed efficiently. We next evaluated the transcriptional activity of the circular forms of HIV-1 DNA bearing two long terminal repeats, since these episomes were reported to exhibit an intrinsic molecular stability. Our results support the notion that these circular species of HIV-1 DNA are naturally transcribed during HIV-1 infection, thereby participating in virus replication.
Upon entry of human immunodeficiency virus type 1 (HIV-1) into a susceptible cell, RNA genomes are reverse transcribed into DNA molecules, which can either integrate into the host cell genome (36) or accumulate in the nucleus as circular DNA bearing one or two long terminal repeats (LTR) (1, 20). Viral integrase activity is essential for a productive infection (12, 13, 22, 34, 37, 39). To account for such a requirement, it is widely considered that the integrated form of HIV-1 DNA is the sole template for transcription of viral genes whereas circular forms are thought to represent a dead end for the virus (10).
However, the question of the transcriptional activity of the unintegrated HIV-1 DNA species has been raised, and many studies have suggested that they could support transcription. Indeed, following transfection of synthetic DNA molecules mimicking the unintegrated forms of HIV-1 DNA, infectious viruses were detected (7). By using HIV-1 integrase-defective viruses, other studies have reported synthesis of viral proteins and/or particles upon infection, albeit transiently and at a low level (8, 25, 27, 32, 37, 39-41). In addition, expression of viral genes in the absence of measurable integrated viral DNA upon infection with HIV-1 integrase-competent viruses was reported, suggesting that expression from unintegrated viral DNA molecules does not require integration inhibition (40, 41). Nevertheless, the presence of undetected integrated HIV-1 DNA cannot be excluded in the latter studies. Moreover, possible transcription from illegitimately integrated HIV-1 DNA should be considered when using catalytically defective HIV-1 integrase mutants since host cell DNA recombination machinery may integrate some viral DNA molecules (16).
In this report, we addressed the question of unintegrated HIV-1 DNA expression in the setting of an infection with an integrase-competent virus. We first monitored levels of various viral DNA and RNA species in a single-round infection assay using highly sensitive quantification methods. In addition, we investigated specifically the transcriptional activity of the two-LTR circles. Our results demonstrate that these unintegrated HIV-1 DNA species are naturally transcribed during infection.
MATERIALS AND METHODS
Cells and viruses.
The CEM lymphoid cell line was cultured in RPMI 1640 containing 10% fetal calf serum. 293T and HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. HIV-1 BRU virus stocks were prepared by transfecting 293 T cells with the HIV-1LAI molecular clone pBRU-2 (29). To produce infectious HIV-1 R7 Δenv viruses, the pR7 Neo Δenv vector (gift from U. Hazan) and an expression vector encoding the glycoprotein G of vesicular stomatitis virus (VSV-G) were cotransfected into 293T cells. The pR7 Neo Δenv vector was constructed by deleting the envelope coding sequence of the HIV-1 R7 Neo genome (14), in which a neomycin resistance gene replaces HIV-1 nef. Transfection assays were carried out using the calcium phosphate method. Viral supernatants were filtered through a 0.45-μm-pore-size-filter and frozen at −80°C.
Measurement of p24gag antigen production.
HIV-1 p24gag antigen contents in viral inocula and culture supernatants were determined by enzyme-linked immunosorbent assay (Perkin-Elmer Life Sciences, Paris, France).
Viral infections.
CEM cells were infected by VSV-G-pseudotyped HIV-1 R7 viruses with 35 ng of p24gag antigen per 106 cells. One hour after infection, cells were washed in a large volume of phosphate-buffered saline (PBS), exposed to trypsin (25 μg/ml) for 1 min at 37°C, and washed once with medium and twice with PBS supplemented with RNase A (40 μg/ml). Then cells were resuspended and maintained in fresh medium. When required, cells were treated for 2 h in the presence of 100 μM zidovudine (AZT; Sigma, Saint Quentin Fallavier, France) before infection and then infected, washed, and cultured all in the presence of 100 μM AZT to inhibit reverse transcription (RT). One to 3 million cells were collected at each time point. To digest residual transfection plasmid, collected cell samples were washed in PBS and incubated with 750 to 1,500 U of DNase I (Invitrogen, Cergy Pontoise, France) in a buffer comprising 20 mM Tris-Cl (pH 8.3), 50 mM KCl, and 2 mM MgCl2 for 1 h at room temperature. Cells were washed in PBS, and dry cell pellets were frozen at −80°C until use.
HeLa cells were infected by incubation with VSV-G-pseudotyped HIV-1 R7 virus for 3 h by using 35 ng of p24gag antigen per 106 cells and then washed three times with PBS and cultured in fresh medium in the presence of 3 μM aphidicolin until 24 h postinfection. Infected cells were then either maintained again for 72 h in the presence of fresh aphidicolin or washed thoroughly and cultured without drug during the same period. At each time point, HeLa cells were collected following exposure to trypsin and treated as described above to eliminate residual transfection plasmids.
DNA and RNA isolation.
Total cell DNA was extracted with a QIAamp blood DNA minikit (QIAGEN, Courtaboeuf, France). Total cell RNA was extracted with the RNeasy minikit (QIAGEN); the procedure involved an “on-column” DNase I digestion step performed according to the manufacturer's instructions. To extract RNA from extracellular virions, 300 μl of culture supernatant was mixed with 5 × 106 uninfected cells to act as an RNA carrier and the mixture was lysed immediately by adding 600 μl of β-mercaptoethanol-containing RTL buffer, which is supplied with the RNeasy minikit (QIAGEN). Then, RNA was recovered according to the manufacturer's instructions for total cell RNA extraction.
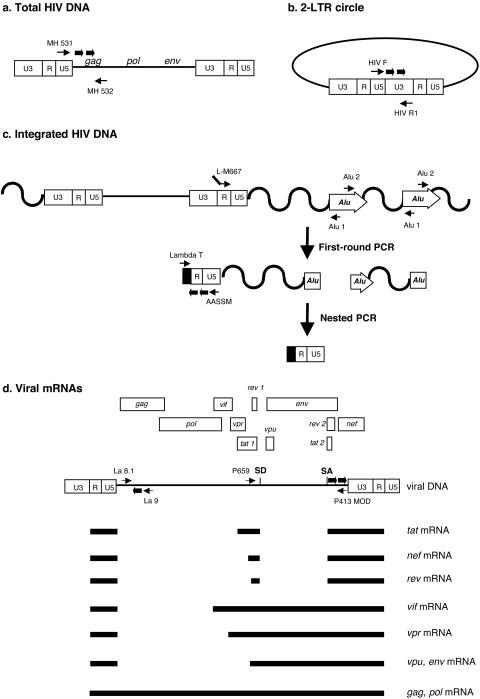
Quantification of total HIV-1 DNA, two-LTR circles, and integrated HIV-1 DNA.
Quantifications were performed by real-time PCR on a Light Cycler instrument (Roche Diagnostics, Meylan, France) using the second-derivative-maximum method provided by the Light Cycler quantification software, version 3.5 (Roche Diagnostics). Sequences of primers and probes are given in Table 1. Total HIV-1 DNA copy number was determined by quantification of viral DNA molecules that achieved the second strand transfer of RT using previously described primers (5) that annealed in the U5 region of the LTR (MH 531) and in the 5′ end of the gag gene (MH 532) (Fig. 1a). Two-LTR circles were amplified with primers spanning the LTR-LTR junction (HIV F and HIV R1) (Fig. 1b). U5-gag sequences and two-LTR junctions were amplified in duplicate from 1/50 of total cell DNA. Reaction mixtures contained 1× Light Cycler Fast Start DNA master hybridization probes (Roche Diagnostics), 4 mM MgCl2, 300 nM forward and reverse primers, and 200 nM (each) fluorogenic hybridization probe, in a final volume of 20 μl. PCR cycle conditions for two-LTR circle and total HIV-1 DNA amplifications are given in Table 2. Copy numbers of total HIV-1 DNA and two-LTR circles were determined in reference to a standard curve prepared by amplification of quantities ranging from 10 to 105 copies of cloned DNA with matching sequences.
TABLE 1.
Primer and probe sequencese
| Primer or probef | Sequenced (5′-3′) | Target |
|---|---|---|
| MH 531 | TGTGTGCCCGTCTGTTGTGT | Total HIV-1 DNA |
| MH 532 | GAGTCCTGCGTCGAGAGAGC | |
| MH FL* | CCCTCAGACCCTTTTAGTCAGTGTGGAAa | |
| MH LC* | TCTCTAGCAGTGGCGCCCGAACAGb | |
| HIV F | GTGCCCGTCTGTTGTGTGACT | 2-LTR circle and U5-U3 RNA |
| HIV R1 | ACTGGTACTAGCTTGTAGCACCATCCA | |
| HIV FL* | CCACACACAAGGCTACTTCCCTGAa | |
| HIV LC* | TGGCAGAACTACACACCAGGGCb | |
| L-M667 | ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG | Integrated HIV-1 DNA (first-round PCR) |
| Alu 1 | TCCCAGCTACTGGGGAGGCTGAGG | |
| Alu 2 | GCCTCCCAAAGTGCTGGGATTACAG | |
| Lambda T | ATGCCACGTAAGCGAAACT | Integrated HIV-1 DNA (second-round PCR) |
| AA55M | GCTAGAGATTTTCCACACTGACTAA | |
| LTR FL* | CACAACAGACGGGCACACACTACTTGAa | |
| LTR LC* | CACTCAAGGCAAGCTTTATTGAGGCb | |
| La 9 | GACGCTCTCGCACCCATCTC | Unspliced HIV-1 RNA |
| La 8.1 | CTGAAGCGCGCACGGCAA | |
| La TM* | TAGCCTCCGCTAGTCAAAATTTTTGGCGTXTc | |
| P659 | GACTCATCAAGTTTCTCTATCAAA | Multiply spliced HIV-1 RNA |
| P413MOD | AGTCTCTCAAGCGGTGGT | |
| EnvMOD FL* | GGATCCTTAGCACTTATCTGGGACGATCa | |
| EnvMOD LC* | GCGGAGCCTGTGCCTCTTCAGCTb | |
| CD3F | GGCAAGATGGTAATGAAGAAATGG | CD3ɛ RNA |
| CD3R | AGGGCATGTCAATATTACTGTGGTT | |
| CD3TM* | TGGTATTACACAGACACCATATAAAGTCTCCATCXTCTGGc | |
| CYC F | CATCTGCACTGCCAAGACTGAG | Cyclophilin A RNA |
| CYC R1 | AGGGAACAAGGAAAACATGGAA | |
| CYC FL* | CCTCCACCCCATTTGCTCGCAGTAa | |
| CYC LC* | CCTAGAATCTTTGTGCTCTCGCTGCAGTb |
Modified with fluorescein at the 3′ end.
Modified with LC red 640 dye at the 5′ end and phosphorylated at the 3′ end.
Modified with 6-carboxyfluorescein at the 5′ end and phosphorylated at the 3′ end.
X, 5-carboxytetramethylrhodamine group.
Primers and probes were purchased from TIB MOLBIOL (Berlin, Germany).
*, probe sequence.
FIG. 1.
Real-time PCR and RT-PCR strategies for the quantification of various HIV-1 DNA and RNA species. Primer and probe locations for amplification of total HIV-1 DNA (a), two-LTR circles (b), and integrated HIV-1 DNA (c) are shown. (d) Schematic representation of the genetic organization of the HIV-1 genome and of some viral transcripts. Unspliced viral RNA is quantified by amplification of a gag sequence that is present only in the full-length viral RNA species. Primers encompassing major donor (SD) and acceptor (SA) splice sites involved in the formation of tat, rev, and nef mRNA are used to quantify multiply spliced viral transcripts. Thin arrows, primers; thick arrows, probes.
TABLE 2.
PCR and RT-PCR cycle conditions
| Target | Conditions for:
|
||
|---|---|---|---|
| RT | Denaturation | PCR cycles | |
| Total HIV-1 DNA | —a | 95°C, 8 min | 95°C, 10 s, 60°C, 10 s, 72°C, 6 s for 50 cycles |
| 2-LTR circle | — | 95°C, 8 min | 95°C, 10 s, 66°C, 10 s, 72°C, 10 s for 15 cycles, then 35 cycles with the annealing temp decreased by 0.5°C/cycle to 59°C |
| Integrated HIV-1 DNA (first-round PCR) | — | 95°C, 8 min | 95°C, 10 s, 60°C, 10 s, 72°C, 170 s for 12 cycles |
| Integrated HIV-1 DNA (second-round PCR) | — | 95°C, 8 min | 95°C, 10 s, 60°C, 10 s, 72°C, 9 s for 50 cycles |
| Unspliced HIV-1 RNA | 61°C, 25 min | 95°C, 2 min | 95°C, 10 s, 60°C, 40 s for 50 cycles |
| Multiply spliced HIV-1 RNA | 61°C, 25 min | 95°C, 4 min | 95°C, 5 s, 54°C, 10 s, 72°C, 8 s for 50 cycles |
| U5-U3 HIV-1 RNA | 61°C, 25 min | 95°C, 2 min | 95°C, 5 s, 60°C, 10 s, 72°C, 10 s for 50 cycles |
| CD3ɛ RNA | 61°C, 20 min | 95°C, 2 min | 95°C, 15 s, 60°C, 40 s for 50 cycles |
| Cyclophilin A RNA | 61°C, 30 min | 95°C, 2 min | 95°C, 1 s, 56°C, 10 s, 72°C, 15 s for 50 cycles |
—, not applicable.
Integrated HIV-1 DNA quantification was performed by real-time Alu-LTR nested PCR (Fig. 1c) as described elsewhere (4). Briefly, in the first-round PCR, Alu-LTR sequences were amplified in duplicate from 1/50 of total cell DNA in a 20-μl reaction mixture comprising 1× Light Cycler Fast Start DNA master hybridization probes (Roche), 4 mM MgCl2, 100 nM forward primer L-M667, and 300 nM primers Alu 1 and Alu 2. The first-round PCR cycle conditions are described in Table 2. Nested PCR was performed on a 1/10 dilution of the first-round PCR products in a reaction mixture comprising 1× Light Cycler Fast Start DNA master hybridization probes, 4 mM MgCl2, 300 nM forward primer Lambda T, 300 nM reverse primer AA55 M, and 200 nM hybridization probes LTR FL and LTR LC (Table 1). The nested-PCR cycling profile is described in Table 2. The integrated HIV-1 DNA copy number was determined in reference to a standard curve generated by the concomitant two-stage PCR amplification of serial dilutions of an integrated HIV-1 DNA standard (4) mixed with uninfected-cell DNA to yield 50,000 cell equivalents. To control linear amplification arising from the L-M667 primer, we performed, for each sample in duplicate, a whole nested-PCR procedure omitting Alu primers in the first-round PCR. The copy number of integrated HIV-1 DNA was then adjusted by subtracting the copy number quantified in the absence of Alu primers from the copy number measured in the presence of Alu primers.
Cell equivalents in sample DNA were calculated based on the amplification of the β globin gene (two copies per diploid cell) with commercially available materials (Control kit DNA; Roche Diagnostics). Two-LTR circle, total, and integrated HIV-1 DNA levels were expressed as copy numbers per 106 cells.
Quantification of RNA species.
Quantifications of RNA species were performed by real-time one-step RT-PCR with the Light Cycler instrument. Quantifications were achieved using the second-derivative-maximum method provided by the Light Cycler quantification software, version 3.5. Amplification products were detected by using either a hybridization probe format for both multiply spliced and U5-U3 HIV-1 RNA quantifications or a TaqMan probe format for unspliced HIV-1 RNA. Unspliced viral RNA was quantified by amplification of a gag sequence, as it is only present in the full-length form of viral RNA (Fig. 1d). Primers encompassing donor and acceptor splice sites involved in the formation of tat, rev, and nef mRNA were used to quantify multiply spliced viral transcripts (Fig. 1d). Sequences of primers and probes are given in Table 1. RT-PCR experiments were performed on 1/50 of total cell RNA in a 20-μl reaction mixture containing 1× Light Cycler RNA master hybridization probes (Roche Diagnostics), 3.25 mM Mn(II) acetate, and a 500 nM primer pair and 200 nM corresponding fluorogenic probe (Table 1). RT-PCR cycle conditions are described in Table 2. Quantities of the various HIV-1 RNA species were determined in reference to a standard curve prepared by amplification of serial dilutions of in vitro-transcribed RNA (RiboMAX large-scale RNA production system; Promega, Charbonnières, France) containing matching sequences into 10 ng of bacteriophage MS2 RNA (Roche Diagnostics)/μl. Determination of cell equivalents in CEM RNA samples was performed by measuring levels of CD3ɛ mRNA, which were reported to be unaffected by HIV-1 infection, in a lymphoid cell line (38) and using previously described primers and probes (30). Determination of cell equivalents in HeLa RNA samples was performed by measuring levels of cyclophilin A mRNA. Amplifications of CD3ɛ and cyclophilin A mRNA sequences were performed by one-stepRT-PCR using reaction mixtures identical to those described above. Specific RT-PCR cycle conditions are shown in Table 2. Standard curves for CD3ɛ and cyclophilin A mRNA quantifications were generated by amplification of serial dilutions of total cell RNA from a known number of uninfected cells. Copy numbers of HIV-1 RNA species are then expressed as copies per 106 cells.
RESULTS
Viral expression is delayed relative to HIV-1 DNA synthesis, circularization, and integration.
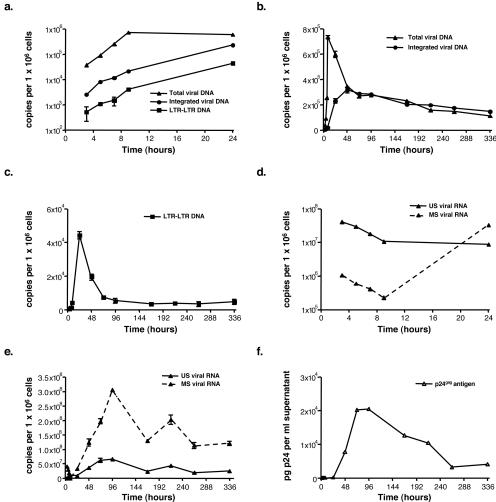
To analyze the kinetics of HIV-1 DNA and RNA synthesis over a single round of viral replication, we infected CEM cells with the VSV-G-pseudotyped HIV-1 R7 virus. Afterwards, cells were washed thoroughly and exposed to trypsin to eliminate viral particles that had not been internalized. Cells were collected at different time points over a 15-day period, and viral DNA and RNA species were quantified by real-time PCR and RT-PCR methods, respectively. Viral DNA was detected as early as 3 h postinfection (p.i.) and already comprised integrated and two-LTR circular (as shown by the presence of an LTR-LTR junction) species of HIV-1 DNA (Fig. 2a). The total HIV-1 DNA level peaked at 9 h p.i. and then decreased steeply until 72 h p.i. to match the integrated HIV-1 DNA level (Fig. 2b). To monitor HIV-1 transcription, we developed quantitative real-time RT-PCR methods to measure unspliced and multiply spliced viral RNA. By 3 h p.i., a high level of unspliced viral RNA was already detectable (Fig. 2d). However, when CEM cells were infected in the presence of AZT, a reverse transcriptase inhibitor, the level of unspliced RNA detected by 3 h p.i. was comparable to that measured in the absence of the inhibitor (data not shown). This finding indicated that these viral transcripts did not originate from an early transcription event, but rather from virus entry. Then, over the next 6 h, we observed a fourfold reduction in levels of unspliced viral RNA (Fig. 2d), which was also observed in the presence of AZT (data not shown). Interestingly, the decline of unspliced viral RNA was concomitant with the increase of total viral DNA. However, the extent of viral DNA synthesis did not account for the dramatic decrease of viral RNA levels. Therefore, our results indicate that the initial and rapid decay of viral RNA upon infection resulted primarily from a degradation process unrelated to RT. We also detected a small amount of multiply spliced viral RNA in infected cells by 3 h p.i., and this RNA species decreased up to 9 h p.i., as did unspliced HIV-1 RNA (Fig. 2d). As unspecific incorporation of a low level of multiply spliced viral RNA into particles is a common feature among retroviruses (24), we hypothesized that the detection of these RNA species early upon infection could indeed reflect the internalization of viral cores but not viral transcription. Consistent with this idea, multiply spliced RNAs were effectively detected within the inoculum and within CEM cells infected in the presence of AZT (data not shown). In conclusion, until 9 h p.i., viral RNA came from internalized particles rather than de novo transcription. Indeed, viral expression was first demonstrated by 24 h p.i., as multiply spliced RNA levels increased. In agreement with previous studies (20, 21), synthesis of multiply spliced RNA was followed by expression of the unspliced HIV-1 RNA species, which occurred by 48 h p.i. (Fig. 2e), concomitant with the release of virus-like particles in supernatant (Fig. 2f). Our results thus indicated that transcription of the viral genome is delayed in comparison to HIV-1 DNA synthesis, circularization, and integration. Since total, two-LTR circle, and integrated HIV-1 DNA levels were maximal by 9, 24, and 48 h p.i., respectively, there was also a delay between the peaks of RT, circularization, or integration and that of viral expression, which happened only at 96 h p.i. (Fig. 2e). These findings further strengthened the notion that transcription of the HIV-1 genome is a late event of the HIV-1 replication cycle, although we cannot exclude the possibility that accumulation of stable viral transcripts may have given rise to such a lag.
FIG. 2.
Kinetic analysis of HIV-1 DNA and RNA synthesis in a single-round infection assay. CEM cells were infected by the VSV-G-pseudotyped HIV-1 R7 virus, and levels of intracellular HIV-1 DNA and RNA species and that of extracellular virus-like particles were monitored over a 15-day period. (a) Dynamics of total and integrated HIV-1 DNA species, together with that of HIV-1 DNA molecules bearing an LTR-LTR junction, during the first 24 h of infection. (b and c) Synthesis and fate of total and integrated HIV-1 DNA (b) and of HIV-1 DNA molecules bearing an LTR-LTR junction (c) over the 15-day period. (d and e) Dynamics of intracellular unspliced (US) and multiply spliced (MS) HIV-1 RNA during the first 24 h p.i. (d) and during the whole culture period (e). (f) Virus-like particles released in supernatant as determined by measuring levels of p24gag antigen. Values are means ± standard errors of the means. The scales in panels a and d are logarithmic. Depicted results were obtained from a representative experiment.
However, VSV-G-mediated virus entry and/or the absence of full-length nef, vpr, and vpu genes within the HIV-1 R7 Neo Δenv genome (HIV-1 HXB2 background) may account for our preliminary observations. To investigate whether the lag between RT, circularization, or integration of viral DNA and transcription is a natural event during HIV-1 replication, we repeated the previous experiment using the wild-type HIV-1 BRU virus. Over the first 24 h upon infection, the kinetics of HIV-1 DNA and RNA synthesis were comparable to those observed with the VSV-G-pseudotyped HIV-1 R7 virus (data not shown). We thus concluded that viral transcription is a true late event of the HIV-1 replication cycle, and we therefore inferred from our data that only the HIV-1 DNA species that persists for a sufficient period of time within an infected cell would be transcribed efficiently.
Evidence for transcription from a DNA molecule comprising an LTR-LTR junction.
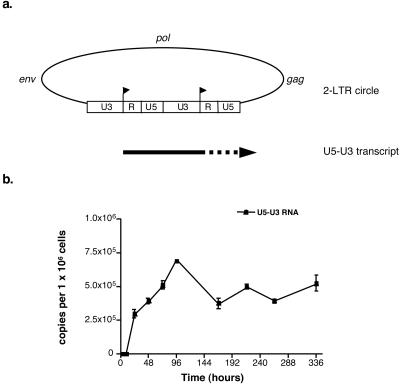
A previous report has shown that the HIV-1 DNA molecules that have not been integrated or circularized upon infection are degraded rapidly (6). Thus, we assumed from our data that unintegrated linear DNA species could not be effective templates for transcription because of their short half-lives. In contrast, the circular species of HIV-1 DNA might be transcribed efficiently, because of their intrinsic molecular stability (6). To test this hypothesis, we developed an assay for the specific detection of viral transcripts promoted from circular forms of viral DNA. We particularly focused on the two-LTR circles of HIV-1 DNA, because they comprise two tandem repeats of the viral promoter in direct orientation, in contrast to the other viral DNA species. Since transcription initiation from such circular molecules might arise from the beginning of each R region, viral transcripts initiated from the upstream start site would contain the U5-U3 sequence (Fig. 3a). To examine such a possibility, we developed a new RT-PCR method to quantify viral RNA containing a U5-U3 sequence and then we attempted to detect such viral transcripts in cell RNA extracts from CEM cells infected by the VSV-G-pseudotyped HIV-1 R7 virus. U5-U3 transcripts were effectively detected in these cells, at times when the synthesis of multiply spliced transcripts became distinguishable, i.e., 24 h p.i. (Fig. 3b). Then, the level of U5-U3 RNA varied with a trend similar to those for the other intracellular RNA species, reaching a maximum level by 96 h p.i. Surprisingly, the U5-U3 RNA level continued increasing from 24 to 96 h p.i., whereas that of the viral DNA species bearing an LTR-LTR junction decreased from 24 h p.i. to reach a steady level at 96 h p.i. (Fig. 2c). The two-LTR circles of HIV-1 DNA are stable DNA species incapable of self-replication, and, consequently, they were shown to be diluted as a function of cell division (6, 31). Therefore, cell division may account for the decrease in the two-LTR circle level observed before 96 h p.i. In contrast, we previously suggested that steady LTR-LTR junction levels from 96 h p.i. until the end of the culture might be explained by the nonspecific integration of some two-LTR circles into the host cell genome (4). Accordingly, our findings suggest that, from 96 h p.i. until the end of the culture, expression of the U5-U3 RNA depended on an integrated molecular form carrying an LTR-LTR junction, rather than on an unintegrated one.
FIG. 3.
Evidence for transcription from a DNA molecule carrying an LTR-LTR junction. (a) Location of transcription start sites within tandem LTR repeats of a two-LTR circle and representation of the U5-U3 viral transcript expressed from the upstream promoter. The U3, R, and U5 sequences within the LTR are indicated. Arrowheads, transcription start sites. (b) Kinetic analysis of U5-U3 RNA synthesis in CEM cells infected by the VSV-G-pseudotyped HIV-1 R7 virus. Values are means ± standard errors of the means. Depicted results were obtained from a representative experiment.
Unintegrated circular species of HIV-1 DNA with two tandem repeats of the LTR are templates for viral expression.
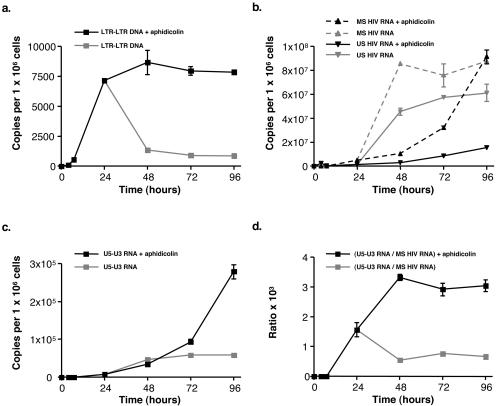
Since our findings did not preclude the possibility that unintegrated two-LTR circles might have generated U5-U3 RNA during the early times of infection, we next investigated whether true two-LTR circles may be templates for transcription. We infected cells again with the VSV-G-pseudotyped HIV-1 R7 virus, but now these cells were cultured in the presence of aphidicolin, a reversible inhibitor of cell DNA polymerases α, δ, and ɛ (18). One day p.i., at times when two-LTR circle formation was achieved, aphidicolin was either removed from the culture medium to induce cell proliferation or maintained for 72 h. This protocol allowed a comparison of U5-U3 RNA levels measured when unintegrated two-LTR circles are diluted and when they persist due to prevention of cell division. This experiment was carried out with a HeLa cell line that was found to be more resistant, in the long term, to aphidicolin-induced cell toxicity than CEM cells (data not shown). As anticipated from a previous study (6), levels of viral DNA carrying an LTR-LTR junction remained constant from 24 to 96 h p.i. when aphidicolin was maintained in the culture medium, whereas this DNA species decreased rapidly when the drug was removed (Fig. 4a). In parallel, we found that levels of multiply spliced and unspliced HIV-1 RNA in aphidicolin-treated cells were reduced, indicating that viral expression was altered in these growth-arrested cells (Fig. 4b). It has been observed previously that replication of the HIV-1 LTR DNA increases transcription from the viral promoter (26). It is thus likely that, in our experiment, the use of aphidicolin to prevent host cell DNA replication lowered transcription from the integrated species of viral DNA compared to that in proliferating cells. In contrast, we observed that levels of U5-U3 RNA were higher in aphidicolin-treated cells than in untreated cells, but only at 72 and 96 h p.i. (Fig. 4c). To take into account that viral expression in aphidicolin-treated cells differed from that in untreated ones, we normalized the quantity of U5-U3 RNA to that of the multiply spliced viral RNA species that are synthesized concurrently. We observed that ratios of U5-U3 RNA to multiply spliced viral RNA were always higher in aphidicolin-treated cells, i.e., under conditions where unintegrated two-LTR circles were not diluted (Fig. 4d). This result demonstrates that the unintegrated form of viral DNA carrying an LTR-LTR junction is a genuine template for transcription. This finding was also supported by the ratios of U5-U3 RNA to unspliced viral RNA (data not shown).
FIG. 4.
Unintegrated two-LTR circles are templates for transcription. HeLa cells were infected by the VSV-G-pseudotyped HIV-1 R7 virus and cultured in the presence of aphidicolin for 24 h. Then, aphidicolin was either maintained in the culture medium or removed to induce cell division. (a) Synthesis and fate of DNA bearing an LTR-LTR junction. (b) Dynamics of intracellular unspliced (US) and multiply spliced (MS) HIV-1 RNA. (c) Dynamics of intracellular U5-U3 HIV-1 RNA. (d) Ratios of U5-U3 RNA to multiply spliced viral RNA. Values are means ± standard errors of the means. Depicted results were obtained from a representative experiment.
DISCUSSION
In a preliminary survey, we attempted to address the contribution of the unintegrated HIV-1 DNA species in viral expression by comparing production kinetics of various HIV-1 DNA and RNA species over a single round of viral replication in a lymphoid cell line. Consistent with recent reports (2, 25), we observed that viral expression was delayed compared to the very early processes of RT, circularization, and integration of HIV-1 DNA. These results indicated that viral DNA molecules should persist for a sufficient period of time within infected cells to be transcribed efficiently. We thus inferred that the unintegrated linear DNA species may not be efficient templates for viral expression, because of their short half-lives (6). However, we considered that the circular molecules of HIV-1 DNA were putative sources for viral expression because of their intrinsic molecular stability (6, 31).
In an effort to examine the potential transcriptional activity of the circular forms of HIV-1 DNA, we took advantage of the existence of two viral promoters in direct orientation in the two-LTR circles of HIV-1 DNA. Indeed, if such molecules were transcribed, the downstream LTR would promote the synthesis of transcripts encoding viral genes (such transcripts are undistinguishable from those expressed from other viral DNA species), whereas the upstream LTR might promote the synthesis of a specific RNA, namely, U5-U3 RNA. We actually detected the latter viral RNA species in a lymphoid cell line infected with HIV-1. However, due to the probable illegitimate integration of a fraction of the two-LTR circles into the cellular genome, we could not demonstrate at first that unintegrated circles had supported their synthesis. To distinguish transcription promoted from unintegrated and integrated molecules, we used the specific feature of the unintegrated two-LTR circles, i.e., they are diluted upon cell division (6, 31). We observed that levels of U5-U3 RNA were higher in growth-arrested cells, i.e., under conditions where unintegrated two-LTR circles are not diluted, than in dividing cells. These findings demonstrated that true unintegrated two-LTR circles are templates for transcription. However, the question arose of whether two-LTR circles can support expression of viral proteins, since the upstream LTR may promote synthesis of only a noncoding RNA. Indeed, transient- or stable-expression assays with plasmids containing two direct repeats of the HIV-1 LTR in tandem, each driving the expression of truncated genes, have previously shown that, in the presence of the HIV-1 Tat protein, the two viral promoters mutually interfered (11) but that they drove equivalent RNA levels (11, 17). Thus, these studies suggested that the level of transcripts promoted from the upstream LTR reflected the transcriptional activity of both promoters. Such an assumption supports the idea that the unintegrated two-LTR circles of HIV-1 are bona fide templates for viral protein synthesis.
Interestingly, it was demonstrated recently that a catalytically defective integrase HIV-1 vector can express a high level of an internally promoted transgene, but only in growth-arrested cells (33). In addition, replication of catalytically defective HIV-1 integrase mutants has been obtained upon infection of primary monocyte-derived macrophages and a human T-cell line (25). These results were achieved by incorporating the DNA replication origin of simian virus 40 (SV40) within the viral genome and by expressing concurrently the SV40 T antigen within infected cells. Consistent with our work, these two reports suggest that dilution of unintegrated viral DNA was responsible for occulting its transcriptional activity. Interestingly, two-LTR circles were the predominant HIV-1 DNA species in these experiments. Altogether, these studies and ours strongly support the idea that two-LTR circular forms of HIV-1 DNA are naturally transcribed during HIV-1 infection, thereby participating in virus replication.
Our results do not exclude the possibility that circular species of HIV-1 DNA bearing a single LTR sequence may be also transcribed during HIV-1 infection. Indeed, these circular viral genomes exhibit a molecular stability comparable to that of the two-LTR circles (31), and they were shown to be transcribed more efficiently in transfection assays (7). In addition, they are about ninefold more abundant than two-LTR circles in infected cells (1, 5). All these findings suggest that one-LTR circles may also be suitable expression templates for the virus and that they may significantly participate in its replication.
As anticipated (25), the demonstration of a transcriptional activity supported by unintegrated HIV-1 circles may offer an alternative to gene therapy approaches using a transgene expression system based on retroviral vector integration, especially when nondividing cells are targeted. Nevertheless, such a strategy requires further investigations to establish that HIV-1 circles can direct the formation of proteins in vivo and even infectious viral particles. If this is confirmed, it would indicate that cells containing viral DNA circles would constitute an additional and stable reservoir for HIV-1, as they are found in vivo (9, 15, 19, 23, 28, 35) and as circles bearing two LTR were shown to persist in peripheral blood mononuclear cells from HIV-1-infected patients, even under the best of our currently available antiretroviral treatments (3).
Acknowledgments
A. Brussel was supported by fellowships from the Agence Nationale de Recherche sur le SIDA and from the Fondation pour la Recherche Médicale. We thank Sidaction for support.
We thank Olfert Landt (TIB MOLBIOL) for the design of primers and hybridization probes. We also thank C. Petit, O. Delelis, S. Pierre, and T. Leste-Lasserre for helpful discussions and G. Langsley and P. Charneau for reviewing the manuscript.
REFERENCES
- 1.Barbosa, P., P. Charneau, N. Dumey, and F. Clavel. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retroviruses 10:53-59. [DOI] [PubMed] [Google Scholar]
- 2.Bell, P., L. J. Montaner, and G. G. Maul. 2001. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 75:7683-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussel, A., D. Mathez, S. Broche-Pierre, R. Lancar, T. Calvez, P. Sonigo, and J. Leibowitch. 2003. Longitudinal monitoring of 2-long terminal repeat circles in peripheral blood mononuclear cells from patients with chronic HIV-1 infection. AIDS 17:645-652. [DOI] [PubMed] [Google Scholar]
- 4.Brussel, A., and P. Sonigo. 2003. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J. Virol. 77:10119-10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 6.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cara, A., A. Cereseto, F. Lori, and M. S. Reitz, Jr. 1996. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J. Biol. Chem. 271:5393-5397. [DOI] [PubMed] [Google Scholar]
- 8.Cara, A., F. Guarnaccia, M. S. Reitz, R. C. Gallo, and F. Lori. 1995. Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not T lymphocytes. Virology 208:242-248. [DOI] [PubMed] [Google Scholar]
- 9.Cara, A., J. Vargas, Jr., M. Keller, S. Jones, A. Mosoian, A. Gurtman, A. Cohen, V. Parkas, F. Wallach, E. Chusid, I. H. Gelman, and M. E. Klotman. 2002. Circular viral DNA and anomalous junction in PBMC of HIV-infected individuals with no detectable plasma HIV RNA. Virology 292:1-5. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J. M., S. H. Hugues, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 11.Eggermont, J., and N. J. Proudfoot. 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 12:2539-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Englund, G., T. S. Theodore, E. O. Freed, A. Engleman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, M., A. Trkola, B. Joos, R. Hafner, H. Joller, M. A. Muesing, D. R. Kaufman, E. Berli, B. Hirschel, R. Weber, and H. F. Gunthard. 2003. Shifts in cell-associated HIV-1 RNA but not in episomal HIV-1 DNA correlate with new cycles of HIV-1 infection in vivo. Antivir. Ther. 8:97-104. [PubMed] [Google Scholar]
- 16.Gaur, M., and A. D. Leavitt. 1998. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J. Virol. 72:4678-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greger, I. H., F. Demarchi, M. Giacca, and N. J. Proudfoot. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 26:1294-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huberman, J. A. 1981. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell 23:647-648. [DOI] [PubMed] [Google Scholar]
- 19.Jurriaans, S., A. de Ronde, J. Dekker, J. Goudsmit, and M. Cornelissen. 1992. Analysis of human immunodeficiency virus type 1 LTR-LTR junctions in peripheral blood mononuclear cells of infected individuals. J. Gen. Virol. 73:1537-1541. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klotman, M. E., S. Kim, A. Buchbinder, A. DeRossi, D. Baltimore, and F. Wong-Staal. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. USA 88:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 25.Lu, R., N. Nakajima, W. Hofmann, M. Benkirane, K. Teh-Jeang, J. Sodroski, and A. Engelman. 2004. Simian virus 40-based replication of catalytically inactive human immunodeficiency virus type 1 integrase mutants in nonpermissive T cells and monocyte-derived macrophages. J. Virol. 78:658-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahreini, P., and M. B. Mathews. 1995. Effects of the simian virus 40 origin of replication on transcription from the human immunodeficiency virus type 1 promoter. J. Virol. 69:1296-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang, S., Y. Koyanagi, S. Miles, C. Wiley, H. V. Vinters, and I. S. Chen. 1990. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature 343:85-89. [DOI] [PubMed] [Google Scholar]
- 29.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 30.Pennington, J., S. F. Garner, J. Sutherland, and L. M. Williamson. 2001. Residual subset population analysis in WBC-reduced blood components using real-time PCR quantitation of specific mRNA. Transfusion 41:1591-1600. [DOI] [PubMed] [Google Scholar]
- 31.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon, B., and I. S. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saenz, D. T., N. Loewen, M. Peretz, T. Whitwam, R. Barraza, K. G. Howell, J. M. Holmes, M. Good, and E. M. Poeschla. 2004. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willard-Gallo, K. E., F. Van de Keere, and R. Kettmann. 1990. A specific defect in CD3 gamma-chain gene transcription results in loss of T-cell receptor/CD3 expression late after human immunodeficiency virus infection of a CD4+ T-cell line. Proc. Natl. Acad. Sci. USA 87:6713-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, Y., and J. W. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]