Abstract
Human cytomegalovirus (HCMV), a member of the herpesvirus family, is a large complex enveloped virus composed of both viral and cellular gene products. While the sequence of the HCMV genome has been known for over a decade, the full set of viral and cellular proteins that compose the HCMV virion are unknown. To approach this problem we have utilized gel-free two-dimensional capillary liquid chromatography-tandem mass spectrometry (MS/MS) and Fourier transform ion cyclotron resonance MS to identify and determine the relative abundances of viral and cellular proteins in purified HCMV AD169 virions and dense bodies. Analysis of the proteins from purified HCMV virion preparations has indicated that the particle contains significantly more viral proteins than previously known. In this study, we identified 71 HCMV-encoded proteins that included 12 proteins encoded by known viral open reading frames (ORFs) previously not associated with virions and 12 proteins from novel viral ORFs. Analysis of the relative abundance of HCMV proteins indicated that the predominant virion protein was the pp65 tegument protein and that gM rather than gB was the most abundant glycoprotein. We have also identified over 70 host cellular proteins in HCMV virions, which include cellular structural proteins, enzymes, and chaperones. In addition, analysis of HCMV dense bodies indicated that these viral particles are composed of 29 viral proteins with a reduced quantity of cellular proteins in comparison to HCMV virions. This study provides the first comprehensive quantitative analysis of the viral and cellular proteins that compose infectious particles of a large complex virus.
Human herpesviruses are large complex enveloped viruses that constitute some of the most important human pathogens. Human cytomegalovirus (HCMV) is a prototypic herpesvirus that encodes over 200 predicted open reading frames (ORFs) (8, 11, 12, 32, 43). The HCMV virion is composed of an icosahedral capsid that contains a linear 230-kbp double-stranded DNA genome with attached proteins and an outer layer of proteins called tegument, surrounded by a cellular lipid layer containing viral glycoproteins (31). Viral and cellular proteins that constitute the infectious HCMV virion have been identified by biochemical and immunological approaches (3, 16). While these studies have yielded information regarding the proteins that compose the capsid, tegument, and envelope, the complete protein composition of the HCMV virion is unknown.
HCMV infection of cells in culture generates three different types of particles, including infectious mature virions described above, noninfectious enveloped particles (NIEPs), and dense bodies. NIEPs are composed of the same viral proteins as infectious virions but lack viral DNA (31). Dense bodies are uniquely characteristic of HCMV infection and are nonreplicating enveloped particles composed primarily of the tegument protein pp65 (UL83). The quantities of these different HCMV particles are dependent on the viral strain and the multiplicity of infection.
The icosahedral capsid of both NIEPs and infectious virions is assembled in the nucleus from five viral proteins encoded by the ORFs UL86, UL85, UL80, UL48-49, and UL46. The capsid is surrounded by the tegument, which is acquired in both the nucleus and cytoplasm of the infected cell. Approximately 20 to 25 known virion-associated tegument proteins have been identified in virion preparations, and many of these proteins are phosphorylated and have unknown functions (31). Cytoplasmic viral capsids containing tegument are enveloped by budding into the trans-Golgi network or a closely apposed cellular compartment, and this acquired lipid bilayer contains nine virally encoded glycoproteins, including gB (UL55), gM (UL100), gH (UL75), gL (UL115), gO (UL74), gN (UL73), gp48 (UL4), gpTRL10, and UL33 (31). Previous studies have determined that there are about 30 to 35 viral proteins that compose HCMV virions (3, 16). In addition to virally encoded structural proteins, a small number of cellular proteins, including CD13 (aminopeptidase N), β2-microglobulin, protein phosphatase I, annexin II, and actin-related protein 2/3 (Arp2/3), have been shown to associate with the HCMV virion (3, 17, 18, 30, 38, 42). The number of viral and cellular proteins that compose an infectious virion will always be controversial and dependent on the stringency of the isolation procedures. Unfortunately, as virions are purified, the particles tend to lose infectivity. This situation makes identification of essential virion proteins difficult to assess except by abundance.
Mass spectrometry (MS)-based proteomic approaches have been applied to analyze the proteomes of a number of organisms, including Deinococcus radiodurans, Plasmodium falciparum, yeast, and adenovirus (9, 14, 24, 28, 41). One MSapproach exploits high-accuracy mass measurements with enhanced dynamic range using liquid chromatography Fourier transform ion cyclotron resonance (LC-FTICR) MS to validate peptide assignments for an organism, from potential mass tags identified using LC-tandem MS (LC-MS/MS). This two-stage approach provides greater confidence in the identifications as well as the basis for subsequent higher sensitivity and throughput measurements without the need for routine MS/MS (35, 36). We have applied the high mass measurement accuracy of LC-FTICR to analyze the proteome of HCMV virions and dense bodies. Additionally, a comparison of the peptide ion intensities was used to evaluate relative abundances of proteins isolated from virion and dense body particles. These analyses resulted in the identification of both new and unknown HCMV ORFs that encode peptides in the virion and demonstrate the utility of this approach.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Normal human dermal fibroblasts (Clonetics) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin-streptomycin-l-glutamine. The HCMV strain AD169 was obtained from the American Type Culture Collection (Rockville, Md.).
HCMV particle purification procedures.
HCMV AD169 particles were purified from the culture medium of infected normal human dermal fibroblasts when the cells displayed maximal cytopathic effect. The cellular supernatants were first clarified by centrifugation at 12,000 × g for 15 min. The clarified medium was layered over a sorbitol cushion (20% d-sorbitol, 50 mM Tris [pH 7.4], 1 mM MgCl2), and virus was pelleted by centrifugation at 64,000 × g for 1 h at 4°C in a Beckman SW28 rotor. The virus pellet was resuspended in TNE buffer (50 mM Tris [pH 7.4], 100 mM NaCl, and 10 mM EDTA). The virus particles were further purified by layering them over a continuous 10-to-50% Nycodenz (Sigma) gradient in TNE buffer and centrifuged at 110,000 × g for 2 h at 4°C in a Beckman SW40.1 rotor. The gradients were fractionated, and the protein composition of each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gradient fractions, containing dense bodies, and virions were diluted in TNE buffer and pelleted by centrifugation at 110,000 × g for 2 h at 4°C and resuspended in TNE buffer. Purity was confirmed by electron microscopy of negatively stained HCMV preparations. In order to assess the protein content of the purified virus and dense bodies, denatured protein preparations were separated on NuPAGE morpholinepropanesulfonic acid (MOPS) gradient gels (Invitrogen, Carlsbad, Calif.) and visualized by Coomassie brilliant blue staining.
Tryptic digestion of HCMV particles.
HCMV particles were denatured by the addition of urea to 8 M and heating to 37°C for 30 min. The sample was then diluted fourfold with 100 mM ammonium bicarbonate (AB), and CaCl2 was added to 1 mM. Methylated, sequencing-grade porcine trypsin (Promega, Madison, Wis.) was added at a substrate-to-enzyme ratio of 20:1 (mass/mass) and incubated at 37°C for 15 h. The digested peptides were dialyzed against a large volume of 100 mM AB.
Cation exchange separation of peptides.
A 250-μg virion particle sample was dialyzed against 100 mM AB, lyophilized to dryness, and trypsin digested as described above. Strong cation exchange chromatography was performed on the peptide sample as previously described (39).
MS/MS analysis of peptides.
Peptide samples were analyzed by high-resolution, reversed-phase capillary LC coupled directly to an electrospray ionization interface with a tandem mass spectrometer as described previously (39). All MS/MS analyses were performed on a Finnigan LCQ ion trap mass spectrometer (Thermo Finnigan, San Jose, Calif.) that was run and operated as described elsewhere (2, 39).
FTICR MS.
For FTICR analysis, unfractionated or fractionated tryptic peptides were resuspended in mobile phase A (0.1% trifluoroacetic acid) and analyzed using reversed-phase capillary LC coupled to an electrospray ionization interface with a FTICR mass spectrometer as previously described (36). In this high-throughput application, a modified and enhanced 9.4T Bruker Apex III FTICR MS instrument was employed for the high mass measurement accuracy peptide measurements (4) coupled with a PAL autosampler (Leap Technologies) and a constant pressure LC system, assembled in-house.
Analysis and quantitation of FTICR results.
The SEQUEST algorithm (13) was used to make preliminary peptide identifications by matching the MS/MS fragmentation spectra with peptides from a combined database comprised of the HCMV.fasta and the human.fasta from the National Center for Biotechnology Information. The human database was modified with the removal of viral proteins and redundant protein entries in the database. An HCMV stop-to-stop (StoS) database was designed to identify peptides from all possible ORFs of 20 amino acids (aa) or longer from HCMV. A peptide was considered tentatively identified with a conservative criteria set developed by Yates (27, 41). Briefly, all accepted SEQUEST results had a ΔCn of 0.1 or greater. Peptides with a +1 charge state were accepted if they were fully tryptic and had a cross-correlation (Xcorr) of at least 1.9. Peptides with a +2 charge state were accepted if they were fully tryptic or partially tryptic and had an Xcorr of at least 2.2. Peptides with +2 or +3 charge states with an Xcorr of at least 3.0 or 3.75, respectively, were accepted regardless of their tryptic state. When a protein was identified by two or fewer unique peptides that met the SEQUEST criteria above, the SEQUEST spectra alignment was manually validated using criteria described elsewhere (27). Analysis of the LC-FTICR experiments was performed using ICR-2LS (25). Each experiment was performed in triplicate, and for identification purposes peptides had to be present in at least two of the triplicate FTICR measurements and/or in multiple experiments.
To generate relative abundances for the proteins, single preparations of virion and dense body particles were analyzed by FTICR in triplicate. Peptides had to be present in at least two of the triplicate FTICR measurements. Abundances of the individual peptides were computed by summing the intensity of the ions from a single scan or multiple scans that matched each peptide, similar to an approach reported by Wang et al. (40). However, because tryptic peptides for a protein do not all have the same FTICR peak abundances, only the most abundant peptides from each protein were used to compute the average abundance for that protein. For this study, peptides from each protein that were in the top 66% in peak abundance were used for analysis. In general the integrated, averaged peptide intensities should correlate with the relative protein mass. Additionally, all peptides used to derive protein abundances were fully tryptic, except for one UL88 peptide.
RESULTS AND DISCUSSION
Identification of HCMV virion proteins.
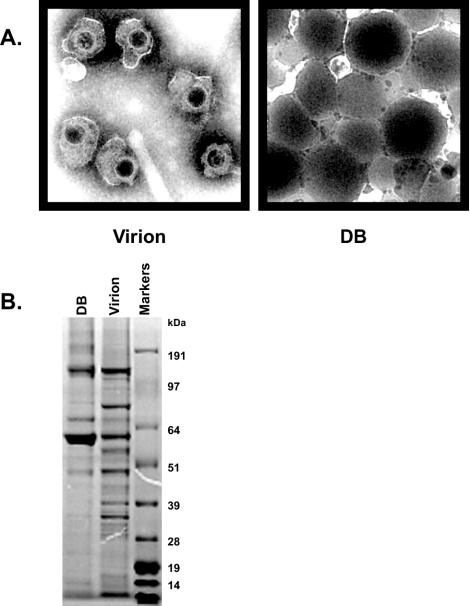
To determine the protein composition of the HCMV virion, viral particles were purified from the supernatant culture fluids of human fibroblast cells infected with HCMV strain AD169. This HCMV strain was selected because of the availability of the complete sequence of the viral genome (8). AD169-infected supernatants were subjected to sequential sedimentation and density ultracentrifugation gradients such that the purity of the preparations was >96% virions as determined by electron microscopy (333 virions, 13 dense bodies, 0 NIEPs, and undetectable cellular organelles counted in 15 individual fields). Examples of the HCMV virion and dense body purified particles used for MS analysis examined by electron microscopy are shown in Fig. 1A, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the same preparations is shown in Fig. 1B.
FIG. 1.
Characterization of HCMV virion and dense body preparations. (A) Electron microscopy of negatively stained HCMV virion and dense body preparations. Magnification, ×8,400. (B) Analysis of the proteins that constitute the purified HCMV particle preparations. Proteins were separated using NuPAGE MOPS gradient gels and visualized by Coomassie blue staining.
Purified virion preparations were digested to yield a complex mixture of polypeptides that was analyzed by a two-stage MS approach. The first stage employed both one-dimensional and two-dimensional LC (multidimensional protein identification technology) coupled to MS/MS (2, 27). The results from the LC-MS/MS analysis were verified and extended by employing high-accuracy mass measurements with LC-FTICR combined with chromatographic elution time information. Using this approach, we identified 59 proteins, including 12 proteins encoded by known HCMV ORFs that were not previously shown to reside in virions (Table 1). The known virion proteins identified included 5 capsid proteins (UL46, UL48-49, UL80, UL85, and UL86), 14 tegument proteins (UL24, UL25, UL26, UL32, UL43, UL47, UL48, UL82, UL83, UL94, UL99, US22, US23, and US24), 11 glycoproteins (TRL10, UL22A, UL41A, UL55, UL73, UL74, UL75, UL77, UL100, UL115, and UL119), 12 proteins involved in DNA replication and transcription (IRS1, TRS1, UL44, UL45, UL54, UL57, UL69, UL72, UL84, UL89, UL97, and UL122), and 2 G-protein-coupled proteins (UL33 and US27) (Table 1). This analysis also identified 12 HCMV-encoded polypeptides not previously associated with the virion, including UL5, UL38, UL50, UL71, UL79, UL93, UL96, UL103, UL132, US23, US24, and TRL14. Nine of these HCMV-encoded polypeptides (UL38, UL50, UL71, UL79, UL93, UL96, UL102, US23, and US24) are required for efficient virus growth in cultured fibroblasts (12, 43). UL4, UL23, UL53, UL56, UL98a, and US28 have previously been reported to be associated with HCMV particles (1, 6, 7, 10, 37). Although peptides corresponding to these viral proteins were detected in our study, the peptides failed to meet the criteria, as outlined in Materials and Methods, for inclusion in our database.
TABLE 1.
HCMV proteins identified by LC-MS/MS and/or by FTICR from virions and dense bodies
| Viral protein group | HCMV ORF | LC-MS/MS
|
FTICR
|
% Cov- erage | |
|---|---|---|---|---|---|
| No. of different peptides | Max XCorr | No. of different peptides | |||
| Virion proteins | |||||
| Capsid | UL46 | 20 | 5.30 | 14 | 44.8 |
| UL48-49 | 8 | 6.52 | 5 | 54.7 | |
| UL80 | 37 | 6.36 | 30 | 35.6 | |
| UL85 | 21 | 6.73 | 22 | 63.1 | |
| UL86 | 149 | 3.97 | 123 | 71.0 | |
| Tegument | UL24 | 8 | 5.06 | 9 | 38.3 |
| UL25 | 60 | 7.04 | 59 | 59.2 | |
| UL26 | 9 | 4.77 | 10 | 53.7 | |
| UL32 | 135 | 3.01 | 100 | 70.5 | |
| UL43 | 7 | 5.50 | 10 | 28.1 | |
| UL47 | 53 | 6.10 | 64 | 57.5 | |
| UL48 | 111 | 4.29 | 109 | 56.8 | |
| UL82 | 70 | 6.39 | 47 | 69.3 | |
| UL83 | 123 | 5.44 | 86 | 92.0 | |
| UL94 | 10 | 5.08 | 12 | 26.4 | |
| UL99 | 8 | 5.87 | 9 | 64.7 | |
| US22 | 2 | 3.16 | 2 | 5.4 | |
| US23 | 1 | 2.61 | 1 | 4.6 | |
| US24 | 1 | 4.83 | 2 | 7.0 | |
| Glycoproteins | RL10 | 5 | 2.36 | 4 | 22.8 |
| TRL14 | *a | 1 | 7.5 | ||
| UL5 | * | 1 | 5.4 | ||
| UL22A | 1 | 5.04 | 1 | 19.4 | |
| UL33 | 4 | 6.11 | 4 | 14.1 | |
| UL38 | * | 1 | 5.7 | ||
| UL41A | 2 | 5.72 | 2 | 25.6 | |
| UL50 | 1 | 2.82 | 4 | 10.6 | |
| UL55 | 21 | 6.16 | 23 | 24.8 | |
| UL73 | 2 | 3.47 | 2 | 6.5 | |
| UL74 | 4 | 5.07 | 4 | 13.5 | |
| UL75 | 21 | 6.15 | 22 | 35.7 | |
| UL77 | 14 | 5.65 | 12 | 31.2 | |
| UL93 | 15 | 5.35 | 14 | 31.7 | |
| UL100 | 13 | 5.24 | 7 | 15.9 | |
| UL115 | 11 | 4.73 | 9 | 47.1 | |
| UL119 | 2 | 2.23 | 1 | 4.6 | |
| UL132 | 8 | 5.89 | 8 | 47.0 | |
| US27 | 4 | 4.25 | 2 | 7.7 | |
| Transcription-repli- cation machinery | IRS1 | 15 | 6.01 | 17 | 25.8 |
| TRS1 | 10 | 6.92 | 23 | 34.7 | |
| UL44 | 1 | 4.32 | 9 | 31.0 | |
| UL45 | 43 | 5.85 | 52 | 52.2 | |
| UL54 | * | 1 | 1.6 | ||
| UL57 | * | 1 | 0.4 | ||
| UL69 | 6 | 4.17 | 7 | 19.0 | |
| UL72 | * | 1 | 4.6 | ||
| UL84 | 1 | 2.50 | 3 | 12.8 | |
| UL89 | * | 1 | 3.1 | ||
| UL97 | 13 | 5.95 | 9 | 32.1 | |
| UL122 | 2 | 4.26 | 4 | 11.7 | |
| Uncharacterized | UL35 | 42 | 6.27 | 40 | 56.1 |
| UL51 | * | 1 | 3.2 | ||
| UL71 | 12 | 6.32 | 11 | 40.4 | |
| UL79 | * | 1 | 10.9 | ||
| UL88 | 14 | 6.8 | 17 | 33.6 | |
| UL96 | 1 | 4.46 | 1 | 19.7 | |
| UL103 | 8 | 5.18 | 8 | 37.0 | |
| UL104 | 9 | 4.68 | 9 | 23.0 | |
| UL112 | 1 | 3.30 | 4 | 4.7 | |
| Dense body proteins | |||||
| Capsid | UL46 | 1 | 3.6 | 6 | |
| UL48-49 | 2 | 5.8 | 1 | ||
| UL80 | 1 | 6.1 | 2 | ||
| UL85 | 4 | 5.0 | 4 | ||
| UL86 | 22 | 5.0 | 19 | ||
| Tegument | UL25 | 17 | 6.3 | 13 | |
| UL26 | 3 | 3.6 | 3 | ||
| UL32 | 11 | 5.4 | 15 | ||
| UL35 | 5 | 5.6 | 9 | ||
| UL47 | 2 | 4.3 | 6 | ||
| UL48 | 7 | 5.4 | 12 | ||
| UL82 | 9 | 5.1 | 6 | ||
| UL83 | 40 | 6.3 | 14 | ||
| UL75 | 4 | 5.6 | 2 | ||
| Transcription-repli- cation machinery | UL45 | 2 | 4.3 | 6 | |
| IRS1 | 3 | 5.6 | 2 | ||
| TRS1 | 1 | 4.7 | 5 | ||
Asterisks indicate proteins whose LC-MS/MS SEQUEST scores were lower than our cutoff values; however, the proteins were identified with the more sensitive FTICR.
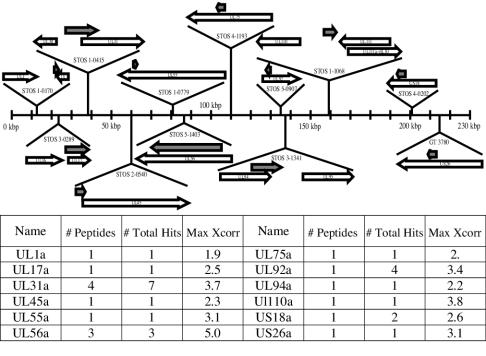
In order to evaluate the presence of small polypeptides in the virion not represented by ORFs in the annotated HCMV genome, we constructed a database of StoS protein coding regions from all reading frames of the HCMV genome that were 20 aa or longer in length. The MS/MS spectra were analyzed against the predicted peptides from this StoS database, and polypeptides corresponding to 12 short ORFs were identified that have not been previously characterized (Fig. 2). Six of these short polypeptides either have very high confidence identifications or were identified from multiple different peptides and are presumably more abundant. None of these new peptides identified corresponded to the candidate ORFs described by Murphy et al. (32), in part because the referenced analysis considered ORFs of ≥50 aa. BLAST analysis revealed that all of the identified StoS sequences were present in the HCMV strains TR, PH, FIX, Merlin, Toledo, and Towne and were between 97 and 100% identical at the DNA level (data not shown). The shortest of the small peptides detected in this study was 22 aa (StoS-1-0779; 66 bp), and the longest detected was 190 aa (StoS-1-0415, 570 bp) in length, overlapping but out of frame with UL31. What is unclear from these studies is whether the proteins encoded by the small ORFs are unique or part of larger proteins.
FIG. 2.
StoS ORF map. The genome of HCMV is represented by the line. Newly identified StoS ORFs are indicated by solid arrows, and ORFs that were recognized previously in AD169 are marked by open arrows.
Relative quantitation of virion proteins.
The intensities of the FTICR spectra of the most abundant peptides for each protein were averaged to determine the relative quantities of viral proteins in preparations of HCMV particles. The relative abundance of each HCMV protein is shown in Table 2 as a percentage of the sum of all the virion proteins. These analyses indicated that the virion is comprised of 50% tegument proteins, 30% capsid proteins, 13% envelope proteins, and 7% undefined proteins. The most abundant protein in the virion was the pp65 tegument protein UL83, which was present in a molar ratio of ~2:1 with UL86 (major capsid protein). These findings are similar to previous observations using different quantitative techniques (3, 21, 23, 34). Similarly, we observed that UL85 (minor capsid protein) was present in a 2:1 molar ratio with UL46 (minor capsid protein-binding protein) and that UL82 (pp71) and UL86 (major capsid protein) were in approximately equimolar amounts. Both of these observations are also consistent with previous reports (21).
TABLE 2.
Average abundances for HCMV proteins associated with virions and dense bodies
| HCMV ORF | Virion abundancea
|
Dense body abundancea
|
||||
|---|---|---|---|---|---|---|
| Avg abundance | No. of peptides used | SD | Avg abundance | No. of peptides used | SD | |
| UL83 | 15.4 | 5 | 5.3 | 60.2 | 2 | 36.6 |
| UL48-49 | 12.6 | 1 | <0.1 | 4.4 | 1 | <0.1 |
| UL100 | 9.2 | 1 | <0.1 | 4.4 | 1 | <0.1 |
| UL32 | 9.1 | 7 | 4.2 | 2 | 1 | <0.1 |
| UL82 | 8.9 | 2 | 5.2 | 1.7 | 5 | 0.3 |
| UL48 | 8.8 | 2 | 4.2 | 0.8 | 2 | 0.4 |
| UL80 | 7.7 | 3 | 4.1 | 1.1 | 1 | <0.1 |
| UL86 | 6 | 11 | 2.4 | 1.5 | 6 | 0.6 |
| UL45 | 4.7 | 1 | <0.1 | 0.9 | 2 | 0.4 |
| UL85 | 2.8 | 4 | 0.8 | 1 | 2 | 0.2 |
| UL25 | 2.2 | 4 | 0.8 | 12.7 | 4 | 5.0 |
| UL46 | 1.5 | 4 | 0.7 | 0.3 | 5 | 0.1 |
| UL47 | 1.5 | 5 | 0.6 | 0.3 | 6 | 0.1 |
| UL55 | 1.4 | 2 | 0.4 | 0.3 | 2 | 0.2 |
| UL94 | 1.2 | 2 | 0.5 | 1 | 1 | <0.1 |
| IRS1 | 0.8 | 1 | <0.1 | 0.3 | 1 | <0.1 |
| UL88 | 0.7 | 4 | 0.2 | 0.1 | 4 | <0.1 |
| UL77 | 0.6 | 1 | <0.1 | ND | NA | NA |
| TRS1 | 0.6 | 2 | 0.3 | 0.3 | 3 | <0.1 |
| UL75 | 0.6 | 5 | 0.3 | 0.6 | 3 | 0.3 |
| UL35 | 0.5 | 5 | 0.2 | 1 | 1 | <0.1 |
| UL115 | 0.5 | 2 | 0.1 | 0.3 | 2 | 0.1 |
| UL119 | 0.5 | 1 | <0.1 | ND | NA | NA |
| UL132 | 0.4 | 3 | 0.2 | 0.2 | 2 | 0.1 |
| UL104 | 0.4 | 2 | 0.2 | ND | NA | NA |
| US27 | 0.2 | 1 | <0.1 | ND | NA | NA |
| RL10 | 0.2 | 2 | <0.1 | >0.1 | 2 | <0.1 |
| UL72 | 0.2 | 1 | <0.1 | ND | NA | NA |
| UL22A | 0.2 | 1 | <0.1 | ND | NA | NA |
| UL97 | 0.1 | 4 | <0.1 | 0.5 | 3 | 0.1 |
| UL71 | 0.1 | 3 | <0.1 | ND | NA | NA |
| UL26 | 0.1 | 2 | <0.1 | 2.4 | 2 | 1.0 |
| US22 | 0.1 | 2 | <0.1 | ND | NA | NA |
| UL103 | 0.1 | 5 | <0.1 | >0.1 | 5 | <0.1 |
| UL24 | 0.1 | 1 | <0.1 | 0.5 | 1 | <0.1 |
| UL44 | 0.1 | 2 | <0.1 | 0.4 | 1 | <0.1 |
| UL33 | 0.1 | 1 | <0.1 | ND | NA | NA |
| UL43 | ND | NA | NA | 0.2 | 3 | <0.1 |
| UL69 | ND | NA | NA | 0.1 | 3 | <0.1 |
| UL93 | ND | NA | NA | >0.1 | 1 | <0.1 |
| UL99 | ND | NA | NA | 0.5 | 2 | 0.2 |
Abundances are based upon a percentage of the total protein present derived from integrated peptide MS peak intensities. ND, not detected; NA, not applicable.
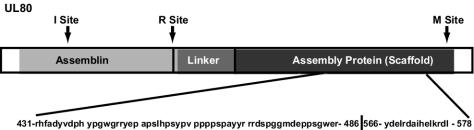
Analysis of the virion preparations also indicated that 8% of the total peptide content was composed of UL80. UL80 encodes a protein that is cleaved into the assembly protein (AP UL80.5) at the carboxyl terminus (UL80, aa 336 to 708) and the protease (assemblin, UL80A) at the amino terminus (UL80, aa 1 to 256). The protease is subsequently autocatalytically cleaved into two more peptides (16, 31). Interestingly, all of the UL80 peptides identified in the virion preparations were from AP (UL80 regions aa 431 to 486 and 566 to 578) (Fig. 3). The AP polypeptide is a major component of immature B capsids that lack viral DNA genome. The AP protein is believed to be lost during the maturation of the HCMV B capsid to the C capsid, which eventually develops into the infectious virion. In contrast, some B capsids form NIEPs. However, examination of the viral preparations by electron microscopy indicated that 96% of the particles were mature electron-dense virions and 4% were dense bodies and there were undetectable amounts of NIEPs, suggesting that AP may be a component of the infectious particle. Consistent with this argument, if NIEPs were contributing to the presence of AP then we would expect that peptides representing assemblin would be present, and this protein was not detected in the virion preparations.
FIG. 3.
Abundant peptides identified as UL80-specific peptides by LC-MS/MS and/or by FTICR associated with virions.
The most abundant HCMV envelope glycoprotein was UL100 (gM), which comprised 10% of the total virion protein. This observation was surprising, since previous reports have suggested that the most predominant envelope glycoprotein is UL55 (gB), reported here to be 10-fold less or 1% of the total virion protein content. The discrepancy in the observations can be accounted for by the highly hydrophobic nature of gM, which is glycoslylated and contains at least eight transmembrane domains, making this protein difficult to detect by gel electrophoresis (26). Although gM complexes with UL73 (gN), only 0.1% of the peptide content detected in our virion preparations was gN. Lastly, UL75 (gH) and UL115 (gL), which exist as a glycoprotein complex (19, 22), were detected at an equal molar ratio. However, UL74 (gO), which is also a member of this complex, was detected at lower levels (<0.1% of the total peptides detected). The unexpected lower levels of gN and gO in virions detected by FTICR may be due to the highly glycosylated state of these molecules, which might prevent complete trypsinization, or due to factors affecting the MS detection (20, 29).
HCMV virion host proteins.
The host protein content of isolated HCMV particles was determined by searching for peptides detected by comparison with peptides predicted from a human-Fasta database. A total of 71 host cell proteins associated with HCMV virions were identified with high confidence, significantly increasing the number of previously identified virion proteins (Table 3; see also Table S1 in the supplemental material) (3, 17, 18, 30, 31, 38, 42). HCMV cellular virion proteins include cytoskeletal proteins, such as α- and β-actin, tubulin, several annexins, α-actinin, and vimentin, as well as cellular proteins involved in translational control, including initiation and elongation factors. Other cellular proteins identified in HCMV virion preparations include clathrin and ADP-ribosylation factor 4. These proteins are involved in vesicular trafficking in the endoplasmic reticulum and Golgi, suggesting a role for these proteins in viral envelopment and/or egress. In addition to the above cellular virion proteins, four isoforms of the signal transduction protein 14-3-3 were also identified in HCMV preparations, together with other signaling proteins such as RasGAP, casein kinase 2, and β2-GTO-binding regulatory protein. The cellular protein β2-microglobulin was previously reported to be a component of the HCMV virion (18, 38). However, this cellular protein was present in low amounts in virion preparations and was eliminated from the cellular protein list, based on the conservative criteria employed in this study. The latter observation, as well as the absence of other cellular proteins in HCMV purified virion preparations, suggests that the cellular proteins identified in this study are part of the infectious virion. The presence of these cellular proteins in virion preparations may be attributed to copurification of cellular components in virion preparations, purified virions sticking to cellular proteins, or that the cellular proteins are constituents of the viral particles. However, we believe that the majority of the cellular proteins in virion preparations are integral parts of the particle, since some of these cellular proteins have already been identified in virion preparations and examination of the viral preparations by electron microscropy indicated undetectable amounts of cellular organelles and debris.
TABLE 3.
Selected host proteins identified by category
| Category | Proteins |
|---|---|
| ATP binding | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 1, sodium-potassium-transporting ATPase |
| Ca+binding | Annexin I, annexin V, annexin VI, annexin A2, calreticulin, α-actinin 1 |
| Chaperone | Cyclophilin A, glucose-regulated protein, heat shock protein 70 kDa, heat shock protein 90 kDa, tumor rejection antigen |
| Cytoskeleton | α-Actin, β-actin, cofilin, filamin, keratin, moesin, α-tubulin, β-tubulin, vimentin |
| Enzymes | Aminopeptidase N, transketolase, vinculin |
| Glycolysis | Enolase, glyceraldehyde-3-phosphate dehydrogenase, lactate dehydrogenase, phosphoglycerate kinase, serine/threonine protein phosphatase PP1, triosephosphate isomerase |
| Protein transport | ADP-ribosylation factor 4, clathrin, polyubiquitin 3, β-RAB GDP dissociation inhibitor |
| Signal transduction | α1-Casein kinase 2, 14-3-3 protein (four isoforms) |
| Transcription-translation | Eukaryotic translation elongation factor 2, eukaryotic translation elongation factor 1, enhancer protein, eukaryotic translation initiation factor 4A |
Identification and relative quantitation of proteins associated with HCMV dense bodies.
The LC-FTICR and LC-MS/MS analyses of dense body preparations identified 31 viral proteins that included 5 capsid proteins, 9 tegument proteins, 4 glycoproteins, and 4 proteinsinvolved in virus transcription and replication (Tables 1 and 2; see also Table S3 in the supplemental material). Ten of these proteins, UL25, UL32, UL35, UL45, UL47, UL48, UL82, UL83, UL86, and TRS1, were all identified with more than five peptides from each protein. UL83 (pp65) was the predominant protein present in dense bodies, representing 60% of the relative protein, similar to previous studies (3, 21). Likewise, UL25 has been found to be more abundant in dense bodies than in virions (3). In this study, UL25 represented 13% of the total protein in dense bodies but only 2.2% of the total protein of the virion, a roughly fivefold increase in relative abundance. Only one other protein, UL26, was present at a significantly increased level in dense bodies (2.4%) compared to virions (0.1%). UL26 was previously reported as more abundant in dense bodies (3), but in contrast it was not detected in virions in the earlier work. Additionally, the tegument phosphoprotein UL32 (pp150) is decreased in dense bodies compared to virions from 9.1% of the virion protein mass to 2% of the dense body protein mass. While the amount of UL32 in the virion was lower than previously estimated, our analysis indicated that pp150 is preferentially incorporated into virions rather than dense bodies (5). Only a small number of host cell proteins were identified associated with dense bodies, including glyceraldehyde-3-phosphate dehydrogenase, annexin A2, β-actin, and the heat shock 70-kDa protein (see Table S2 in the supplemental material). A possible explanation for this observation is that dense body particles egress from the host cell in a fundamentally different manner than virion particles.
Similar to the findings of Baldick and Shenk (3), the dense body particles isolated here appeared more complex than previously indicated (21), with 21 viral proteins identified. Additionally, the presence of five nucleocapsid proteins, UL46, UL48-9, UL80, UL85 and UL86, in the dense body preparations was perhaps surprising, given that dense bodies are reported to lack a nucleocapsid (15, 33). However, at least two of these proteins, UL85 and UL86, have previously been detected in dense bodies (3). The five capsid proteins combined represent a relatively small portion of the total dense body protein (~7.8%), whereas these same five proteins represent ~24% of the total protein isolated from virion particles.
According to our analysis, the glycoproteins encoded by UL55, UL100, and UL132 were decreased in relative abundance by four-, two-, and twofold, respectively, while UL75 and UL115 were equivalent in comparison to the abundance of these glycoproteins in virions. The total percentage of viral glycoproteins in the dense body preparations was 5.2%, compared to 13% in virion preparations. The discrepancy may reflect the variation in size as well as high tegument composition (85.5%) of the dense bodies. The other glycoproteins identified in virion preparations (TRL10, UL22A, UL33, UL77, UL119, and US27) were not detected in the dense body preparations. The fact that dense bodies are infectious suggests that the full complement of virion glycoproteins necessary for the particle to enter the cell is present in these particles. These observations suggest that the other glycoproteins not detected in dense preparations are either below our detection limits or are not essential for dense body infectivity.
Conclusions.
Using state-of-the art proteomic techniques, we have identified the viral and cellular proteins associated with purified preparations of HCMV particles and dense bodies and we have determined their relative abundance. Previous studies suggested that the HCMV virion contains about 30 to 40 proteins. The data presented here indicate a significantly higher number of viral structural genes, 59, including polypeptides encoded by 12 small ORFs which were not previously predicted to encode polypeptides. Additionally, a large number of host cellular genes were identified as associated with the virion, indicating a greater role for cellular proteins in the ontogeny of the virus. The function of these newly identified virion proteins and their contribution to structure and infectivity are unknown and the focus of future studies.
Supplementary Material
Acknowledgments
We thank the NIH National Center for Research Resources (RR18522) and the Environmental Molecular Science Laboratory (a U.S. Department of Energy user facility located at the Pacific Northwest National Laboratory) for support of portions of this research. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC06-76RLO-1830. This work was supported in part by a Public Service grant from the National Institutes of Health (AI 21640) (J.A.N.).
We also acknowledge Rick Zangar, Josh Adkins, and Joel Pounds for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adair, R., E. R. Douglas, J. B. Maclean, S. Y. Graham, J. D. Aitken, F. E. Jamieson, and D. J. Dargan. 2002. The products of human cytomegalovirus genes UL23, UL24, UL43 and US22 are tegument components. J. Gen. Virol. 83:1315-1324. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, J. N., S. M. Varnum, K. J. Auberry, R. J. Moore, N. H. Angell, R. D. Smith, D. L. Springer, and J. G. Pounds. 2002. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell Proteomics 1:947-955. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belov, M. E., G. A. Anderson, M. A. Wingerd, H. R. Udseth, D. C. Tang, K. R. Prior, M. A. Swanson, E. F. Buschbach, E. Stritmatter, R. J. Moore, and R. D. Smith. 2004. An automated high performance capillary liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometer for high-throughput proteomics. J. Am. Soc. Mass Spectrom. 15:212-232. [DOI] [PubMed] [Google Scholar]
- 5.Benko, D. M., R. S. Haltiwanger, G. W. Hart, and W. Gibson. 1988. Virion basic phosphoprotein from human cytomegalovirus contains O-linked N-acetylglucosamine. Proc. Natl. Acad. Sci. USA 85:2573-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogner, E., M. Reschke, B. Reis, T. Mockenhaupt, and K. Radsak. 1993. Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology 196:290-293. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C. P., D. H. Vesole, J. Nelson, M. B. Oldstone, and M. F. Stinski. 1989. Identification and expression of a human cytomegalovirus early glycoprotein. J. Virol. 63:3330-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Chelius, D., A. F. Huhmer, C. H. Shieh, E. Lehmberg, J. A. Traina, T. K. Slattery, and E. Pungor, Jr. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J. Proteome Res. 1:501-513. [DOI] [PubMed] [Google Scholar]
- 10.Dal Monte, P., S. Pignatelli, N. Zini, N. M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005-1012. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eng, J., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 14.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419:520-526. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, W. 1983. Protein counterparts of human and simian cytomegaloviruses. Virology 128:391-406. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 17.Giugni, T. D., C. Soderberg, D. J. Ham, R. M. Bautista, K. O. Hedlund, E. Moller, and J. A. Zaia. 1996. Neutralization of human cytomegalovirus by human CD13-specific antibodies. J. Infect. Dis. 173:1062-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy, J. E., J. A. McKeating, and P. D. Griffiths. 1987. Cytomegalovirus strain AD169 binds beta 2 microglobulin in vitro after release from cells. J. Gen. Virol. 68:777-784. [DOI] [PubMed] [Google Scholar]
- 19.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, M. T., and T. Compton. 1999. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J. Virol. 73:3886-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 22.Kaye, J. F., U. A. Gompels, and A. C. Minson. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693-2698. [DOI] [PubMed] [Google Scholar]
- 23.Landini, M. P., B. Severi, G. Furlini, and L. Badiali De Giorgi. 1987. Human cytomegalovirus structural components: intracellular and intraviral localization of p28 and p65-69 by immunoelectron microscopy. Virus Res. 8:15-23. [DOI] [PubMed] [Google Scholar]
- 24.Lasonder, E., Y. Ishihama, J. S. Andersen, A. M. Vermunt, A. Pain, R. W. Sauerwein, W. M. Eling, N. Hall, A. P. Waters, H. G. Stunnenberg, and M. Mann. 2002. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419:537-542. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. W., S. J. Berger, S. Martinovic, L. Pasa-Tolic, G. A. Anderson, Y. Shen, R. Zhao, and R. D. Smith. 2002. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. USA 99:5942-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehner, R., H. Meyer, and M. Mach. 1989. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J. Virol. 63:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 28.Lipton, M. S., L. Pasa-Tolic, G. A. Anderson, D. J. Anderson, D. L. Auberry, J. R. Battista, M. J. Daly, J. Fredrickson, K. K. Hixson, H. Kostandarithes, C. Masselon, L. M. Markillie, R. J. Moore, M. F. Romine, Y. Shen, E. Stritmatter, N. Tolic, H. R. Udseth, A. Venkateswaran, K. K. Wong, R. Zhao, and R. D. Smith. 2002. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 99:11049-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michelson, S., P. Turowski, L. Picard, J. Goris, M. P. Landini, A. Topilko, B. Hemmings, C. Bessia, A. Garcia, and J. L. Virelizier. 1996. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell-derived PP2A. J. Virol. 70:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegalovirus and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 32.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarov, I., and I. Abady. 1975. The morphogenesis of human cytomegalovirus. Isolation and polypeptide characterization of cytomegalovirions and dense bodies. Virology 66:464-473. [DOI] [PubMed] [Google Scholar]
- 34.Schmolke, S., H. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, R. D., G. A. Anderson, M. S. Lipton, C. Masselon, L. Pasa-Tolic, Y. Shen, and H. R. Udseth. 2002. The use of accurate mass tags for high-throughput microbial proteomics. Omics 6:61-90. [DOI] [PubMed] [Google Scholar]
- 36.Smith, R. D., G. A. Anderson, M. S. Lipton, L. Pasa-Tolic, Y. Shen, T. P. Conrads, T. D. Veenstra, and H. R. Udseth. 2002. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics 2:513-523. [DOI] [PubMed] [Google Scholar]
- 37.Spaete, R. R., R. C. Gehrz, and M. P. Landini. 1994. Human cytomegalovirus structural proteins. J. Gen. Virol. 75:3287-3308. [DOI] [PubMed] [Google Scholar]
- 38.Stannard, L. M. 1989. Beta 2 microglobulin binds to the tegument of cytomegalovirus: an immunogold study. J. Gen. Virol. 70:2179-2184. [DOI] [PubMed] [Google Scholar]
- 39.Varnum, S. M., C. C. Covington, R. L. Woodbury, K. Petritis, L. J. Kangas, M. S. Abdullah, J. G. Pounds, R. D. Smith, and R. C. Zangar. 2003. Proteomic characterization of nipple aspirate fluid: identification of potential biomarkers of breast cancer. Breast Cancer Res. Treat. 80:87-97. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W., H. Zhou, H. Lin, S. Roy, T. A. Shaler, L. R. Hill, S. Norton, P. Kumar, M. Anderle, and C. H. Becker. 2003. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75:4818-4826. [DOI] [PubMed] [Google Scholar]
- 41.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 42.Wright, J. F., A. Kurosky, E. L. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.