Abstract
Toll/interleukin-1 receptor (TIR) domains in Toll-like receptors are essential for initiating and propagating the eukaryotic innate immune signaling cascade. Here, we investigate TirS, a Staphylococcus aureus TIR mimic that is part of a novel bacterial invasion mechanism. Its ectopic expression in eukaryotic cells inhibited TLR signaling, downregulating the NF-kB pathway through inhibition of TLR2, TLR4, TLR5, and TLR9. Skin lesions induced by the S. aureus knockout tirS mutant increased in a mouse model compared with wild-type and restored strains even though the tirS-mutant and wild-type strains did not differ in bacterial load. TirS also was associated with lower neutrophil and macrophage activity, confirming a central role in virulence attenuation through local inflammatory responses. TirS invariably localizes within the staphylococcal chromosomal cassettes (SCC) containing the fusC gene for fusidic acid resistance but not always carrying the mecA gene. Of note, sub-inhibitory concentration of fusidic acid increased tirS expression. Epidemiological studies identified no link between this effector and clinical presentation but showed a selective advantage with a SCCmec element with SCC fusC/tirS. Thus, two key traits determining the success and spread of bacterial infections are linked.
Author Summary
Pathogenic microbes have evolved elaborate strategies to manipulate host defenses to establish and spread in the host population. One such mechanism involves disruption of the immune signaling cascade orchestrated by the Toll-like receptors (TLRs), which sense microbial attack. TLR signaling elicits a proinflammatory response that controls immune cell recruitment to infected tissues. Here, we show that Staphylococcus aureus, an opportunistic human pathogen, expresses a host defense–like protein, TirS, that actively perturbs the initial TLR activation stage. Results with isolated human cells and mouse models show that TirS is a broad innate immune inhibitor of TLR-dependent signaling and modulates bacterial virulence, attenuating local inflammation. Moreover, the tirS gene lies near antimicrobial resistance genes for an antibiotic that enhances TirS production, shifting the balance to favor the pathogen and promote disease. Understanding mechanisms by which S. aureus modulates the immune response may lead to novel approaches for preventing and treating infection.
Introduction
The innate immune system constitutes the first line of host defense against invading microbial pathogens in multicellular organisms. Key components of the innate immune response are pattern recognition receptors, which recognize a wide range of conserved bacterial structures, collectively called pathogen-associated molecular pattern and initiate an intracellular signaling immune cascade [1]. The Toll-like receptor/interleukin (IL)-1 receptor (TLR/IL-1R) superfamily, which comprises Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs), is required for many host innate immune responses and characterized by the presence of Toll/interleukin-1 receptor (TIR) domains cytoplasmically located on each TLR [2]. The TIR domain is critical for protein–protein interactions between TLRs with the corresponding TIR-containing adaptors. These interactions activate specific transcription factors such as nuclear factor-κB (NF-κB), which regulates the expression of various inflammatory mediators [3,4]. The TIR domain therefore plays a pivotal role in signaling from these receptors, and their importance in immune regulation has made them the subject of intense study.
The TLR signaling pathway is a key target of pathogen mechanisms of host immune system evasion [4]. Indeed, microbes can target various levels of the TLR signaling pathway, from modification of pathogen-associated molecular patterns to modifications in the immune signaling cascade. A potential host evasion mechanism involving TLRs came to light with the identification of bacterial TIR homologues. The majority of studies on bacterial TIR proteins have focused on their potential role as virulence factors that directly subvert host TLR signaling. For example, TIR-like protein A (TlpA) from Salmonella enterica serovar Enteriditis reduces NF-κB activation by a TLR4, IL-1R, and MyD88-dependent pathway and modulates IL-1β secretion during infection [5]. TcpC in the uropathogenic Escherichia coli CFT073 and Btp1/BtpA/TcpB in Brucella species suppress TLR2- and TLR4-mediated activation of NF-κB by targeting MyD88 [6,7]. A second TIR-containing protein in Brucella ssp. (BtpB) was reported to be a potent inhibitor of TLR signaling, probably via MyD88 as well [8].
The presence of a putative TIR–domain–containing protein in Staphylococcus aureus was suggested through a data search analysis [5] before being recently confirmed [9]. S. aureus is an important human pathogen that causes a wide variety of community and healthcare-associated infections [10]. This bacterium has a proven ability to adapt to the selective pressure of antibiotics. S. aureus was initially methicillin-sensitive (MSSA) but isolates resistant to this antibiotic were identified soon after its introduction (MRSA, or methicillin-resistant S. aureus) [11]. S. aureus becomes resistant to methicillin mainly by the acquisition of the methicillin-resistant gene mecA, which occurred first in hospital settings and now takes place in the community and in livestock [12,13]. The mecA gene is carried on a particular class of mobile genetic elements prevalent in staphylococci, the staphylococcal chromosomal cassette (SCC), designated as SCCmec [14]. Askarian et al. [9] characterized the TIR domain protein TirS in the SCC476 element of the methicillin-susceptible S. aureus strain MSSA476. SCC476 is integrated at the same site on the chromosome as SCCmec elements in MRSA [15]. TirS interferes with the TLR2-induced MAPK and NF-κB signaling pathway and enhances bacterial survival within the host [9].
In the present work, we report that TirS is spread among 12% of MRSA and MSSA strains. In an attempt to describe the genetic context of tirS (for staphylococcal TIR gene) in S. aureus, we fully sequenced the SCC element of representative bacterial strains. In all MRSA and MSSA lineages, the tirS gene was invariably located within this mobile genetic element and co-located with the fusC and mecA (for the MRSA strains) antibiotic resistance genes. Interestingly, our results show that sub-inhibitory concentration of fusidic acid induced overexpression of tirS. We also confirm previous findings that tirS expression induces a negative regulation of the TLR signaling pathway. Our results with a mouse model of skin infection support that TirS modulates bacterial virulence through attenuation of host inflammatory responses during infection. This work is the first description of a TIR homolog protein carried by a mobile genetic element conferring resistance to antibiotics, suggesting a potential selective advantage. Indeed, these features may contribute to the ability of S. aureus to survive and establish a critical population size.
Results
tirS distribution and molecular epidemiology in S. aureus lineages
To assess the prevalence of tirS in various MRSA and MSSA lineages, a series of 226 well-characterized clinical isolates from more than 27 clonal complexes (CCs) or sequence types were subjected to tirS-specific PCR. Among the 226 strains examined, 28 (12.4%) yielded positive tirS amplification (Table 1). tirS was detected in MRSA and MSSA strains belonging to only 3 CCs: CC1, CC5, and CC8. In detail, the tirS gene was detected in 18/18 MRSA strains, CC5 Geraldine clone; 1/6 MRSA strains, CC5 pediatric clone; 1/2 MRSA strains, CC1; 6/9 MSSA strains, CC1; and 2/10 MSSA strains, CC8. Of further interest was our finding of a perfect association between tirS gene amplification and the MRSA Geraldine clone.
Table 1. Distribution of tirS gene among MSSA and MRSA clinical strains.
| CC/ST ab | SCCmec bc | Clone name b | No. of strains tested | No. of positive PCR tests for tirS /no. of strains tested (%)d |
|---|---|---|---|---|
| CC1 | - | NA | 9 | 6/9 (67) |
| IV | USA400 | 2 | 0/2 (0) | |
| IV | WA MRSA-1/57 | 2 | 0/2 (0) | |
| V | Bengal Bay/WA MRSA-60 | 2 | 0/2 (0) | |
| V | Other | 2 | 1/2 (50) * | |
| CC5 | - | NA | 9 | 0/9 (0) |
| I | Geraldine | 18 | 18/18 (100) * | |
| IV | Pediatric | 6 | 1/6 (16) * | |
| VI | New pediatric | 2 | 0/2 (0) | |
| V | WA MRSA-11/80 | 2 | 0/2 (0) | |
| II | New York Japan | 1 | 0/1 (0) | |
| II | EMRSA-3/Rhine-Hesse | 3 | 0/3 (0) | |
| CC8 | - | NA | 6 | 2/6 (40) |
| I | North German/Iberian | 3 | 0/3 (0) | |
| I | Ancestral | 2 | 0/2 (0) | |
| III | Vienna/Hungarian/Brazilian | 2 | 0/2 (0) | |
| IV | Lyon /EMRSA-2 | 6 | 0/6 (0) | |
| IV | EMRSA-14/WA MRSA-5 | 2 | 0/2 (0) | |
| IV | USA300 | 4 | 0/4 (0) | |
| IV | Other | 2 | 0/2 (0) | |
| IV | MRSA-44 | 2 | 0/2 (0) | |
| IV | USA700 | 2 | 0/2 (0) | |
| V | MRSA-91 | 1 | 0/1 (0) | |
| CC6 | IV | WA MRSA 51 | 2 | 0/2 (0) |
| CC9 | - | NA | 1 | 0/1 (0) |
| CC10 | - | NA | 1 | 0/1 (0) |
| CC12 | - | NA | 3 | 0/3 (0) |
| CC15 | - | NA | 5 | 0/5 (0) |
| CC20 | - | NA | 2 | 0/2 (0) |
| CC22 | - | NA | 8 | 0/8 (0) |
| IV | EMRSA-15/Barnim/Middle Eastern | 5 | 0/5 (0) | |
| CC30 | - | NA | 20 | 0/20 (0) |
| IV | Southwest pacific | 3 | 0/3 (0) | |
| ST34 | - | NA | 4 | 0/4 (0) |
| ST36 | II | EMRSA-16 | 2 | 0/2 (0) |
| CC45 | - | NA | 8 | 0/8 (0) |
| IV | Berlin | 2 | 0/2 (0) | |
| IV | Other | 2 | 0/2 (0) | |
| CC59 | - | NA | 3 | 0/3 (0) |
| IV | USA1000 | 2 | 0/2 (0) | |
| V | Taiwan | 2 | 0/2 (0) | |
| V | Other | 1 | 0/1 (0) | |
| CC80 | IV | European | 4 | 0/4 (0) |
| CC88 | IV | WA MRSA-2 | 2 | 0/2 (0) |
| V | Other | 2 | 0/2 (0) | |
| ST93 | IV | Queensland Clone | 2 | 0/2 (0) |
| CC97 | - | NA | 5 | 0/5 (0) |
| CC121 | - | NA | 5 | 0/5 (0) |
| CC130 | XI | MRSA-XI | 2 | 0/2 (0) |
| CC152 | - | NA | 5 | 0/5 (0) |
| CC182 | - | NA | 3 | 0/3 (0) |
| ST188 | - | NA | 2 | 0/2 (0) |
| ST398 | - | NA | 9 | 0/9 (0) |
| IV | LA-MRSA | 4 | 0/4 (0) | |
| ST1755 | - | mecC+ | 2 | 0/2 (0) |
| Other | - | NA | 13 | 0/13 (0) |
a CC: clonal complex; ST: sequence type
b CC/ST, SCCmec, and MRSA clone name were identified using the identibac S. aureus Genotyping Kit.
c—: absence of SCCmec
d tirS gene detected by specific PCR as described in the Material and Methods
* SCCmec element sequencing
We next examined the molecular epidemiology of tirS in human staphylococcal infections. S. aureus strains were isolated from clinical specimens of individuals presenting skin and soft tissue infections, community-acquired pneumonia, bacteremia, infective endocarditis, or nasal colonization (asymptomatic bacterial carriage). No significant association was detected between specific disease and the presence of tirS. Among the 28 tirS-positive clinical samples, 10 were isolated from healthy patients (asymptomatic portage; 36%), 8 from patients with cutaneous infection (29%), 5 from patients with pneumonia (18%), 3 from patients with osteomyelitis (11%), 1 from a patient with bacteremia (4%), and 1 from a patient with infective endocarditis (4%).
Genomic context of the tirS gene in MRSA and MSSA
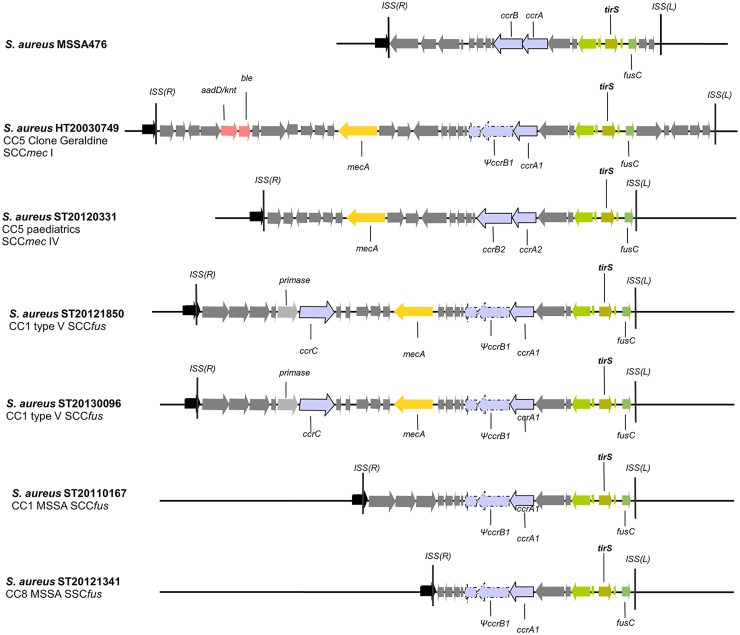
Because the tirS gene was previously described on a staphylococcal chromosomal cassette SCC476 [9], we performed whole-genome shotgun sequencing of six representative MRSA and MSSA strains positive for the presence of the tirS gene. These included four MRSA lineages: the prototype Geraldine clone strain HT20030749 (CC5; SCCmec I), strain ST20120331 (CC5, SCCmec IV), and strains ST20121850 and ST20130096 (CC1; SCCmec V fusC+) (ENA database project study accession number PRJEB12840; sample accessions ERS1070204 to ERS1070207). Two MSSA strains tirS-positive were also added to the study: strain ST20110167 (CC1) and strain ST20121341 (CC8) (sample accessions ERS1434451 and ERS1434452). As reference to the comparison with our sequenced strains, we used the genome of strain MSSA476, in which tirS was recently described [9]. Whole-genome alignment and search for the site-specific insertion sequences (ISS) typical of SCC-like cassette insertions showed that tirS is invariably present within the SCC element of all six MRSA and MSSA genomes analyzed (Fig 1). Moreover, this gene is found in a highly conserved region within the J1 region (between the ccr complex and the 5' ISS(L)), consisting of five open reading frames (ORFs) that include the tirS gene but also the fusC gene, responsible for resistance to fusidic acid. This region is present as well in MRSA as in the MSSA strains analyzed (Fig 1).
Fig 1. Characterization of the genomic context of the tirS region in S. aureus MRSA and MSSA strains.
Localization of the tirS gene and surrounding context in 4 tirS-positive MRSA strains and 2 tirS-positive MSSA strains. For all 6 strains, the tirS gene was found within the SCC element. Schematic diagrams represent the genomic comparison of the 6 SCC regions of both MRSA and MSSA strains used in our study and the SCC476 element of strain MSSA476 for which the tirS gene was first described [9]. Predicted ORFs are marked in the direction of transcription as arrows. The highly conserved tirS region, consisting of the tirS gene and 4 surrounding genes among which we find the fusC gene (fusidic acid resistance), is represented by green arrows. Light rose arrows represent genes coding for antibiotic resistance, orange arrows represent the mecA gene conferring resistance to beta-lactams and blue-gray arrows represent the site specific recombinases (ccrAB or ccrC) of SCC elements. The SCC elements ISS(L) and ISS(L)) are also represented as black vertical bars. Abbreviations: ccrAB = cassette chromosome recombinase A and B; fusC = fusidic acid resistance gene; mecA = methicillin resistance gene; ccrC: cassette chromosome recombinase C; ble = bleomycin resistance gene; aaD/knt = kanamycin nucleotidyltransferase (resistance gene).
Furthermore, BLASTn searches using the tirS and its surrounding four ORFs (“tirS region”), as annotated in the strain MSS476 genome (GenBank: BX571857.1), against the GenBank nucleotide collection (nr/nt) revealed the presence of the tirS region in 14 MRSAs, 2 other MSSAs, and a methicillin-sensitive Staphylococcus hominis strain. Unexpectedly, in all 17 staphylococci, the tirS region was conserved and located in SCC elements in the vicinity of the site-specific recombinase of type ccrAB. For 15 of the S. aureus strains in which the tirS region was detected, a whole-genome shotgun sequence was available, and we observed that they belonged to four distinct clonal complexes: CC1 (2 MRSA and 1 MSSA), CC5 (7 MRSA), CC8 (1 MSSA), and CC22 (4 MRSA).
TirS production
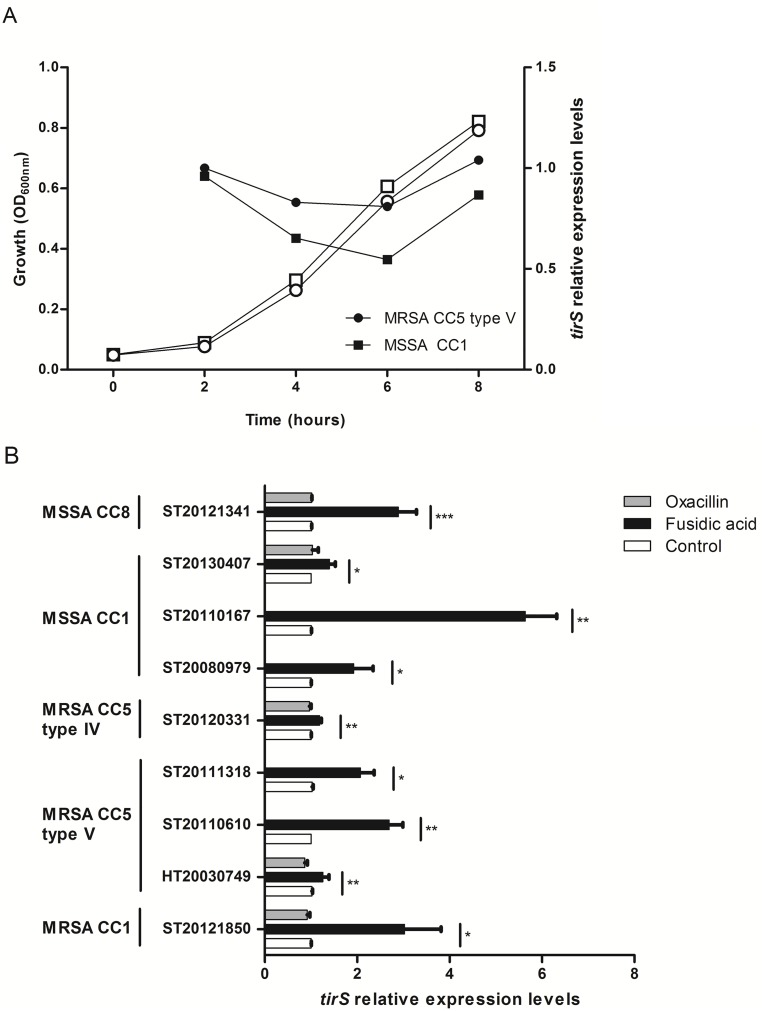
To investigate TirS production by S. aureus, tirS expression was examined by quantitative real-time reverse transcription PCR (RT-qPCR) during bacterial growth in one MRSA (clone Geraldine, CC5) and one MSSA (CC1) tirS-positive strains. The analysis of the tirS expression kinetics showed stable expression of tirS during growth with a marginal increase at the end of the exponential phase (Fig 2A). The difference in tirS expression and the standardization gene hu was generally around seven cycles.
Fig 2. Impact of bacterial growth and antibiotic on tirS expression.
(A) The expression of tirS was quantified by RT-qPCR and normalized to the level of hu expression from total RNA extracts prepared from bacterial cultures at 2, 4, 6, and 8 h. MHB medium condition at 2 h was used as a reference. The figure shows an average of 2 independent experiments. White symbols represent bacterial growth, black symbols represent tirS expression. (B) Expression of tirS on 2-h bacterial cultures and 30 min of antibiotic treatment. Bacterial strains without stress were used as reference (relative quantity = 1) to estimate the relative quantity of tirS mRNA. White bars correspond to negative control (no antibiotic), grey bars to bacteria exposed to oxacillin, and black bars to bacteria exposed to fusidic acid. Data represent mean ± SEM of three to six independent assays. * p < 0.05; ** p < 0.01; *** p < 0.001.
Because tirS is located in the SCC element, we investigated the effect on tirS expression of adding a sub-minimum inhibitory concentration (sub-MIC) of oxacillin (1/2 MIC) and fusidic acid (1/4 MIC) during the exponential phase of S. aureus growth in 9 strains. Addition of fusidic acid appeared to significantly modulate tirS expression both in MSSA and MRSA strains (Fig 2B). tirS was upregulated up to six-fold with fusidic acid compared to its expression without antibiotic stress and level of induction by fusidic acid was strain dependent. In contrast, no difference in levels of expression of tirS was observed with exposure to sub-MIC of oxacillin.
TirS interferes with TLR signaling
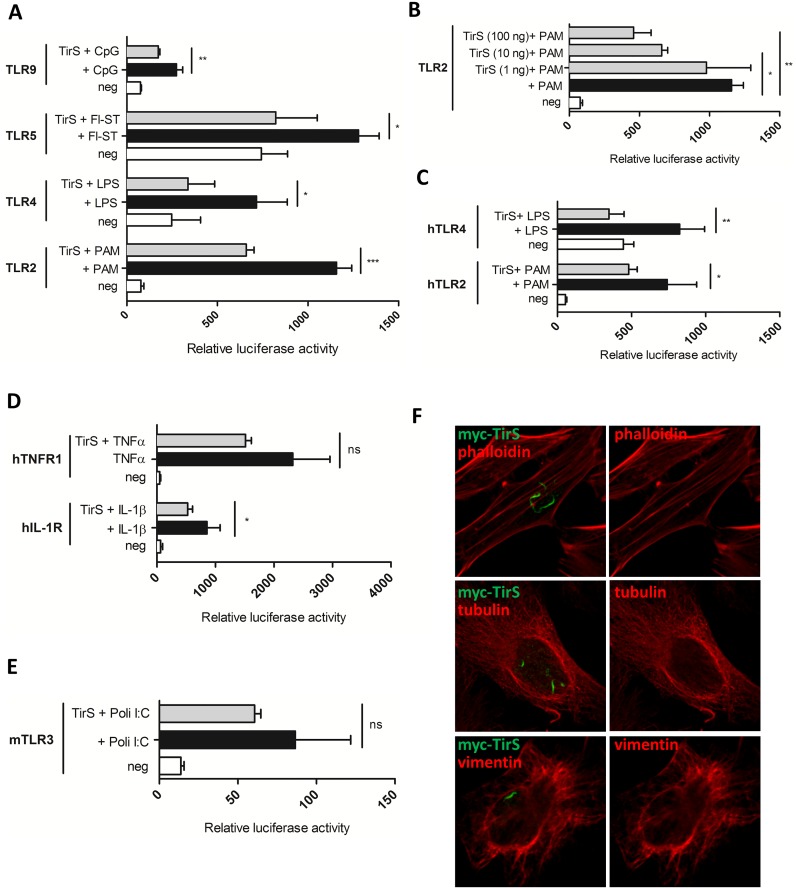
TirS belongs to the family of bacterial TLRs [9]. We therefore investigated the ability of TirS to specifically interfere with TLR signaling using an in vitro NF-κB-dependent luciferase reporter system. Although a variety of TLR receptors have been described, in humans the most relevant for recognition of bacterial molecules are TLR2, TLR4, TLR5, and TLR9. Ectopic expression of tirS in HEK293T cells transfected with the luciferase reporter vector and various TLRs resulted in reduced TLR2 activation and to a lesser extent, reduced activation of TLR9, TLR4, and TLR5 (Fig 3A). As a control, we confirmed that this inhibitory effect of TirS on murine TLR2 was dose-dependent by carrying out the transfections with increasing amounts of the expression vector encoding TirS (Fig 3B). Inhibition of human TLR2 and TLR4 pathways following addition of the appropriate ligands was also observed (Fig 3C). These results suggest that TirS may, at least partly, target a common molecule between these pathways such as the adaptor molecule MyD88. Consistently, reduction of IL-1R following IL-1β stimulation was also observed in the presence of TirS (Fig 3D). In contrast, although much lower levels of activation can be obtained for TLR3, which is independent of MyD88, we did not observe any significant effect of TirS on this pathway (Fig 3E). This was also the case for endogenous TNF receptor (TNFR) (Fig 3D). Overall, these results are in agreement with data from Askarian et al. [9] that previously described TirS inhibition of TLR2, MyD88 and TIRAP dependent pathways in vitro.
Fig 3. TirS interferes with TLR and IL-1R signaling.
(A) HEK293T cells were transiently transfected for 24 h with the luciferase reporter vector and murine TLR2, TLR4, TLR5, or TLR9 in the presence or absence of TirS (50 ng). Cells were then stimulated with the appropriate ligand (PAM, LPS, Fl-ST, and CpG) for 6 h before measurement of luciferase activity. White bars correspond to negative control, black bars to cells stimulated with the appropriate ligand, and gray bars to cells transfected with TirS and stimulated with the ligand. Data represent the means ± SEM of the relative luciferase activity and were obtained from duplicates of 3 independent experiments. (B) Luciferase activity of murine TLR2 following transfection with increasing amounts of vector encoding tirS (1, 50, 100 ng) in order to obtain different levels of expression and for (C) human TLR2 and TLR4. (D) Effect of TirS on TNFR and IL-1R activation following TNF-α or IL-1β stimulation, respectively and (E) TLR3 following stimulation with poli:IC. (F) HeLa cells were transfected with myc-TirS for 10 h and labeled for myc (green) and different components of the cytoskeleton: actin (top panels, phalloidin), microtubules (medium panels, tubulin), or intermediate filaments (bottom panels, vimentin). ns: not significant, * p < 0.05, ** p < 0.01, and *** p < 0.001.
In addition to its ability to interfere with TLR signaling, the bacterial TIR effector protein from Brucella BtpA targets and modulates microtubules [16] through a WxxxE motif [17] as well as the recently identified TIR protein from Bacillus anthracis referred to as BaTcp [18]. Since the WxxxE is also present in TirS, we investigated the intracellular localization of ectopically expressed TirS by confocal microscopy on HeLa cells transfected with myc-TirS and indirect immuno-fluorescence staining with anti-myc antibodies. We found that myc-tagged TirS accumulated in filament-like structures of irregular shapes within the host cytosol (Fig 3F). Similar results were observed for GFP-TirS, suggesting that this localization was not dependent on the tag (S1 Fig). In addition, TirS showed no co-localization with cytoskeleton components as observed after labeling with either phalloidin for actin, tubulin for microtubules, or vimentin for intermediate filaments (Fig 3F), suggesting a different targeting than previously described for BtpA and BaTdp.
TirS modulates S. aureus virulence
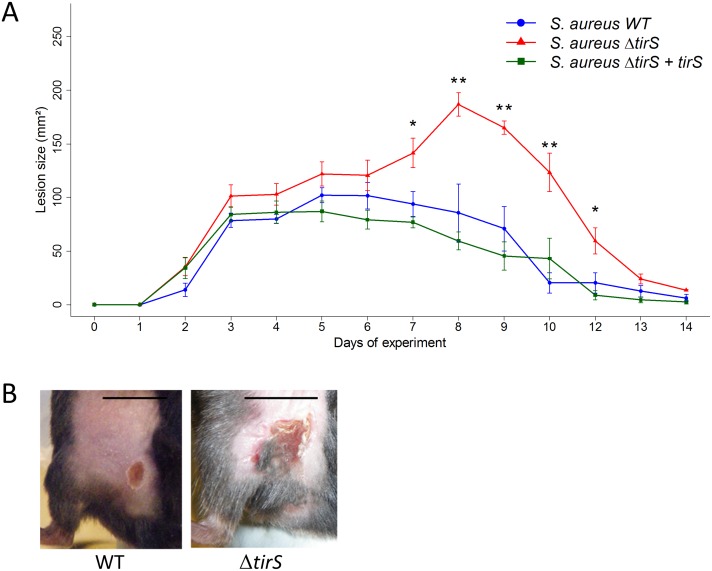
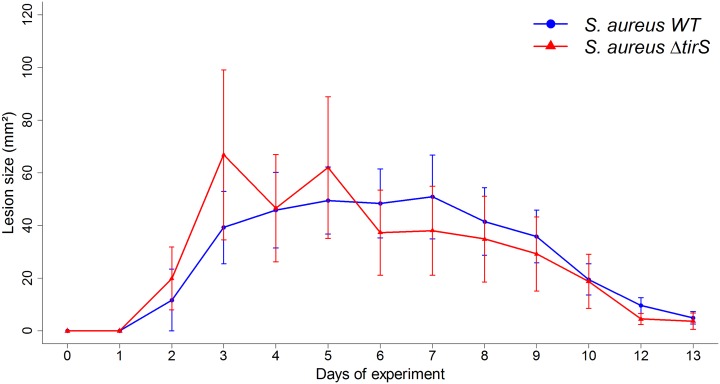
To further investigate the role of TirS during infection, we carried out in vivo studies in mice. Because S. aureus is the leading cause of skin infection, we developed a subcutaneous model of infection in mice with the inoculation of S. aureus clone Geraldine-wild type (WT) strain, a deleted for the tirS gene (ΔtirS) strain, or a ΔtirS strain restored for tirS gene in a chromosomic position (ΔtirS + tirS). First, we confirmed that bacterial growth was not affected by genetic manipulation of TirS by assessing growth in two different media (brain–heart infusion (BHI), tryptic soy broth (TSB)) (S2 Fig). Then, wild-type (WT) C57Bl/6 mice were inoculated with the three strains. Cutaneous infection resulted in the development of visible lesions by day 1 that healed by day 14 regardless of the strain (Fig 4A and 4B). As control, injection of sterile phosphate buffered saline (PBS) did not induce any skin lesions in mice. Infection with ΔtirS strain resulted in larger lesions (~2.5-fold) compared with the WT strain (p < 0.01). These differences appeared around day 6 and persisted for about 4 days before being resolved at the same time for both strains. Similar lesion sizes were observed in mice infected with the S.aureus restored strain compared with the WT strain (p = 0.9), confirming the role of TirS on the observed results.
Fig 4. S. aureus deleted for tirS induced larger skin lesions in WT C57Bl/6 mice.
(A) Data are presented as mean total lesion size (mm2) ± SEM and are representative of 3 independent experiments with at least 4 mice/group. * p < 0.05; ** p < 0.01 (B) Photographs of representative lesions at day 7 after staphylococcal infection. Black bars indicate 10 mm. Abbreviations: WT = wild-type; ΔtirS = deleted for the tirS gene; ΔtirS + tirS = ΔtirS strain chromosomally restored for tirS gene.
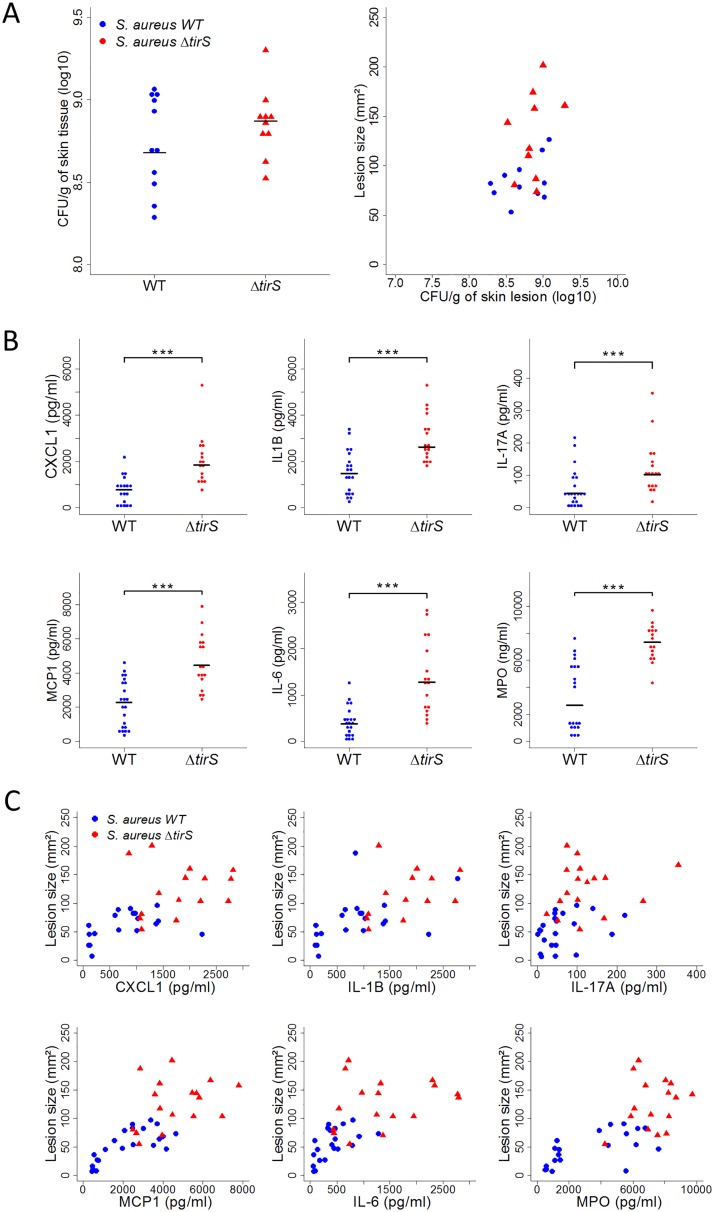
To better understand the factors that might be important in determining the size of skin lesions in the mouse model, animals were sacrificed between 5 and 9 days after infection induced by MRSA clone Geraldine WT and ΔtirS strains to the sample lesion. First, we enumerated the bacterial load in the skin lesion. No significant differences in colony-forming unit (CFU) counts between the strains were observed (Fig 5A). Moreover, lesion size was not correlated with bacterial burden in either the WT or Δ-tirS strain (S. aureus WT: r2 = 0.39; p = 0.2, S. aureus ΔtirS: r2 = 0.35; p = 0.2) (Fig 5B). This finding raised the possibility that the inflammatory response was more important in determining lesion severity, as assessed by lesion size, than bacterial burden in the lesions.
Fig 5. TirS induces reduced inflammation despite similar bacterial counts in skin lesions.
(A) Bacterial burden in skin lesions of mice infected with S. aureus WT (n = 11) or S. aureus ΔtirS (n = 10) and correlation between bacterial burden and lesion size. Data were obtained 5 and 7 days after inoculation from 2 independent experiments. (B) Levels of IL-1β, IL-6, IL-17, CXCL1, MCP1 (pg/ml), and myeloperoxidase (MPO) activity (ng/ml) from lesional skin at 7, 8, and 9 days after inoculation. Data were obtained from 16 to 22 mice infected with S. aureus WT or S. aureus ΔtirS from 4 independent experiments (all data shown). (C) Correlation between levels of IL-1β, IL-6, IL-17, CXCL1, MCP1, MPO, and lesion size from lesional skin at 7, 8, and 9 days after inoculation. Data were obtained from 16 to 22 mice infected with S. aureus WT or S. aureus ΔtirS from 4 independent experiments (all data shown). Each symbol represents an animal and the median values are marked by horizontal bold lines. Abbreviations: WT = wild-type; ΔtirS = deleted for the tirS gene. *** p < 0.001.
To follow up on these observations, we sought to measure neutrophil and macrophage activity with the quantification of levels of myeloperoxidase (MPO) and a panel of inflammatory cytokines in the skin lesions of mice infected with MRSA clone Geraldine WT and ΔtirS strains. Interestingly, the levels of MPO and IL-1β, IL-6, IL-17, CXCL1, and MCP1 were significantly lower in the murine skin lesions developed after the WT strain inoculation compared with the ΔtirS strain (Fig 5B). Correlation between skin lesion size and the levels of MPO, IL-1β, IL-6, IL-17, CXCL1, and MCP1 was statistically confirmed for each of these inflammatory markers (MPO: r2 = 0.71, p < 0.001; CXL1: r2 = 0.55, p < 0.001; IL-1β: r2 = 0.61, p < 0.001; IL-17A: r2 = 0.45, p < 0.01; MCP1: r2 = 0.74, p < 0.001; IL-6: r2 = 0.64, p < 0.001) (Fig 5C). Therefore, the smaller lesions observed after experimental inoculation with the bacterial WT strain were associated with lower levels of chemokines and less inflammation but not with decreased bacterial burden. These data support the notion that the local inflammatory response is a key determinant of lesion size. These results are consistent with a role for TirS in the control of the inflammatory response during S. aureus infection in vivo.
Finally, to evaluate the contribution of the TIR-containing adaptor protein MyD88, a key component of the TLR signaling pathway implicated in proinflammatory mechanisms [19], MyD88-deficient mice from the C57BL/6 genetic background were subcutaneously inoculated with MRSA clone Geraldine WT and ΔtirS strains. An identical infection protocol and bacterial concentration were used as in WT mice. As control, injection of sterile PBS did not induce any skin lesions in mice. Interestingly, there was no significant difference in lesion size between MyD88-deficient mice infected with the WT or ΔtirS S. aureus strain (Fig 6), suggesting that MyD88 might play a role in virulence of the MRSA clone Geraldine used. These results confirm in vitro previously reported data showing that TirS inhibits signaling in a MyD88-dependent manner [9].
Fig 6. S. aureus deleted for tirS induced similar skin lesion size in MyD88-deficient mice.
Data are presented as mean total lesion size (mm2) ± SEM and are representative of 2 independent experiments with at least 4 mice/group. Abbreviations: WT = wild-type; ΔtirS = deleted for the tirS gene.
Discussion
Bacterial strategies for innate immune evasion involve manipulation of the TLR signaling by TIR homologues such as TirS for S. aureus [20]. In this work we report the localization of the tirS in different SCC elements and its role in the control of the inflammatory response during S. aureus infection. Using a mouse model of S. aureus skin infection, we evaluated the role of TirS on S. aureus virulence. We show that S. aureus Geraldine deleted for the tirS gene exhibited superior virulence compared to the WT strain, as attested by the size of the skin lesion. Of note, bacterial counts in the skin lesions did not differ between the mutant and the WT strains and did not correlate with clinical severity (i.e., lesion size). This finding suggests that bacterial burden may not be the primary driver of lesion severity and argues that lesion severity may be due, at least in part, to the associated inflammatory response. In support of this hypothesis, we found a correlation between lesion size and levels of the proinflammatory cytokines, as well as neutrophil activity (as assessed by MPO levels). These findings are consistent with previous studies, which underscores that the severity of skin infection is often driven by the inflammatory response to the invading pathogen as much or more than by the direct effects of the pathogen itself [21–23]. This inference suggests that the attenuation of skin inflammation observed with S. aureus WT strain, compared to its tirS mutant, was driven by modulation of the inflammation resulting from TirS action. Such a conclusion is concordant with the fact that the ectopic expression of TirS in eukaryotic cells appeared to temper stimuli-induced TLR2-, TLR4-, TLR5-, and TLR9-mediated NF-kB activation. Accordingly, a previous and independent work has reported a negative interference of TirS with the TLR2 signaling pathway [9]. These results can be directly linked to our in vivo observations in mice, explaining the modulation of virulence during S. aureus infection by tirS [4,20]. Here the bacterial TIR effector has been shown to induce attenuation of virulence during infection because of the downregulation of the innate immune pathway.
In addition to control of inflammatory responses, some TIR domain-containing proteins such as BtpA or BaTcp have been shown to target and modulate microtubules [18,24] through a WxxxE motif that is also present in TirS and involved in cross-talk between the TLR and GTPase signaling pathways [16]. We found that ectopically expressed TirS accumulated in filament-like structures of irregular shapes within the host cytosol but that do not co-localize with typical cytoskeleton components (microtubules, actin or intermediate filaments). This observation is different from those previously described for other bacterial TIR proteins from Brucella [8,16] and BaTdp from B. anthracis [18]. The role of TirS accumulation in filament-like structures in the modulation of S. aureus pathogenicity remains to be explored.
The host interacting partner of TirS has yet to be identified. MyD88 is a general adaptor protein that plays an important role in Toll/IL-1 receptor family signaling. In vitro results of Askarian et al. [9] showed that TirS interferes with TLR2 and both MyD88 and TIRAP pathways in vitro. Here we report that in vitro, TirS is a potent inhibitor of not only TLR2 but also TLR4, TLR5, and TLR9. All of these proteins are dependent on MyD88, a general adaptor protein that plays an important role in the Toll/IL-1 receptor family signaling, which argues for an interaction of TirS with MyD88. Consistently, we found that S. aureus carrying tirS induced no increased virulence in a MyD88 knockout mice model. Taken together, these results suggest that TirS modulates an inflammatory response at the site of the infection through the MyD88 adaptor protein. Preliminary work from our lab has not been able to purify TirS, which is highly insoluble and failed to detect an interaction by co-immunoprecipitation assays. Further work is now required to understand the molecular mechanism by which TirS interacts and/or competes with MyD88.
As yet, there is no clear consensus on the mechanism by which TIR proteins enter host cells and localize to the host cell cytoplasm. From deduced amino acid sequences, no recognizable signal sequence for secretion was detected in TIR proteins, including TirS. In the case of TlpA from Salmonella enterica, for which no direct evidence of secretion has been reported, the suggested mechanism is a role for type III or type IV secretion systems (T3SS or T4SS) that can directly inject TIR effectors into the host cells [5]. TcpC of E. coli has been reported to be secreted into the media of cultured bacterial cells and subsequently taken up into host macrophages to interfere with TLR-mediated tumor necrosis factor induction [17]. In the case of Brucella, BtpA and BtpB are translocated into host cells in a manner partially dependent on the T4SS [8]. As observed with other TIR protein genes, the tirS gene is not co-located with genes encoding for a secretion system in S. aureus. However, TirS has been reported to be secreted into the media through an unknown mechanism [9]. Further studies are needed to clarify this issue, perhaps by performing real-time imaging and co-localization studies.
A comparison of different SCC elements in various S. aureus genetic backgrounds highlights the invariable presence of tirS within the SCC fusC/tirS mobile genetic element, sometimes included in the SCCmec elements in the J1 region. As proposed previously [9, 25], the finding that tirS region was invariably present within an SCC element suggests that similar to mecA and fusC transmission, SCC-mediated horizontal transfer is the major mechanism of tirS dissemination. Moreover, horizontal transfer of the tirS region is also suggested with the nearby presence of site-specific recombinases of the ccrAB type in all SCC elements carrying the tirS gene [25]. Of further interest, all published sequences of SCC fusC/tirS have identified four conserved additional genes. Future work should attempt to assign functional roles of these putative proteins and evaluate their involvement in the regulation or modulation of tirS and fusC transcription.
The co-location of mecA and fusC genes in this genetic element suggested that, in addition to antibiotic resistance genes, the presence of tirS may also confer a selective advantage for S. aureus. However, our observation indicated that a sub-inhibitory concentration of fusidic acid but not oxacillin induced overexpression of tirS in all strains tested. These results were unexpected because we observed the opposite outcome for Panton Valentine leukocidine and alpha-toxin expression with the same antibiotics: overexpression by oxacillin and inhibition by fusidic acid [26]. The acquisition by S. aureus of a gene encoding for factors that modulate virulence in the SCCmec element is not exceptional. The pls gene encoding for a large surface protein with an LPXTG peptidoglycan-anchoring sequence is a part of the type I SCCmec element [27]. This protein reduces S. aureus adherence and invasiveness [28]. Pls was also described in SCCmec III [29] but never in MSSA. Conversely, genes encoding for phenol soluble modulins alpha 1 to 3 (PSM-α1–3; small peptides with an amphipathic α-helical structure and strong surfactant-like properties that induce the production of proinflammatory cytokines and recruit, activate and lyse neutrophils) were first described in MSSA [30] before being found to be part of SCCmecII, SCCmecIII, and SCCmecVIII (PSM-α-mec) [31,32]. To our knowledge, is it not yet known whether antibiotics whose resistance is encoded by these SCC elements modulate the expression of Pls and/or PSM-mec.
A number of bacteria expressing TIR-containing proteins have been described, but as far as we know, only one study references their prevalence throughout a bacterial species [24]. In this work, we report the presence of tirS gene in 12.4% of a clinical MSSA and MRSA S. aureus strain collection. The TirS effector was not shown to be associated with a specific clinical human presentation by molecular epidemiology studies. For the Geraldine clone that always holds the tirS gene, previous observation did not identify a particular association with disease severity [33,34]. By contrast, the first nosocomial outbreak due to the Geraldine clone was recently reported, emphasizing its efficiency in being transmitted and easily spread within health care settings [35]. Indeed, this clone has been reported to be both a community-acquired and hospital-acquired MRSA. Thus, the clone Geraldine SCCmec element may provide a selective advantage in both settings: in the hospital because of a higher antimicrobial resistance compared to drug-susceptible WT S. aureus strains, and in the community because of its enhanced inhibition of the innate immune response. Similarly, different groups recently described the emergence in the community and in the hospital of ST1, ST45, and ST149 MRSA fusC positive strains in England and of fusC positive ST5 MRSA in New Zealand that also are tirS positive [36,37]. These epidemiological observations highlight the selective advantage of S. aureus in carrying the SCCmec element with SCC fusC/tirS.
In summary, we identify for the first time a bacterial TIR homolog protein genetically linked to an antimicrobial agent resistance determinant in the genetic mobile element SCC, thus providing a molecular connection between two key traits determining the successful outcome and spread of bacterial infections. Moreover, TIR homolog protein production was modulated by one antibiotic, fusidic acid, for which the resistance is encoded in a conserved region, which includes the tirS gene, and located within these SCCmec and non-mec SCC elements. This result expands knowledge about bacterial TIR homologs that constitute an ingenious strategy of pathogenic bacteria to evade the host immune system. The current state of knowledge strongly suggests that TIR effectors should be considered as potential key effectors of host defense, which emphasizes that further research is required to elucidate the precise mechanism of action of these interesting molecules. From the clinical point of view, the identification of the critical role of TirS signaling for modulating the immune response to a site of infection raises the possibility that this pathway could be locally targeted to engage the host’s own immune responses in the treatment of a microbial infection.
Materials and Methods
Ethical statements
Isolates were obtained as part of routine diagnostic testing and were analyzed anonymously. All data were collected in accordance with the European Parliament and Council decision for the epidemiological surveillance and control of communicable disease in the European community [38,39]. Ethical approval and informed consent were not required.
All mouse protocols were carried out in strict accordance with the Directive 2010/63/EU revising Directive 86/609/EEC on the protection of animals used for scientific purposes. This directive was translated in the French regulation as the Décret N°2013–118 of February 2013 under the jurisdiction of the Ministry of Education, Research and Technology. The initial research project had been approved by the local Animal Ethic Evaluation Committee CECCAPP (Comité d’Evaluation Commun au Centre Léon Bérard, à l’Animalerie de transit de l’ENS, au PBES et au laboratoire P4) with the references ENS_2014_025 and ENS_2014_052 and subsequently authorized by the French Ministry of Education, Research and Technology.
Bacterial strain characterization
A subset of 226 strains from the collection of the Centre National de Référence des Staphylocoques (Lyon, France), composed of 103 strains of the main community and hospital-acquired MRSA clones, and 123 strains of MSSA were used in this study. They were sent to the laboratory for detection of toxin production in the context of nasal colonization, skin and soft tissue infection, pneumonia, bacteremia, or endocarditis. The S. aureus HT20030749 strain belonging to the clone Geraldine was isolated from blood culture of patient with bone–joint infection.
All strains were genotyped as previously described. Briefly, bacterial DNA was extracted according to the manufacturer's recommended protocol using commercial extraction kits (Qiagen). The diagnostic DNA microarrays, identibac S. aureus Genotyping (Alere) used for this study, as well as related procedures and protocols, have been previously described in detail [40]. This microarray covers 332 different target sequences corresponding to approximately 185 distinct genes and their allelic variants. The assigning of isolates to CCs was determined by the comparison of hybridization profiles with those previously characterized by using multilocus sequence typing reference strains [40].
Illumina Sequencing
Genomic DNA were extracted from each isolate using a QIAcube extraction kit (Qiagen). The Nextera XT DNA preparation kit (Illumina) was used to generate sequencing libraries from 1 ng of DNA. Whole-genome sequencing was finally done with an Illumina HiSeq (Illumina, San Diego, CA, USA) to generate 150-bp paired-end reads.
De novo assembly
For each isolate, the raw paired-end reads were assembled using a modified version of the A5-miseq open-source pipeline [41]. This pipeline implements a complete sequencing data processing workflow from raw read cleaning to de novo assembly. The first task of the read cleaning involves removing the regions of the raw reads that are contaminated by the adapters during the Nextera XT protocol using the Trimmomatic program [42]. Then, the reads are filtered and trimmed according to quality and length criteria using Trimmomatic and the preprocess function of String Graph assembler (SGA) [43]. Finally, the correct function of SGA is used to correct errors in the reads by a k-mer frequency-based method. After being quality filtered and error corrected, the reads were assembled by the IDBA-UD500 program, which implements an iterative De Bruijn graph built with several values of k-mer lengths, from low to high values, instead of a single value as for most de novo assemblers [44]. The reads are then mapped against the assembly using BWA-MEM [45] to polish the contigs at every position where basecalls differ between the mapping and the assembly. The scaffolding implemented at the end of the original A5-miseq pipeline was not performed because it provides no gain in the subsequent analysis of marker detection.
Characterization of SCCmec V and tirS regions
First, the three MRSA genomes were aligned against the strain MSSA476 genome using progressiveMauve with default parameters [46]. Then, identification of SCC was done by searching for the ISS of these elements [47,48], both at the end of the ORF X / rlmH gene and further downstream. The newly detected SCCmec elements were annotated using the RAST Server (http://rast.nmpdr.org/), and blastn searches were performed (http://blast.ncbi.nlm.nih.gov/) against the nucleotide collection (nr/nt) using the megablastn algorithm and the organism filter for S. aureus (taxid:1280).
tirS amplification
Oligonucleotide primers tirS-For-CTTCAAAAAGAGCAGTCTAGG and tirS-Rev-CTTCAACACTCACTTTATGCC were designed according to tirS sequence. After amplification for 30 cycles (30 s of denaturation at 94°C, 30 s of annealing at 53°C, and 30 s of extension at 72°C), the PCR products were resolved by electrophoresis through 1.5% agarose gels (Sigma, Saint Quentin Fallavier, France). This step was followed by SYBR Safe DNA (Life Technologies) staining and analysis. To assess the specificity of tirS amplification, PCR products were subjected to DNA sequencing (Biofidal, Lyon, France). S. aureus HT20030749 and RN4220 strains were used as positive and negative amplification controls, respectively.
Transcription of tirS
tirS expression was analyzed using RT-qPCR. MRSA ST20121850 (CC1), MSSA ST20130407, ST20110167, ST20080979 (CC1), MSSA ST20121341 (CC8), MRSA ST20120331 (CC5, type IV), MRSA ST20111318, ST20110610, HT20030749 (CC5, type V) were grown in fresh MHB at 37°C, after a 1:100 dilution of overnight cultures. For kinetics analysis, total RNA of two representative strains (MRSA HT20030749, CC5 and MSSA ST20130407, CC1) was purified at 2, 4, 6, and 8 h of growth as previously described [49]. Total RNA was extracted with the RNeasy Plus (Qiagen) kit including a gDNA eliminator column and an additional DNAse treatment (Qiagen). RNA quality and quantity were determined by Bioanalyzer (Agilent) using the RNA Nano chips and quantified using the ND-8000 (NanoDrop Technologies). Absence of DNA contamination was checked by using tirS-specific primers and probe at optimal concentrations (assessed as previously described [50] without the reverse-transcription step). The final concentrations were 0.2 μM for primers and probe (tirS-F-CTATTTGGCATAAAGTGAGTGTTGAAG, tirS-R-AAATCACTTGTATTCAATGCATACTTATCT, and tirS-P-CGTGCATACAACCCATAT labeled with NED at the 5’ and 3’-minor groove binder), and reactions were performed in a one-step RT-PCR enzymatic mixture (Agilent Technologies, Brilliant II QRT-PCR Master Mix Kit) in a final volume of 20 μl in the CFX96 system (Bio-Rad) and following manufacturer’s instructions. Differences in Ct values between tested transcripts and hu signals were used for normalization purposes and based on the MHB medium condition at 2 h as a reference. The fold change was expressed as the inverse exponential of the difference between MHB Ct (reference) and the stress condition Ct. This assay was also used on 2-h cultures to assess gene expression values of tirS in various stress conditions such as the presence of 1/2 MIC of oxacillin for MSSA and 5 μg/ml for MRSA or 1/4 MIC of fusidic acid for MSSA and MRSA [49] on total RNA purified after 30 min of exposition. MICs were determined using CLSI recommendations [50], and the stress experiments were performed with drug concentrations showing minor impact on growth rate following control experiments (S3 Fig).
Construction of the tirS-deleted mutant
S. aureus RN4220 (lab strain collection) was used for plasmid amplification and genetic manipulations because it is a nitroso-guanidine-induced mutant capable of accepting E. coli DNA [51]. To delete the tirS gene, we performed allelic replacement using double crossover recombination as previously described [52]. Using the primers and restriction enzymes listed in S1 Table, we generated two fragments of 930 bp 5′ and 1029 bp 3′ of tirS. These fragments were ligated in 5′ and 3′ of the chloramphenicol acetyl transferase gene [53] and inserted into pMAD [54]. Plasmid and inserts were checked by PCR and sequencing using the primers listed in S1 Table. Plasmid was introduced by electroporation in RN4220 and then in HT20030749 (Geraldine strain). We performed double crossover recombination yielding to deletion of tirS in HT20030749 (Geraldine strain). The mutant strain obtained was called ΔtirS. Gene deletion was checked by PCR and sequencing using specific primer hybridizing with internal and external positions of the deleted region (S1 Table). Insert presence was checked with primers IngDNA_F and vG_CDS_1_R or IngDNA _F and vG_cat_1_R, yielding a negative PCR and a 2017-bp amplicon for the correct construct.
Construction of the tirS restored strain
Total DNA and plasmid DNA were prepared with Qiagen kits (QIAamp DNA Mini Kit and QIAprep Spin Miniprep Kit) after lysostaphin lyses for S. aureus. When necessary, transformation of E. coli DH5α (Promega, Madison, USA) was performed by treatment with CaCl2, and S. aureus strains were transformed by electroporation (Bio-Rad gene pulser). The tirS-deleted strain was complemented by inserting the tirS gene sequence downstream from the leukocidin promoter P-lukS in the bacterial chromosome using sequences homologous to sequence NWMN_0029 and NWMN_0030 of Newman strain (GenBank: AP009351.1) for chromosomal recombination. The region of NWMN_0029 and NWMN_0030 was amplified using primers New29-523 and New30-2371, restricted by EcoRI and SalI and cloned on a pBluescript vector (Stratagene) to obtain plasmid pLUG37. The tirS gene sequence was amplified and cloned between the P-lukS promoter region and lukF-PV transcriptional terminator of the Panton-Valentine leukocidin genes (respectively amplified using primers phi259/phi748 and phi2648/phi2819) on pBluescript. The whole DNA fragment obtained by SmaI was cloned in pLUG37 in the natural EcoRV restriction site between the NWMN39 and NWMN30 sequences. From the resulting plasmid, the DNA fragment corresponding to NMWN30-PlukS-tirS-term lukF-NWMN29 obtained by EcoRI-SalI restriction was cloned in the pMAD vector (pLUG1158). The resulting chromosomic restored strain was called ΔtirS + tirS. Expression of tirS in the restored strain was confirmed by RT-PCR. Oligonucleotides primers for PCR and DNA cloned subfragments are detailed in S2 Table.
Luciferase activity assay
HEK293T cells (American Type Culture Collection (ATCC), USA) were transiently transfected using Fugene (Roche) for 24 h, according to the manufacturer’s instructions, for a total of 0.4 μg of DNA consisting of 50 ng TLR plasmids, 200 ng of pBIIXLuc, a reporter plasmid containing luciferase under the control of two Igκ-κB sites [55], 5 ng of control Renilla luciferase (pRL-null, Promega), and 50 ng of myc-TirS expression vector, unless stated otherwise. In the case of TLR4, MD2 was co-transfected for efficient detection of LPS. When indicated, an increasing amount of vector (ng) was used for the transfections to obtained different levels of expression of TirS. In all cases, the total amount of DNA was kept constant by adding empty vector. Negative control corresponds to empty vector alone (pCDNA3.1). Where indicated, cells were treated with E. coli LPS (1 μg/ml), Pam2CSK4 (100 ng/ml), CpG ODN1826 (1 μM), and Flagellin Fl-ST (1 μg/ml), all obtained from Invivogen, for 6 h, and then cells were lysed and luciferase activity measured using the Dual-Glo Luciferase Assay System (Promega). In the case of IL-1R and TNFR, endogenous detection was monitored following 6 h of stimulation with IL-1β (100 ng/ml) or TNF-α (100 ng/ml). The tirS construct was obtained with the following primers tirS-fw-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTCAGTATTAGAAACTAAATTAAAAAG and tirS-rev-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAATTCTTAGAATTAACGATTACTTG and then cloned in the gateway (Life Technologies) entry vector and subsequently in the pDEST-Myc (Life Technologies) to create an N-terminal myc tag fusion with TirS.
Immunofluorescence microscopy
HeLa cells (ATCC, USA) were transfected with myc-tirS using Fugene (following manufacturer’s instructions) for 10 h. Cells were either fixed in 3% paraformaldehyde, pH 7.4, at room temperature for 15 min or placed in ice-cold methanol for 5 min. Cells were then permeabilized for 10 min with 0.1% saponin in PBS, followed by blocking for 1 h with 2% bovine serum albumin and 10% horse serum in PBS with 0.1% saponin. Primary antibodies were incubated for 1 h followed by three washes in PBS, 1 h incubation for secondary antibodies, two washes in PBS, and one wash in water before being mounted with Prolong Gold. Primary antibodies used were rabbit anti-myc (Abcam) at 1/1000, with either mouse anti-beta tubulin (TUB 2.1) at 1/250 or mouse anti-vimentin (V9) at 1/100 (both Sigma). Secondary antibodies used: donkey anti-rabbit Alexa 488, donkey anti-mouse Alexa 555 (Life Technologies) at 1/1000. The actin cytoskeleton was labeled with phalloidin 568 (Life Technologies). Samples were examined on a Zeiss 710 laser scanning confocal microscope for image acquisition. Images of 1024 × 1024 pixels were then assembled using ImageJ and Adobe Photoshop 7.0.
Growth curves of S. aureus strains
S. aureus Geraldine WT and ΔtirS strains were grown in fresh BHI medium or TSB medium at 35°C, after dilution of overnight cultures to OD600 = 0.03. A thermostated microplate reader (TECAN M200 Infinite Pro) was used to follow bacterial growth by measuring OD600 every 15 min for 24 h. As controls, specific wells were inoculated with medium only. Experiments were done in triplicate.
Murine model of S. aureus subcutaneous infection
Bacterial isolates and growth
S. aureus Geraldine WT, ΔtirS, and ΔtirS + tirS strains were used in a murine model of skin infection. For preparation of the inoculum used for subcutaneous inoculation, the bacteria were grown into BHI medium at 37°C for 8 h with constant shaking (200 rpm). They were washed twice and resuspended in sterile PBS, aliquoted to a final concentration of 1.107 bacteria/ml, and stored at -80°C until used. For determination of bacterial titers, samples were serially diluted, plated on agar, and incubated overnight at 37°C. Viability of the inocula was confirmed by colony counts with each experiment. Moreover, to ensure that the results of experiments were consistent, hemolysis phenotype was checked using blood agar plates (Trypcase soy agar + 5% sheep blood, bioMérieux, France).
Mouse strains
Mice on a C57BL/6 genetic background were used in all experiments. Six-week-old WT female mice were purchased from Charles River, France. Male and female MyD88-deficient mice were kindly provided by L. Genestier (from S. Akira’s lab; [56]). All mice were maintained in pathogen-free conditions in a biosafety level 2 facility at the Plateau de Biologie Expérimentale de la Souris (PBES, Ecole Normale Supérieure de Lyon, Lyon, France).
Skin lesion model
One day prior to infection, mice were prepared for inoculation. Animals first were anesthetized with 2% isoflurane, and a flank was shaved with electric clipper and hair remover cream (Nair, Church & Dwight Co. Inc., Princeton, NJ) on the shaved flank. On the day of infection, mice were infected subcutaneously with 100 μl of the bacterial suspension (1.106 CFUs) in the shaved area. This inoculum was determined in preliminary studies to produce consistent skin lesions. Mice were returned to their cages and observed to awaken. All mice had free access to food and water throughout the duration of the experiments. Animals were weighed at 24 h intervals for 14 days. The area of lesions was measured daily using an electronic caliper and calculated with the formula A = π × (L/2) × (W/2) (mm2). PBS injection was used for controls.
Bacterial recovery and cytokine quantification in skin lesions
To determine the number of CFUs at the site of infection, a second set of mice was inoculated as described above. On days 5 to 9 after infection, mice were euthanized by cervical dislocation; the lesion and the surrounding tissues were removed and transferred in sterile tubes. Tissue samples were weighed, homogenized in 1 ml of PBS (gentleMACS Dissociator, Miltenyi Biotec, Germany), diluted in sterile PBS, and plated on selective agar (ChromID S. aureus, bioMérieux, France). Enumeration of CFUs was performed 24 h later. For determination of cytokine and MPO levels, lesion homogenates were centrifuged and the supernatant removed and immediately stored at -80°C. Cytokine levels were determined using Luminex assays (Bio-Techne) according to the manufacturer’s instructions. The amount of MPO in the skin lesions was quantified using a commercially available ELISA kit (Bio-Techne).
Statistical analysis
The data were analyzed using R software (http://www.r-project.org). Student’s t test, Wilcoxon test, or one-way ANOVA followed by multiple comparisons tests (Tukey) were used to compare tirS expression, luciferase activity, lesion size, cytokine levels, and bacterial CFUs between the groups. Correlations were assessed by Spearman’s correlation. In all experiments, values of * p < 0.05, ** p < 0.01, and *** p < 0.001 were considered statistically significant.
Supporting Information
(PDF)
(PDF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We gratefully acknowledge Valérie Vogel and David Hernandez for help concerning bioinformatics analysis, and Myriam Girard, Christophe Ginevra and Hélène Meunier for technical assistance. We thank Christopher Montgomery and Lloyd Miller for mouse model consultation. We also thank Jacqueline Marvel for helpful discussions. We are grateful to Laurent Genestier for helpful discussions and for providing MyD88-deficient mice and to the staff members at PBES, with a particular thanks to Nadine Aguilera for their support in the animal facility.
Data Availability
ENA database project study accession number PRJEB12840; sample accessions ERS1070204 to ERS1070207 and ERS1434451, ERS1434452.
Funding Statement
This work was supported by the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program "Investissements d'Avenir" (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR) and a FINOVI Young Researcher Grant and by the Swiss National Science Foundation grant 31003A_153474. SP was supported by the Innovative Medicines Initiative Joint Undertaking under the Combatting Bacterial Resistance in Europe (COMBACTE) grant agreement no. 115523. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition by innate immunity. Cell. 2006; 124: 783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 2.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–65. 10.1146/annurev.biochem.76.060305.151318 [DOI] [PubMed] [Google Scholar]
- 3.Narayanan KB, Park HH. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis. 2015;20:196–209. 10.1007/s10495-014-1073-1 [DOI] [PubMed] [Google Scholar]
- 4.Rana RR, Zhang M, Spear AM, Atkins HS, Byrne B. Bacterial TIR-containing proteins and host innate immune system evasion. Med Microbiol Immunol. 2013;202(1):1–10. 10.1007/s00430-012-0253-2 [DOI] [PubMed] [Google Scholar]
- 5.Newman R, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74(1):594–601. 10.1128/IAI.74.1.594-601.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLOS Pathog. 2008;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirl C, Miethke T. Microbial Toll/interleukin 1 receptor proteins: A new class of virulence factors. Int J Med Microbiol. 2010;300:396–401. 10.1016/j.ijmm.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 8.Salcedo SP, Marchesini MI, Degos C, Terwagne M, Von Bargen K, Lepidi H, et al. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol. 2013;3(July):28 10.3389/fcimb.2013.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Askarian F, Van Sorge NM, Sangvik M, Beasley FC, Henriksen JR, Sollid JUE, et al. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-KB signaling. J Innate Immun. 2014;6(4):485–98. 10.1159/000357618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowy FD. Staphylococcus aureus Infections. N Engl J Med. 1998;339:520–32. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 11.Jevons MP. ‘Celbenin’- Resistant Staphylococci. Br Med J. 1961;1:124–5. [Google Scholar]
- 12.Wulf MWH, Sørum M, Van Nes A, Skov R, Melchers WJG, Klaassen CHW, et al. Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect. 2008;14(1):29–34. 10.1111/j.1469-0691.2007.01873.x [DOI] [PubMed] [Google Scholar]
- 13.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying panton-valentine leukocidin genes: Worldwide emergence. Emerg Infect Dis. 2003;9(8):978–84. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000. June;44(6):1549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101(26):9786–91. 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felix C, Kaplan Türköz B, Ranaldi S, Koelblen T, Terradot L, O’Callaghan D, et al. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Commun Signal. 2014;12:53 10.1186/s12964-014-0053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14(4):399–406. 10.1038/nm1734 [DOI] [PubMed] [Google Scholar]
- 18.Carlsson E, Thwaite JE, Jenner DC, Spear AM, Flick-Smith H, Atkins HS, et al. Bacillus anthracis TIR Domain-Containing Protein Localises to Cellular Microtubule Structures and Induces Autophagy. PLoS One. 2016; 10.1371/journal.pone.0158575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, et al. Dual function of MyD88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest. 2010;120(10):3663–7. 10.1172/JCI42771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuire VA, Arthur JSC. Subverting Toll-Like Receptor Signaling by Bacterial Pathogens. Front Immunol. 2015;6:607 10.3389/fimmu.2015.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casadevall A, Pirofski LA. The damage-response framework of microbial pathogenesis and infectious diseases. Nat Rev Microbiol. 2003;1(October):17–24. 10.1007/978-0-387-09550-9_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery CP, Daniels MD, Zhao F, Spellberg B, Chong AS, Daum RS. Local Inflammation Exacerbates the Severity of Staphylococcus aureus Skin Infection. PLoS One. 2013;8(7):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, et al. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. MBio. 2012;3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radhakrishnan GK, Harms JS, Splitter GA. Modulation of microtubule dynamics by a TIR domain protein from the intracellular pathogen Brucella melitensis. Biochem J. 2011;439(1):79–83. 10.1042/BJ20110577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Zmasek CM, Cai X, Godzik A. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev Comp Immunol. Elsevier Ltd; 2011;35(4):461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumitrescu O, Badiou C, Bes M, Reverdy M, Vandenesch F, Etienne J, et al. Effect of antibiotics, alone and in combination, on Panton–Valentine leukocidin production by a Staphylococcus aureus reference strain. Clin Microbiol Infect. 2004;14(4):384–8. [DOI] [PubMed] [Google Scholar]
- 27.Juuti K, Ibrahem S, Virolainen-Julkunen A, Vuopio-Varkila J, Kuusela P. The pls Gene Found in Methicillin-Resistant Staphylococcus aureus Strains Is Common in Clinical Isolates of Staphylococcus sciuri. J Clin Microbiol. 2005;43(3):1415–9. 10.1128/JCM.43.3.1415-1419.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain M, Schäfer D, Juuti KM, Peters G, Haslinger-Löffler B, Kuusela PI, et al. Expression of Pls (plasmin sensitive) in Staphylococcus aureus negative for pls reduces adherence and cellular invasion and acts by steric hindrance. J Infect Dis. 2009;200:107–17. 10.1086/599359 [DOI] [PubMed] [Google Scholar]
- 29.Deurenberg RH, Vink C, Oudhuis GJ, Mooij JE, Driessen C, Coppens G, et al. Different Clonal Complexes of Methicillin-Resistant Staphylococcus aureus Are Disseminated in the Euregio Meuse-Rhine Region. Antimicrob Agents Chemother. 2005;49(10):4263–71. 10.1128/AAC.49.10.4263-4271.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Queck SY, Jameson-Lee M, Villaruz AE, Bach THL, Khan BA, Sturdevant DE, et al. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol Cell. 2008;32(1):150–8. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queck SY, Khan BA, Wang R, Bach THL, Kretschmer D, Chen L, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLOS Pathog. 2009;5(7):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monecke S, Engelmann I, Archambault M, Coleman DC, Coombs GW, Cortez De Jäckel S, et al. Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol Cell Probes. 2012;26:99–103. 10.1016/j.mcp.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 33.Durand G, Bes M, Meugnier H. New methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital-and community-acquired infections. J Clin Microbiol. 2006;44(3):847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dauwalder O, Lina G, Durand G, Bes M, Meugnier H, Jarlier V, et al. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J Clin Microbiol. 2008;46(10):3454–8. 10.1128/JCM.01050-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leroyer C, Lehours P, Tristan A, Boyer F, Marie V, Elleau C, et al. Outbreak in newborns of methicillin-resistant Staphylococcus aureus related to the sequence type 5 Geraldine clone. Am J Infect Control. Elsevier Inc; 2016;44(2):10–2. [DOI] [PubMed] [Google Scholar]
- 36.Baines SL, Howden BP, Heffernan H, Stinear TP, Carter GP, Seemann T, et al. Rapid emergence and evolution of Staphylococcus aureus clones harbouring fusC-containing Staphylococcal Cassette Chromosome elements. Antimicrob Agents Chemother. 2016; 60(4):2359–65. 10.1128/AAC.03020-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellington MJ, Reuter S, Harris SR, Holden MTG, Cartwright EJ, Greaves D, et al. Emergent and evolving antimicrobial resistance cassettes in community-associated fusidic acid and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2015;45:477–84. 10.1016/j.ijantimicag.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Commission of the European Communities. Commission decision of 22 December 1999 on the communicable diseases to be progressively covered by the community network under decision number 2119/98/EC of the European Parliament and of the Council. Off J Eur Communities. 2000;(L28/50).
- 39.The European Parliament and the Council of the UE. Decision number 2119/98/EC of the European Parliament and of the Council of 24 September 1998: setting up a network for the epidemiological surveillance and control of communicable diseases in the community. Off J Eur Communities. 1998;(L 268/1).
- 40.Monecke S, Luedicke C, Slickers P, Ehricht R. Molecular epidemiology of Staphylococcus aureus in asymptomatic carriers. Eur J Clin Microbiol Infect Dis. 2009;28:1159–65. 10.1007/s10096-009-0752-2 [DOI] [PubMed] [Google Scholar]
- 41.Coil D, Jospin G, Darling AE. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31(4):587–9. 10.1093/bioinformatics/btu661 [DOI] [PubMed] [Google Scholar]
- 42.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40(Web Server issue):W622–7. 10.1093/nar/gks540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22(3):549–56. 10.1101/gr.126953.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng Y, Leung HCM, Yiu SM, Lv MJ, Zhu XG, Chin FYL. IDBA-tran: A more robust de novo de Bruijn graph assembler for transcriptomes with uneven expression levels. In: Bioinformatics. 2013. 29(13):i326–34. 10.1093/bioinformatics/btt219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–176010. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darling AE, Mau B, Perna NT. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito T, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel Type V Staphylococcal Cassette Chromosome. Antimicrob Agents Chemother. 2004;48(7):2637–51. 10.1128/AAC.48.7.2637-2651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noto MJ, Archer GL. A Subset of Staphylococcus aureus Strains Harboring Staphylococcal Cassette Chromosome mec (SCCmec) Type IV Is Deficient in ccrAB-Mediated SCCmec Excision. Antimicrob Agents Chemother. 2006;50(8):2782–8. 10.1128/AAC.00032-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaume M, Hernandez D, Farinelli L, Cile Deluen C, Linder P, Gaspin C, et al. Cartography of Methicillin-Resistant S. aureus Transcripts: Detection, Orientation and Temporal Expression during Growth Phase and Stress Conditions. PLoS One. 2010;5(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Natl Comm Clin Lab Stand Wayne, PA. 2003 (CLSI Document M100–S13).
- 51.Kreiswirth B, Löfdahl S, Betley M, O’Reilly M, Schlievert P, Bergdoll M, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–12. [DOI] [PubMed] [Google Scholar]
- 52.Fischer A, Kambara K, Meyer H, Stenz L, Bonetti E-J, Girard M, et al. GdpS contributes to Staphylococcus aureus biofilm formation by regulation of eDNA release. Int J Med Microbiol. 2014;304:284–99. 10.1016/j.ijmm.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 53.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel Cassette-Based Shuttle Vector System for Gram-Positive Bacteria. Appl Environ Microbiol. 2004;70(10):6076–85. 10.1128/AEM.70.10.6076-6085.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnaud M, Chastanet A, Débarbouillé M. New Vector for Efficient Allelic Replacement in Naturally Nontransformable, Low-GC-Content, Gram-Positive Bacteria. Appl Environ Microbiol. 2004;70(11):6887–91. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kopp E, Ghosh S. Inhibition of NF-k-B by sodium salicylate and aspirin. Science. 1994;265:956–9. [DOI] [PubMed] [Google Scholar]
- 56.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9(143.–). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
ENA database project study accession number PRJEB12840; sample accessions ERS1070204 to ERS1070207 and ERS1434451, ERS1434452.