Abstract
DNA viruses can modulate the activity of cellular acetyltransferases to regulate virus gene expression and to affect cell cycle progression in order to support virus replication. A role for protein acetylation in regulating the nuclear export of the bipartite geminivirus DNA genome was recently suggested by the findings that the viral movement protein NSP, which shuttles the viral genome between the nucleus and the cytoplasm, interacts with a novel Arabidopsis acetyltransferase, AtNSI, and the increased expression of AtNSI enhances susceptibility to Cabbage leaf curl virus infection. To further investigate the interaction of NSP and AtNSI and to establish the importance of this interaction in virus infections, we used a reverse yeast two-hybrid selection and deletion analysis to identify NSP mutants that were impaired in their ability to bind AtNSI. These mutants identified a 38-amino-acid region of NSP, to which no function had so far been assigned, as being necessary for NSP-AtNSI interaction. Three NSP missense mutants were analyzed in detail and were found to be comparable to wild-type NSP in their levels of accumulation, nucleocytoplasmic shuttling, DNA binding, and cooperative interaction with the viral cell-to-cell movement protein MP. Despite this, Cabbage leaf curl virus that expressed each mutated NSP was defective in its ability to infect Arabidopsis, exhibiting lower levels of infectivity than the wild-type virus, and delayed systemic spread of the virus and attenuated disease symptoms. Our data demonstrate the importance of the interaction of NSP with AtNSI for virus infection and pathogenicity.
Successful infection by a virus depends on its ability to express its gene products, replicate, and move to adjacent cells. For plant viruses, this last step requires the action of virus-encoded movement proteins to coordinate the replication of the viral genome with its cell-to-cell transport and to overcome the barrier of the plant cell wall (27, 50). Bipartite geminiviruses (Begomovirus) such as Cabbage leaf curl virus (CLCV) and Squash leaf curl virus (SqLCV), with their single-stranded DNA (ssDNA) genomes that replicate in the nucleus, accomplish this through the cooperative interaction of two movement proteins: the nuclear shuttle protein NSP and the cell-to-cell movement protein MP. NSP is a ssDNA binding protein responsible for transporting replicated viral genomes between the nucleus and the cytoplasm (39, 46, 56). MP traps these complexes in the cytoplasm and directs them to and across the cell wall (36, 47, 57). In the newly invaded cells, NSP-DNA complexes are released, and NSP targets the viral genome back to the nucleus to initiate new rounds of replication and infection (46).
Although it is well recognized that movement proteins facilitate cell-to-cell transport of the viral genome by modifying plasmodesmata (37), the complex transwall channels that connect adjacent plant cells, little is known about how movement proteins reach plasmodesmata or the host factors that regulate movement protein function. Subcellular localization studies have shown that SqLCV and CLCV MPs target to the cortical endoplasmic reticulum (ER) (47, 57; J. D. Lewis and S. G. Lazarowitz, unpublished data) and that the Tobacco mosaic virus (TMV) 30-kDa movement protein can associate with microtubules and actin filaments as well as with the ER (19, 31, 33). Gillespie and colleagues (16), in a recent study on the accelerated movement of a TMV mutant encoding a functionally enhanced 30-kDa movement protein [MP(R3)], reported that in contrast to the prevailing view that the interaction of the 30-kDa protein with microtubules is crucial for cell-to-cell transport of the viral RNA genome (19, 31, 33), movement protein interactions with the cortical ER are important and cell-to-cell trafficking of TMV can occur independently of microtubules. The interaction with microtubules appears to downregulate the levels of TMV movement protein, acting to draw the 30-kDa protein away from the ER and target it for proteasome degradation via a well-recognized cellular pathway (16). Consistent with this, Kragler and colleagues (26) found that the TMV 30-kDa protein interacts with a microtubule-associated protein which does not seem to be required for movement but does appear to negatively regulate the process.
A number of groups have used protein-protein interaction assays to identify host proteins that are potentially relevant to virus movement. Using a gel-overlay assay or a yeast two-hybrid screen, two groups independently identified a cell wall pectin methylesterase (PME) based on its interaction with the TMV 30-kDa protein (9, 11). Yeast two-hybrid screens have also been used to identify a variety of host proteins that interact with a range of viral movement proteins. These have included DnaJ family members that interact with Tomato spotted wilt virus NSm (48); an apparent plant homolog of the rat vesicular trafficking protein PRA1 reported to interact with the Cauliflower mosaic virus (CaMV) gene I-encoded protein (21); a protein containing RGD motifs characteristically found in several extracellular animal proteins that interact with several members of the integrin family of cell surface matrix receptors, which was reported to interact with the Turnip crinkle virus p8 protein (29); a homeodomain-containing protein reported to interact with Tomato bushy stunt virus P22 (10); three ankyrin-repeat-containing proteins that can interact with the smallest (12K) of the triple gene block movement proteins encoded by Potato virus X (12); a receptor-like protein kinase that binds to Tomato golden mosaic virus and Tomato crinkle leaf yellows virus NSP (30); and a plant-specific acetyltransferase, AtNSI, that interacts with CLCV and SqLCV NSP (32). However, only two of these studies demonstrated the relevance of the reported interaction to virus movement and infectivity in vivo. TMVs expressing deleted forms of the 30-kDa protein impaired in the ability to bind the identified PME are defective in cell-to-cell movement and are not infectious, and TMV systemic spread is delayed in tobacco plants when PME expression is suppressed (8, 9). CLCV infectivity is enhanced on transgenic 35S::AtNSI Arabidopsis lines that overexpress AtNSI (32).
The nuclear trafficking of NSP requires two classic basic nuclear localization sequences (NLS-A and NLS-B) (46) and a leucine-rich nuclear export sequence (NES) of the type found in rapidly shuttling nuclear proteins such as human immunodeficiency virus Rev and Xenopus laevis TFIIIa (Fig. 1) (56). NSP function is also highly regulated by posttranslational modifications and through its interactions with other viral proteins, which include the viral coat protein (CP) as well as MP (24, 39, 41). SqLCV CP can bind, without encapsidating, replicated progeny ssDNA genomes to sequester them away from the nuclear replication pool and make them available for NSP-mediated export from the nucleus (41). It is this synergy between CP and NSP that AtNSI, through its interaction with NSP, appears to regulate.
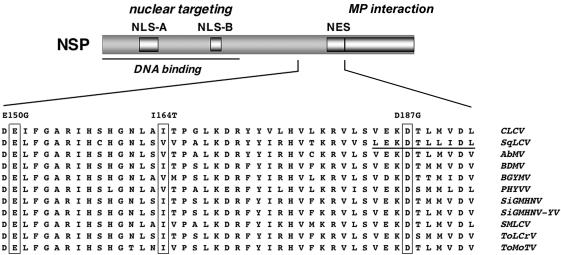
FIG. 1.
Functional regions of NSP. (Top) Functional domain map of NSP showing locations of the bipartite (NLS-A) and simian virus 40 large T antigen-type (NLS-B) NLSs, the NES, and the DNA binding and MP interaction domains. (Bottom) Thirty-eight amino acid region of CLCV NSP implicated in AtNSI binding (this study) aligned with the corresponding region from NSP encoded by the bipartite geminiviruses SqLCV, Abutilon mosaic virus (West India) (AbMV), Bean dwarf mosaic virus (BDMV), Bean golden yellow mosaic virus (BGYMV), Pepper huasteco yellow vein virus (PHYVV), Sida golden mosaic Honduras virus (SiGMHNV), Sida golden mosaic Honduras virus yellow vein strain (SiGMHNV-YV), Squash mild leaf curl virus (SMLCV), Tomato leaf crumple virus (ToLCrV), and Tomato mottle Taino virus (ToMoTV). The NES in the SqLCV sequence is underlined. Three residues identified by missense mutations in this study are boxed.
AtNSI is a nuclear acetyltransferase that is unique to and highly conserved among plants (32). Although the sequence of its acetyltransferase domain places AtNSI in the GCN5 family of nuclear histone acetyltransferases, it is distantly related to these transcriptional coactivators and other characterized nuclear histone acetyltransferases. Notably, AtNSI lacks a bromodomain and other regions that are required for binding to histones and, consistent with this, AtNSI does not act as a transcriptional coactivator when tested in a nucleosome-templated assay in vitro, using two different promoters (32). Among the bipartite geminivirus nuclear proteins, AtNSI specifically acetylates CLCV CP in vitro, although CP does not stably interact with AtNSI. These findings, and the increased efficiency of CLCV infection on transgenic Arabidopsis lines that overexpress AtNSI, suggest that AtNSI activity is important for NSP to displace CP and bind progeny genomes for export from the nucleus (32). We have proposed that NSP bound to newly replicated viral ssDNA recruits AtNSI to a ternary complex to acetylate genome-bound CP. This would disrupt CP-ssDNA binding and allow NSP to displace CP as NSP cooperatively binds the viral genome to export it from the nucleus (32).
To identify the region of NSP that interacts with AtNSI and to establish that this interaction is required for virus infection, we used a reverse yeast two-hybrid selection and deletion analysis to identify CLCV NSP mutants that were defective in AtNSI binding. We report here that the 38-amino-acid (aa) region between CLCV NSP residues 150 and 187 is necessary for AtNSI binding. Three NSP missense mutants that were impaired in their ability to bind AtNSI were analyzed in detail and found to be comparable to wild-type NSP in their stability in plant cells, nucleocytoplasmic shuttling, DNA binding properties, and ability to interact with CLCV MP. Nevertheless, CLCV that was engineered to express each of these mutated forms of NSP showed reduced levels of infection on Arabidopsis, accumulated lower levels of viral DNA in systemic leaves, and produced delayed and attenuated symptoms, and the extent of these defects generally correlated with the measured defects in NSP binding to AtNSI. Our studies establish the important role of NSP-AtNSI interaction and AtNSI activity in bipartite geminivirus infection and pathogenicity.
MATERIALS AND METHODS
Yeast reverse two-hybrid assay.
The AtNSI coding region was fused to that of the GAL4 activation domain (GAD) in pBI771 to create pGAD-AtNSI. pGBD-NSP contained the NSP coding region fused to the coding region for the GAL4 DNA binding domain in pBI880 (44). N- and C-terminal truncations of NSP were generated by PCR amplification, with pGBD-NSP as a template and primers designed to make in-frame fusions between internal NSP sites and the GAL4 DNA. These constructs were cotransformed with pGAD-AtNSI into Saccharomyces cerevisiae strain PJ69-4A (MATa gal4 gal80 his3Δ200 leu2-3,112 trp1-Δ901 LYS2::GAL1-HIS3 ade2::GAL2-ADE2 met::GAL7-lacZ) and then plated on dropout medium (15). Histidine prototrophs were recovered and further analyzed in medium lacking adenine or histidine and supplemented with 5 mM 3-aminotriazole. Analyses of β-galactosidase (β-Gal) activity were performed by use of a filter lift assay (34).
Single amino acid changes that disrupted the interaction of NSP and AtNSI were identified in a reverse two-hybrid screen using yeast strain MaV203 (MATα leu2-3,112 trp1-901 his3Δ200 ade2-101 cyh2r can1r gal4Δ gal80Δ GAL1::lacZ HIS3UASGAL1::HIS3@LYS2 SPAL10UASGAL1::URA3; Invitrogen) as previously described (6, 51, 53). Mutagenic PCR was performed without MnCl2, using pBI880-NSP as a template and with primers annealing ∼200 bp upstream and downstream of the NSP coding region to allow for homologous recombination. Transformants were plated on synthetic complete medium lacking leucine and tryptophan (SC-L-T) containing 0.1% 5-fluoroorotic acid (5-FOA), and a library was generated and further characterized. Selected clones were grown on SC-L-T, and DNAs were extracted (2) and retransformed into yeast strains MaV203 and PJ69-4A. The clones were analyzed by plating on SC-L-T with 0.1% 5-FOA (MaV203) or on SC-H-L-T with or without 5 mM 3-aminotriazole (PJ69-4A). Liquid β-Gal assays were performed with strain PJ69-4A as reported by Montano (34). Cultures grown to saturation in dropout medium were serially diluted 10-fold in sterile water, and 5 μl of each dilution was assayed.
GST pull-down and ssDNA binding assays.
Wild-type or mutated NSP, each cloned as a transcriptional fusion to the T7 promoter in pGEM7Z(+), were labeled with [35S]methionine by coupled in vitro transcription and translation, using the TnT reticulocyte lysate system according to the manufacturer's instructions (Promega). For in vitro protein binding, 5 μl of the appropriate [35S]Met-labeled NSP protein was diluted with a solution containing 20 mM HEPES (pH 7.4), 150 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, 0.1% Nonidet P-40, and 0.1% gelatin (35) to a final volume of 150 μl and then incubated with 2 μg of purified glutathione S-transferase (GST) or GST-AtNSI tethered to glutathione-Sepharose for 2 h at 4°C with gentle agitation (32). After four 10-min washes with 1 ml of phosphate-buffered saline-dithiothreitol (35), the samples were denatured in sodium dodecyl sulfate and resolved by electrophoresis on 12% polyacrylamide gels (SDS-12% PAGE) (45) and detected by autoradiography using En3Hance (Perkin-Elmer).
In vitro binding reactions of NSP and its mutants to ssDNA-cellulose (USB) were performed as previously described (39).
Transient expression studies in protoplasts and leaves.
To determine the subcellular localization of the NSP missense mutants, we generated N-terminal green fluorescent protein (GFP) fusions by PCR to translationally fuse the coding sequence for each mutant to that for GFP in the vector pCKGFPS65C, which drives transcription from the enhanced 35S promoter (42). Each fusion was constructed with an NH2-Gly-Gly-Gly-Gly-Ser-COOH linker between NSP-HIS6 and GFP. p35S::MPB was created by using NcoI and BamHI to insert the coding region for MP in place of that for GFP in pCKGFPS65C. For NSP accumulation studies, the NSP-HIS6 coding sequence was cloned in place of that for GFP in pCKGFPS65C by the use of NcoI and XbaI to create p35S::NSP-HIS6.
Arabidopsis leaf protoplasts were prepared and transfected by using polyethylene glycol (PEG) as described previously (1). For preparations of Nicotiana benthamiana protoplasts, leaf strips (∼2 mm long) were floated on 8 mM CaCl2 in 0.66 M mannitol for 30 min and digested overnight by gentle shaking (30 rpm on a platform shaker) in 0.2% cellulase and 0.05% macerozyme in the dark. Cells were then released from undigested tissue by increasing the speed to 80 rpm for 10 min and were collected by low-speed centrifugation in an IEC clinical centrifuge (model CL) (2 min, 210 × g). The protoplasts were resuspended in 8 mM CaCl2 in 0.66 M mannitol, layered over a cushion of 21% sucrose in water, and centrifuged for 8 min at 440 × g, after which the intact cells were collected from the interface (S. M. Gray, personal communication). After a wash in 8 mM CaCl2 in 0.66 M mannitol, the protoplasts were washed, for 5 min each, in 0.53, 0.46, and finally 0.4 M mannitol and then transfected by using PEG (1). All of these and subsequent centrifugation steps were performed for 2 min in an IEC CL centrifuge at 70 × g. For visualization of GFP fusion proteins, live protoplasts were placed on glass slides under coverslips and were imaged with a Leica TCS SP2 confocal laser scanning microscope.
For agroinfiltration of N. benthamiana leaves, the complete NSP expression cassette was excised from each p35S:NSP-HIS6 construct by digestion with HindIII (see above) and then was cloned into pCB301 (59). These clones were transformed into Agrobacterium tumefaciens strain GV3101 (25) by electroporation. The cultures were grown to an optical density of 0.5, and agroinfiltration was performed as previously described (54). Samples were collected after 72 h and processed as described by Weigel and Glazebrook (58). To determine the levels of NSP accumulation, protoplasts were collected after 24 h and resuspended in Tris-EDTA containing 50 mM NaCl. After the addition of SDS-containing loading buffer and heating for 5 min at 100°C, the samples were resolved by SDS-PAGE on 12.5% gels, blotted onto nitrocellulose, and detected with an anti-His6 monoclonal antibody (Invitrogen) by chemiluminescence (Amersham Biosciences) (38, 45).
Infectivity assays.
The CLCV A and B components, including two common regions, were PCR amplified from pGEM7Zf(+) vectors (20), and each of these was cloned separately into the XbaI and KpnI sites of the binary vector pCB301 (59). For the introduction of each NSP missense mutation, the 546-bp AclI-HpaI fragment from NSPE150G, NSPI164T, or NSPD187G was cloned in place of the corresponding fragment in wild-type NSP. The resulting constructs were transformed into A. tumefaciens strain GV3101 (25) by electroporation and were used to inoculate Arabidopsis ecotype Col-0 12-day-old seedlings. For the inoculation procedure, the first two true leaves were cut transversally with a razor blade and dipped for 5 min into a suspension of GV3101, which were overnight cultures concentrated 50× and mixed to contain the appropriate CLCV A and B component clones. Plants were grown at 22°C under a 16 h-8 h light-dark cycle. DNAs were extracted from systemically infected leaves by use of a DNeasy plant kit (Qiagen) as directed by the manufacturer and were quantified by semiquantitative PCR using NSP- or AtNSI-specific (internal control) primers (58).
DNA sequence analysis. DNA sequence analysis was performed by the Cornell University BioResource Center on an Applied Biosystems Automated 3730 DNA analyzer.
RESULTS
A 38-aa region of NSP is needed for AtNSI binding.
Based on mutational analyses and transient expression assays in tobacco protoplasts, NSP has a striking domain structure in which each functional region can act independently (46, 56). With SqLCV NSP as a model, five functional elements have been identified. Two essential basic NLSs, a bipartite NLS (residues 21 to 42) and a simian virus 40 large T antigen-type NLS (residues 81 to 96), are located in the N-terminal region of NSP within the DNA binding domain (residues 39 to 109) (22, 46). The C-terminal 55 aa contains the domain required for interaction with MP (residues 200 to 254), just upstream of which is the essential NES (residues 184 to 194) (46, 56). Given the high degree of sequence similarity among NSP proteins encoded by bipartite geminiviruses, the same functional elements can potentially be identified in CLCV NSP (Fig. 1). To date, no functions have been mapped to the central region of NSP located between residues 109 and 184.
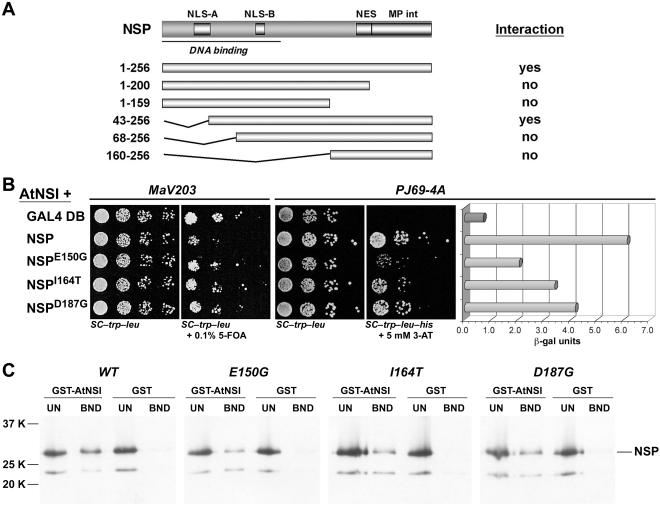
To initially identify the regions of NSP that interact with AtNSI, we tested N- and C-terminal truncations of CLCV NSP for their ability to interact with AtNSI in a classic yeast two-hybrid interactive screen or an in vitro GST pull-down assay. Both assays gave the same results. In the yeast two-hybrid screen, both full-length NSP and NSP lacking the first 42 aa (NSP43-256), when fused to the GAL4 DNA binding domain, interacted with AtNSI fused to the GAD (Fig. 2A). None of the other NSP truncations interacted with AtNSI in this assay, indicating that the N-terminal 42 aa of NSP were dispensable for interacting with AtNSI. The same results were obtained using the in vitro GST pull-down assay, in which full-length NSP and NSP43-256 were the only forms that specifically interacted with GST-AtNSI tethered to glutathione-Sepharose resin (Fig. 2A).
FIG. 2.
Mutations within NSP residues 150 to 187 affect AtNSI interaction. (A) NSP truncations tested for interaction with AtNSI in a yeast two-hybrid assay or an in vitro GST-AtNSI pull-down assay. The results of both assays were identical (interaction column). (B) Interactions of wild-type CLCV NSP or NSP missense mutants with AtNSI (AtNSI+) as determined in a yeast reverse two-hybrid assay (S. cerevisiae strain MaV203) and the classic two-hybrid assay (growth in and β-Gal assays of extracts from S. cerevisiae strain PJ69-4A). NSPE150G, NSPI164T, and NSPD187G were defective in their interaction with AtNSI based on the increased growth of MaV203 coexpressing pGAD-AtNSI in the presence of 0.1% 5-FOA and the decreased growth on SC-Trp-Leu-His and decreased β-Gal activity of PJ69-4A coexpressing pGAD-AtNSI. GAL4 DB, the GAL4 binding domain empty bait vector, a control that shows the level of MaV203 growth in the presence of 0.1% 5-FOA and the complete lack of histidine prototrophy for PJ69-4A. (C) Binding of wild-type NSP and NSP missense mutants to GST-AtNSI in vitro. CLCV NSP, NSPE150G, NSPI164T, and NSPD187G were labeled with [35S]Met by coupled in vitro transcription and translation, and equal amounts were incubated with a standardized amount of GST-AtNSI or GST alone, each tethered to glutathione-Sepharose. After being washed, bound NSP was analyzed by SDS-PAGE on 12.5% gels. Seventeen percent of the unbound fractions (UN) and all of the bound (BND) fractions were analyzed. The positions of molecular weight markers are indicated. Full-length NSP migrates at ∼33K. The faster migrating band corresponds to an internal in-frame AUG start within the CLCV NSP coding sequence.
To more precisely map the region within NSP that is required for interaction with AtNSI, we used a reverse yeast two-hybrid system in which disruption of a known protein-protein interaction is detected based on selection against the expression of a deleterious gene, in this case URA3, which confers sensitivity to 5-FOA when yeast cells are grown in the presence of uracil (28, 53). An advantage of this approach is its unbiased nature. By screening a PCR-based randomly mutagenized library of CLCV NSP, we could identify single amino acids that are required for the interaction with AtNSI. In addition, weakly interacting as well as noninteracting mutants can be obtained.
We randomly mutagenized NSP fused to the GAL4 DNA binding domain in pBI880 by PCR, using Taq polymerase in the absence of MnCl2 and primers that annealed ∼200 nucleotides upstream and downstream of the NSP insert. Gap repair transformation with linearized pBI880 was then used to introduce the PCR products into yeast strain MaV203, which contained the AtNSI coding region fused to the GAD in plasmid pBI771 (pBI771-AtNSI) (6), and mutants were selected by growth on dropout medium containing 0.1% 5-FOA. To confirm the results, we transformed plasmid DNAs isolated from surviving colonies into MaV203 harboring pBI771-AtNSI and retested them for growth on 0.1% 5-FOA. The isolated DNAs were also transformed into yeast strain PJ69-4A harboring pBI771-AtNSI to quantify the degree to which the NSP-AtNSI interaction was impaired based on the levels of β-Gal activity, and each NSP insert was sequenced to identify the mutation(s).
We initially screened for NSP mutants that had completely lost the ability to interact with AtNSI, based on the most vigorous growth of MaV203 on 5-FOA in the reverse yeast two-hybrid screen and only background levels of β-Gal activity in the classic interactive yeast two-hybrid screen with strain PJ69-4A. These screens generated only truncations or deletions of NSP. We therefore concentrated on characterizing NSP mutants that were defective, but not fully null, in their interaction with AtNSI, as evidenced by their decreased but detectable levels of β-Gal activity. We recovered three NSP missense mutants that were impaired in their ability to interact with AtNSI, specifically NSPE150G, NSPD187G, and the double mutant NSPF29S/I164T (Fig. 1 and 2). Subcloning of each mutation in NSPF29S/I164T showed that NSPI164T was responsible for the impaired interaction with AtNSI (Fig. 2). Based on their β-Gal activities as well as the relative levels of growth of PJ69-4A on selective media, NSPE150G was the most defective mutant in its ability to interact with AtNSI, followed by NSPI164T and NSPD187G, with the last being the least defective in its interaction with AtNSI (Fig. 2B).
These findings were confirmed biochemically with an in vitro GST pull-down assay in which [35S]Met-labeled in vitro-transcribed and -translated wild-type NSP, or each NSP missense mutant, was incubated with GST-AtNSI or GST tethered to glutathione-Sepharose resin (Fig. 2C). In this more direct in vitro binding assay, in which equal amounts of NSPE150G, NSPI164T, and NSPD187G were directly compared with wild-type NSP for their ability to bind to a standardized amount of AtNSI, all three missense mutants exhibited notably reduced levels of binding to AtNSI. These in vitro binding results correlated qualitatively with those of the yeast two-hybrid screen and β-Gal assays in showing that NSPE150G, NSPI164T, and NSPD187G were each defective in binding AtNSI. These findings, combined with the analysis of NSP truncations, showed that the 38-residue region of NSP from aa 150 to 187 was necessary for binding to AtNSI. The mutations in NSPE150G and NSPI164T lie within a region of the protein to which no function had so far been assigned (residues 120 to 183) (46, 56). The mutation in NSPD187G is located within the NES (aa 184 to 193), although it affects a residue that has not been shown to be essential for NES function (see below) (13, 56).
CLCV expressing NSPE150G, NSPI164T, or NSPD187G exhibits lower levels of infectivity and attenuated symptoms on Arabidopsis.
To demonstrate that the NSP-AtNSI interaction is necessary for virus infection in vivo, we introduced each of the missense mutations in NSPE150G, NSPI164T, and NSPD187G into the CLCV B component, and the resulting mutated viral DNAs were coinoculated with the wild-type CLCV A component onto 12-day-old Arabidopsis seedlings (Col-0) by agroinoculation (20). Each of the NSP mutants showed lower levels of infection on Arabidopsis (Table 1; Fig. 3A) than wild-type CLCV, which had an average infectivity level of 97%, as assessed by the appearance of disease symptoms. CLCV-NSPI164T always gave somewhat higher levels of infection (85% on average) than CLCV-NSPE150G and CLCV-NSPD187G, which had average infectivity levels of 78 and 75%, respectively.
TABLE 1.
Infectivities of CLCV NSP mutants on Arabidopsis plants
| NSP mutation | No. of symptomatic plants/no. of inoculated plants (%)
|
|||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | % infectivity (SD) | |
| None (wild type) | 43/45 (96) | 40/41 (98) | 44/45 (98) | 97 (1.23) |
| E150G | 33/45 (73) | 30/42 (71) | 40/45 (89) | 78 (9.58) |
| I164T | 36/45 (80) | 38/44 (86) | 42/45 (93) | 86 (6.67) |
| D187G | 32/45 (71) | 30/42 (71) | 37/45 (82) | 75 (6.33) |
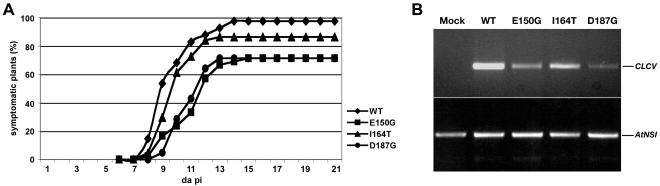
FIG. 3.
CLCV expressing missense mutant NSPE150G, NSPI164T, or NSPD187G is less infectious than wild-type (WT) CLCV on Arabidopsis (Col-0). (A) Percentages of symptomatic plants appearing at different days postagroinoculation with wild-type or mutated CLCV, as indicated. The results for one typical experiment are shown (see Table 1). (B) Semiquantitative PCR of DNA extracts from Arabidopsis leaves systemically infected with wild-type CLCV or CLCV NSP missense mutants, as indicated. CLCV DNA was amplified with NSP-specific primers; AtNSI DNA (loading control) was amplified with gene-specific primers. Mock, results for DNA extracts from Arabidopsis plants inoculated with GV3101 containing the empty binary vector.
The relative decreases in the levels of infection for the three NSP missense mutants were confirmed by using semiquantitative PCR to quantify the relative amounts of viral DNA that accumulated in systemically infected leaves at 15 days postinoculation. Consistent with the observed relative infectivity levels, CLCV-NSPE150G, CLCV-NSPI164T, and CLCV-NSPD187G each accumulated lower levels of viral DNA in systemically infected leaves than did wild-type CLCV (Fig. 3B). Among the three NSP missense mutants, CLCV-NSPI164T accumulated the highest levels of DNA in systemically infected leaves, followed by CLCV-NSPE150G and CLCV-NSPD187G, which correlated with their relative levels of infection (Fig. 3). To demonstrate that each of the identified NSP missense mutations was responsible for the observed infectivity defects, we also PCR amplified the entire NSP coding region from total DNA extracts of systemically infected leaves, using gene-specific primers, and sequenced the products. No second site mutations were present in any of the three NSP mutants. Thus, the individual missense mutations in NSPE150G, NSPI164T, and NSPD187G were each responsible for the infectivity defects that we observed.
Correlating with their relative defects in infectivity levels, each of the NSP mutants also produced attenuated disease symptoms (Fig. 4). For each mutant, the degree of attenuation in symptoms was correlated with the relative decrease in systemic levels of viral DNA (Fig. 3B). CLCV-NSPI164T, which was the most infectious of the mutants based on both its efficiency of infection and its systemic accumulation of viral DNA, produced the most severe symptoms, although these were clearly milder than those produced by wild-type CLCV. CLCV-NSPD187G produced the mildest symptoms, with the symptoms produced by CLCV-NSPE150G being intermediate in severity between those produced by CLCV-NSPI164T and CLCV-NSPD187G (Fig. 4). Fitting with their striking attenuation of disease symptoms and lower infectivity levels, CLCV-NSPE150G and CLCV-NSPD187G also exhibited delays of 1 to 2 days in the initial appearance of disease symptoms (Fig. 3A).
FIG. 4.
CLCV expressing missense mutant NSPE150G, NSPI164T, or NSPD187G produces attenuated symptoms on Arabidopsis (Col-0). The images show pots of Arabidopsis plants at 21 days postagroinoculation with wild-type CLCV (WT) or CLCV expressing each NSP missense mutant, as indicated. Mock, Arabidopsis plants 21 days after inoculation with GV3101 containing the empty binary vector.
NSPE150G, NSPI164T, and NSPD187G have wild-type stabilities, DNA binding properties, and nuclear accumulation.
The infectivity defects of CLCV-NSPE150G, CLCV-NSPI164T, and CLCV-NSPD187G could have been due to decreased levels of accumulation of the mutated NSP proteins or to defects in DNA binding or nuclear targeting rather than to their defective interactions with AtNSI. To distinguish between these possibilities, we used transient expression assays in N. benthamiana protoplasts and leaves, or Arabidopsis protoplasts, to examine the stability and nuclear targeting of each NSP mutant. We also tested the ability of each mutated NSP to bind ssDNA in vitro.
The relative levels of NSP accumulation were determined by transient expression assays in N. benthamiana-transfected protoplasts or agroinfiltrated leaves in which the wild-type or mutated forms of NSP were each expressed as a transcriptional fusion to the CaMV 35S promoter and detected by Western blot analyses of cell extracts. Since our available NSP antisera were raised against SqLCV NSP and weakly cross-reacted with CLCV NSP, we cloned each NSP construct as a translational fusion to a C-terminal His6 tag and used an anti-His6 monoclonal antibody for these studies. We did not use GFP-NSP fusions for these analyses as the GFP moiety could have stabilized the mutated forms of NSP. In both of these transient assays, the levels to which CLCV NSPE150G-HIS6, NSPI164T-HIS6, and NSPD187G-HIS6 accumulated were comparable to the levels of accumulation of wild-type CLCV NSP-HIS6, and there was no direct correlation between these relative levels of accumulation and the infectivity levels for each NSP mutant (Fig. 5A and data not shown).
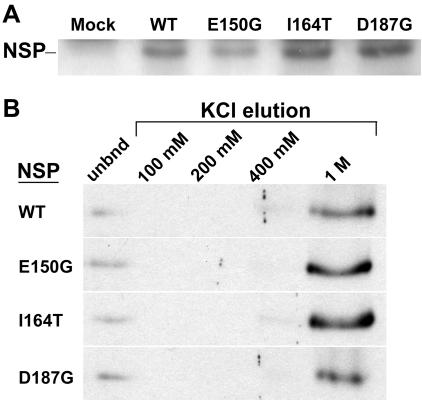
FIG. 5.
Stabilities and DNA binding properties of wild-type CLCV NSP (WT) and missense mutants NSPE150G, NSPI164T, and NSPD187G. (A) Immunoblot of extracts from N. benthamiana leaves that were agroinfiltrated with wild-type NSP-HIS6, NSPE150G-HIS6, (E150G), NSPI164T-HIS6 (I164T), NSPD187G-HIS6 (D187G), or GV3101 containing the empty binary vector (mock). The extracts were analyzed by SDS-PAGE on 12.5% gels, blotted onto nitrocellulose, and probed with an anti-His6 monoclonal antibody. (B) Equal amounts of wild-type CLCV NSP and missense mutants NSPE150G, NSPI164T, and NSPD187G, labeled with [35S]Met by coupled in vitro transcription and translation, were incubated with a standardized amount of denatured calf thymus ssDNA bound to cellulose in Z-50 buffer and then eluted by sequential washing with Z buffer containing increasing concentrations of KCl, as indicated. Equal amounts (5%) of the unbound (unbnd) fractions and each eluted NSP fraction were analyzed by SDS-PAGE on 12.5% gels.
To determination the ability of NSPE150G, NSPI164T, and NSPD187G to bind ssDNA, equal amounts of [35S]Met-labeled in vitro-transcribed and -translated wild-type NSP, or each NSP missense mutant, was incubated with ssDNA-cellulose in DNA binding buffer Z (50 mM Tris-HCl [pH 8.0], 12.5 mM MgCl2, 1 mM EDTA, 0.1% Nonidet P-40, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) containing 50 mM KCl (Z-50 buffer), and the bound NSP was eluted by sequential washing in Z buffer containing increasing concentrations of KCl (39, 41). As shown in Fig. 5B, NSPE150G, NSPI164T, and NSPD187G were indistinguishable from wild-type NSP in their binding affinities for ssDNA. All of the proteins bound a standardized amount of ssDNA-cellulose to the same extent (∼90% of the input). A minor amount of wild-type or each mutated NSP eluted in 400 mM KCl, with the vast majority of the bound protein eluting in 1 M KCl (Fig. 5B).
To assess the ability of NSPE150G, NSPI164T, and NSPD187G to target the nucleus, we fused the N terminus of each missense mutant and wild-type NSP in frame to GFP and transiently expressed each protein from the CaMV 35S promoter in transfected N. benthamiana or Arabidopsis (Col-0) protoplasts. Examination by confocal laser scanning microscopy (CLSM) showed that in contrast to free GFP, which was found throughout the cells, CLCV GFP-NSPE150G, GFP-NSPI164T, and GFP-NSPD187G all accumulated in the nuclei of both cell types, and based on the total number of fluorescing cells, they did so to the same extent as wild-type CLCV GFP-NSP (Fig. 6A and data not shown). No differences in the timing or apparent level of nuclear accumulation of each NSP missense mutant compared to wild-type NSP were detected. Fifty to sixty percent of transfected N. benthamiana protoplasts and ∼15 to 20% of transfected Arabidopsis protoplasts showed nuclear accumulation of the wild type or each mutated GFP-NSP fusion, with nuclear fluorescence first appearing at 14 to 16 h posttransfection and peaking at ∼24 h posttransfection. Both N- and C-terminal NSP fusions to GFP behaved the same in this assay, completely accumulating in the nucleus.
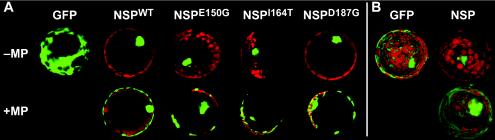
FIG. 6.
Subcellular localization of GFP or N-terminal GFP fusions of wild-type CLCV NSP (NSPWT), NSPE150G, NSPI164T, or NSPD187G when expressed alone (−MP) or together with (+MP) CLCV MP in transiently transfected N. benthamiana protoplasts. (A) CLSM of typical individual protoplasts. (B) Superimposed CLSM Z series for GFP or wild-type CLCV GFP-NSP, showing nuclear localization of GFP-NSP when expressed alone (top) and its relocalization to the cortical ER when coexpressed with CLCV MP (bottom). The GFP distribution was not altered by the coexpression of MP. Green, GFP fluorescence; red, chlorophyll autofluorescence showing the locations of chloroplasts.
NSPE150G, NSPI164T, and NSPD187G exit from the nucleus and interact with CLCV MP.
In addition to entering the nucleus, NSP has to exit from the nucleus and interact with MP. Defects in either of these stages could be responsible for the infectivity defects of the three NSP missense mutants. The former was of particular concern for NSPD187G since the mutation was within the NSP NES (56).
To show that NSPE150G, NSPI164T, and NSPD187G were not defective in either nuclear export or their interaction with MP, we used the ability of MP, when it is coexpressed with NSP in protoplasts, to trap NSP in the cytoplasm and redirect it to the cortical ER to examine both steps simultaneously (46, 47). Previous studies of SqLCV have shown that NSP mutants that are defective in nuclear export or in the ability to interact with MP will remain in the nucleus when coexpressed with wild-type MP (46, 56). N. benthamiana protoplasts were cotransfected with plasmids that expressed CLCV MP and either wild-type CLCV GFP-NSP, or CLCV GFP-NSPE150G, GFP-NSPI164T, or GFP-NSPD187G, and were examined at different times posttransfection for the accumulation of mutated or wild-type GFP-NSP at the cortical ER due to the presence of MP. Protoplasts singly transfected with each GFP-NSP construct served as controls. As early as 16 to 24 h posttransfection, the wild type and each of the mutated CLCV GFP-NSP proteins were seen to be redirected from the nucleus to the cortical ER in the presence of CLCV MP (Fig. 6A). No differences were observed in the timing of this interaction or in the relative levels to which wild-type CLCV GFP-NSP, or GFP-NSPE150G, GFP-NSPI164T, or GFP-NSPD187G accumulated at the cortical ER when coexpressed with MP (Fig. 6A and data not shown). In earlier SqLCV studies, the interaction of NSP with MP in the cortical region of transfected cells was detected by indirect immunofluorescence staining of fixed protoplasts, in which the ER structure was not well preserved (46, 56). The use of GFP-NSP fusions to monitor the interactions allowed us to clearly demonstrate the interaction of CLCV GFP-NSP with CLCV MP at the cortical ER by examining a superimposed confocal Z series taken of live protoplasts (Fig. 6B).
Thus, CLCV NSPE150G, NSPI164T, and NSPD187G are not defective in nuclear export, nor are they defective in their ability to interact with CLCV MP. These findings, combined with the facts that NSPE150G, NSPI164T, and NSPD187G behave like wild-type CLCV NSP in their stabilities and nuclear accumulation in plant cells and their in vitro ssDNA binding properties and that each individual missense mutation was responsible for the observed infectivity defects, lead us to conclude that the NSP-AtNSI interaction is necessary for CLCV infection.
DISCUSSION
Bipartite geminivirus cell-to-cell movement is a highly regulated process in which the nuclear shuttle protein NSP and its interactions with a variety of both cellular and viral proteins are of key importance. NSP, which has the characteristic properties of a ssDNA binding protein (39), interacts with the nuclear import and export machinery of the cell to move the viral ssDNA genome between its nuclear replication site and the cytoplasm (46, 47), and it is trapped in the cytoplasm and directed to carry the viral genome across the cell wall through a cooperative interaction with MP (36, 43, 47). The balance between replication and nuclear export of the ssDNA genome appears to be controlled through NSP binding to the viral genome and the plant-specific nuclear acetyltransferase AtNSI to target genome-bound CP, which sequesters viral ssDNA away from the replication pool, for acetylation (32, 41). Recent studies are consistent with a model in which NSP, bound to newly replicated ssDNA, recruits AtNSI to a ternary complex to acetylate CP, which would decrease the avidity of CP-ssDNA binding and thus lead to CP displacement as NSP cooperatively binds to the viral genome (32). For the studies reported here, we used a reverse yeast two-hybrid screen as an unbiased approach to identify CLCV NSP missense mutants that were defective in AtNSI binding. We further showed that their defects were limited to AtNSI binding and that each mutant was impaired in symptom production and infectivity in Arabidopsis. These findings now establish that NSP-AtNSI binding is necessary for bipartite geminivirus pathogenicity and infectivity in vivo.
The strength of the reverse yeast two-hybrid selection is its unbiased nature. This allows one to define the binding region within a protein that is involved in a specific interaction (51, 52). Using this system, we identified three missense mutations in NSP—-NSPE150G, NSPI164T, and NSPD187G—-that impaired its ability to interact with AtNSI both in the yeast two-hybrid screen and in vitro, as determined by a GST pull-down assay (Fig. 2B and C). These mutations implicate the region of NSP that spans residues 150 to 187, which strikingly had no previously assigned specific function, as containing the AtNSI binding site. Our analysis of N- and C-terminal truncations of NSP confirmed that the N-terminal 42 residues, which contained the bipartite NLS and part of the ssDNA binding region, were not required for AtNSI binding (Fig. 1 and 2). However, beyond this none of the other NSP truncations could interact with AtNSI. The most likely explanation for this failure is that the majority of deletions caused global misfolding of NSP. In support of this, our previous analyses of SqLCV NSP showed that a C-terminal deletion (Δ195-256) impaired the nuclear targeting of NSP, despite the fact that the NSP NLSs are located within the NSP N terminus at residues 21 to 42 and 81 to 96 (46).
Correlated with their impaired ability to bind AtNSI, the three NSP missense mutants NSPE150G, NSPI164T, and NSPD187G clearly decreased the pathogenicity of CLCV for Arabidopsis: viral disease symptoms were delayed and notably attenuated for these mutants, and the efficiency of virus infection on this host was correspondingly reduced (Fig. 3 and 4; Table 1). Both of these features are consistent with virus movement being impaired, and they have been characteristically observed in analyses of SqLCV MP or NSP mutants (23, 24, 46, 47). The decreased efficiency of virus infection was confirmed for each NSP mutant by quantifying the relative levels of viral DNA in extracts of systemically infected leaves (Fig. 3). The observed decreases in the levels of systemic DNA for each NSP mutant correlated with the degrees to which CLCV symptoms were attenuated and infectivity was decreased, a finding that again is consistent with a defect in virus movement. In addition, the extents to which CLCV symptom production and infectivity levels were affected by NSPE150G, NSPI164T, and NSPD187G were similar both in the degree of the effects on disease appearance and the efficiencies of infection observed on natural hosts when bipartite geminivirus CP was eliminated (3, 5, 14, 18, 23, 24, 40) and in the magnitude of change observed in the efficiency of CLCV infection on Arabidopsis lines that overexpress AtNSI (32). Eliminating virus CP does not abolish infectivity, although it does abolish whitefly transmission: depending on the particular bipartite geminivirus and plant host combination, the effects can vary from a simple delay of 1 to 2 days in the appearance of disease symptoms to a delay in symptom appearance of several days accompanied by attenuated disease symptoms and a decrease in the efficiency of infection by ∼40 to 50% (3, 5, 14, 18, 23, 24, 40). The fact that wild-type CLCV infectivity is enhanced ∼30% on Arabidopsis lines that overexpress AtNSI from a 35S::AtNSI transgene is directly relevant here (32). Our findings that CLCV mutants that expressed NSPE150G, NSPI164T, and NSPD187G produced delayed and attenuated symptoms, and were 15 to 30% less efficient than wild-type CLCV in infecting Arabidopsis, are consistent with these results. Thus, rather than pathogenicity being an all-or-nothing situation, it appears that bipartite geminiviruses may utilize host metabolic or regulatory pathways in a variety of ways to maximize their ability to infect the host, a conclusion that was also suggested by the recent demonstration that the transcriptional transactivator protein TrAP encoded by tomato golden mosaic virus interacts with and inhibits the host kinases SNF1 and ADK (17, 55).
While mutational studies have shown that the yeast two-hybrid screen and associated β-Gal assay can be used to examine defects in a known protein-protein interaction, the results in general correlate qualitatively, but not necessarily quantitatively, with those obtained when the same interaction is studied in vivo in the relevant cell type (for examples, see references 34 and 49). In our examination of NSPE150G, NSPI164T, and NSPD187G, we found that the degree to which each mutant was defective for infectivity in planta correlated qualitatively, but not always quantitatively, with its defect in interacting with AtNSI, as determined with the yeast two-hybrid assay. NSPE150G was the most defective mutant in interacting with AtNSI in this assay and NSPI164T was less defective, and this did directly correlate with NSPE150G being less infectious, accumulating less viral DNA in systemically infected leaves, and producing more attenuated symptoms on Arabidopsis than did NSPI164T (Fig. 2 to 4). However, although NSPD187G appeared to have the least defective AtNSI interaction, as determined by the yeast two-hybrid and β-Gal assays, it was as defective as NSPE150G in its infectivity on Arabidopsis and its systemic accumulation of viral DNA, and it produced somewhat more attenuated symptoms.
Because our heterologous SqLCV NSP antisera reacted weakly with CLCV NSP, we could not readily detect the accumulated levels of NSPE150G, NSPI164T, and NSPD187G in immunoblots of yeast cell extracts. Thus, it is possible that some of these differences were due to different levels of accumulation of each NSP mutant in the yeast two-hybrid assay. The in vitro GST pull-down assay allowed us to examine these defects in binding to AtNSI directly and more quantitatively. In this direct in vitro binding assay, NSPE150G, NSPI164T, and NSPD187G were all similarly defective in their ability to bind AtNSI (Fig. 2C). A truly quantitative in vitro binding assay, such as a filter binding assay, would more precisely examine the relative defects in AtNSI binding for NSPE150G, NSPI164T, and NSPD187G.
Given the large number of interactions in which NSP is involved, it was critical to establish that NSPE150G, NSPI164T, and NSPD187G were specifically defective only in their interaction with AtNSI. Clearly, a missense mutation could have affected the stability of NSP, its interactions with the nuclear import or export machinery of the cell, its cooperative interaction with MP, or its binding to viral genomes, any one of which would affect virus movement and the efficiency of infection. NSPE150G, NSPI164T, and NSPD187G each accumulated to the same level as wild-type NSP when expressed in N. benthamiana leaves or protoplasts (Fig. 5A and data not shown), which indicated that none of the mutations had a significant effect on the stability of NSP in plant cells. Furthermore, NSPE150G, NSPI164T, and NSPD187G were indistinguishable from wild-type NSP in their in vitro ssDNA binding properties (Fig. 5B); thus, defects in DNA binding did not account for the infectivity defects of these missense mutants.
To examine nuclear targeting, we used CLSM to visualize the nuclear accumulation of GFP-NSP fusions in Arabidopsis and N. benthamiana protoplasts. We used these same constructs, coexpressed with MP in protoplasts, to examine both nuclear export and the interaction with MP based on MP trapping NSP in the cytoplasm, an assay that was used to identify the NES in SqLCV NSP (56). An additional advantage of using the latter assay in living protoplasts was that it allowed us to both visualize the interaction with MP based on the fluorescence of the GFP-NSP fusions and clearly image this interaction at the cortical ER (Fig. 6).
Based on the timing of their appearance and the levels of GFP fluorescence observed, we detected no differences in the nuclear accumulation of NSPE150G, NSPI164T and NSPD187G compared to wild-type NSP, nor did we observe differences from wild-type NSP in their nuclear export and interaction with MP based on our coexpression assay (Fig. 6). This last result clearly showed that the mutation in NSPD187G, although it was within the predicted NES in CLCV NSP, was not essential for NES function. These studies further establish that CLCV NSP and MP behave in the same manner as do SqLCV NSP and MP in their subcellular localization to nuclei or the cortical ER, respectively, and their cooperative interaction to trap NSP in the cytoplasm and direct it to the cortical ER (Fig. 6).
In contrast to results from previous studies in which SqLCV NSP was completely redirected to the cortical cytoplasm when it was coexpressed with SqLCV MP in Nicotiana tabacum cv. Xanthi protoplasts (46, 47), similar residual levels of wild-type and mutated nuclear GFP-NSP remained at 24 h in the presence of MP in N. benthamiana protoplasts (Fig. 6). The earlier studies used indirect immunofluorescent staining of fixed protoplasts to localize SqLCV NSP as well as MP, and sufficiently high levels of NSP or MP were first detected at 48 h posttransfection. The transfection rates for the electroporated N. tabacum cv. Xanthi protoplasts were also lower, being ∼10 to 15% compared to the 50 to 60% we obtained here for N. benthamiana protoplasts transfected by the use of PEG. Combined with the increased sensitivity afforded by GFP, the higher transfection efficiency of N. benthamiana protoplasts may have led to higher levels of accumulation of our GFP-NSP fusion proteins, which allowed us to readily detect these fusions between 16 and 24 h posttransfection in these living cells. However, it may also have saturated the nuclear export machinery.
Consistent with NSP and MP accumulating more rapidly and to higher levels in the transfected N. benthamiana cells, it was difficult to image these cells after 36 h, as they rapidly began to lyse probably due to the documented toxicity of MP (39, 47). However, we cannot exclude the possibility that GFP had some effect on the efficiency with which the GFP-NSP fusions were exported from the nucleus. If there was some effect, it was specific to GFP since the nuclear export and interaction with MP for all three NSP missense mutants were indistinguishable from those for wild-type NSP. It is also possible that some, if not all, of the residual nuclear fluorescence was accumulated free GFP cleaved from the fusion proteins, which has been reported in other studies (for example, see reference 4).
Based on these studies, we conclude that NSPE150G, NSPI164T, and NSPD187G are specifically defective in their binding to AtNSI. Consistent with this, none of these three mutations had previously been identified in our extensive analyses of SqLCV NSP in host range or its DNA binding activity, nuclear trafficking, and interaction with MP (22-24, 46, 56). Thus, the effects of NSPE150G, NSPI164T, and NSPD187G in attenuating disease symptoms and decreasing the infectivity of CLCV on Arabidopsis appear to be the consequence of their specific defect in interacting with AtNSI. This establishes a role for NSP-AtNSI interaction in CLCV pathogenesis and the virus's ability to move in and infect Arabidopsis cells. Our earlier studies showed that AtNSI also interacts with SqLCV NSP, which suggested a more general role for this interaction in bipartite geminivirus infection. In this context, it is interesting that NSPE150G, NSPI164T, and NSPD187G identify three residues that are highly conserved among the NSP proteins encoded by a wide range of bipartite geminiviruses (Fig. 1). From the sequences analyzed, only Ile164 is seldom changed to Val, which is also a neutral hydrophobic amino acid; Glu150 and Asp187 are completely invariant. This further supports the conclusion that NSP-AtNSI binding plays an important role in the bipartite geminivirus life cycle.
The findings reported here support and extend our earlier studies on NSP-AtNSI interaction and its role in virus infection. Based on our previous results, CP can synergistically interact with NSP by binding replicated viral ssDNA, without encapsidating it, to sequester the viral genome away from the replication pool and thus make it available for nuclear export (41). We have proposed that NSP, bound to the viral ssDNA genome, recruits AtNSI to a ternary complex to acetylate the genome-bound CP. Analogous to the proposed role of histone acetylation in chromatin remodeling during transcriptional activation (7), this would decrease CP avidity for the viral ssDNA and thus allow NSP to displace CP as NSP cooperatively binds the genome for export from the nucleus (32). Several observations are consistent with this model. AtNSI acetylates CLCV CP in vitro, yet AtNSI and CP do not stably interact themselves. In addition, the overexpression of AtNSI from a 35S::AtNSI transgene enhances the level of CLCV infection of Arabidopsis (32). This would be expected since CP itself does not stably interact with AtNSI; thus, high levels of AtNSI should not affect CP first binding to replicated genomes. However, the high levels of AtNSI would be expected to increase the rate at which it is recruited by NSP to acetylate CP, thereby increasing the rates of CP release and nuclear export of the viral genome. Our model further predicts that NSP mutants such as NSPE150G, NSPI164T, and NSPD187G, which are impaired in their ability to bind AtNSI, would be defective in recruiting AtNSI to displace CP from the viral genome and thus would be less efficient in exporting the viral ssDNA from the nucleus. This defect in movement would be evident as attenuated symptoms and decreased levels of infectivity, as we observed for CLCV-NSPE150G, CLCV-NSPI164T, and CLCV-NSPD187G.
Acknowledgments
We thank the present and former members of our laboratory, in particular Roisin McGarry, Jennifer Lewis, and Yoshimi Barron, for valuable suggestions and stimulating discussions during the course of these studies.
This work was supported by National Science Foundation grant MCB-9982622 to S.G.L. and a PRAXIS XXI (FCT, MCES, Portugal) predoctoral fellowship to M.F.C.
REFERENCES
- 1.Abel, S., and A. Theologis. 1994. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5:421-427. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Azzam, O., J. Frazer, D. De La Rosa, J. S. Beaver, P. Ahlquist, and D. P. Maxwell. 1994. Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204:289-296. [DOI] [PubMed] [Google Scholar]
- 4.Batoko, H., H. Q. Zheng, C. Hawes, and I. Moore. 2000. A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12:2201-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brough, C. L., R. J. Hayes, A. J. Morgan, R. H. Coutts, and K. W. Buck. 1988. Effects of mutagenesis in vitro on the ability of cloned tomato golden mosaic virus DNA to infect Nicotiana benthamiana plants. J. Gen. Virol. 69:503-514. [Google Scholar]
- 6.Burke, T., J. Cook, M. Asano, and J. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., M. Tini, and R. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. H., and V. Citovsky. 2003. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 35:386-392. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M. H., J. Sheng, G. Hind, A. K. Handa, and V. Citovsky. 2000. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desvoyes, B., S. Faure-Rabasse, M.-H. Chen, J.-W. Park, and H. B. Scholthof. 2002. A novel plant homeodomain protein interacts in a functionally relevant manner with a virus movement protein. Plant Physiol. 129:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorokhov, Y. L., K. Makinen, O. Y. Frolova, A. Merits, J. Saarinen, N. Kalkkinen, J. G. Atabekov, and M. Saarma. 1999. A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett. 461:223-228. [DOI] [PubMed] [Google Scholar]
- 12.Fridborg, I., J. Grainger, A. Page, M. Coleman, K. Findlay, and S. Angell. 2003. TIP, a novel host factor linking callose degradation with the cell-to-cell movement of potato virus X. Mol. Plant-Microbe Interact. 16:132-140. [DOI] [PubMed] [Google Scholar]
- 13.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner, W. E., G. Senter, L. Brand, J. S. Elmer, S. G. Rogers, and D. M. Bisaro. 1988. Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J. 7:899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geitz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5:255-269. [Google Scholar]
- 16.Gillespie, T., P. Boevink, S. Haupt, A. G. Roberts, R. Toth, T. Valentine, S. Chapman, and K. J. Oparka. 2002. Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14:1207-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao, L., H. Wang, G. Sunter, and D. M. Bisaro. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes, R. J., and K. W. Buck. 1989. Replication of tomato golden mosaic virus DNA B in transgenic plants expressing open reading frames (ORFs) of DNA A: requirement of ORF AL2 for production of single-stranded DNA. Nucleic Acids Res. 17:10213-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270:1983-1985. [DOI] [PubMed] [Google Scholar]
- 20.Hill, J. E., J. O. Strandberg, E. Hiebert, and S. G. Lazarowitz. 1998. Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: implications for bipartite geminivirus evolution and movement. Virology 250:283-292. [DOI] [PubMed] [Google Scholar]
- 21.Huang, Z., V. M. Andrianov, Y. Han, and S. H. Howell. 2001. Identification of Arabidopsis proteins that interact with the cauliflower mosaic virus (CaMV) movement protein. Plant Mol. Biol. 47:663-675. [DOI] [PubMed] [Google Scholar]
- 22.Ingham, D. J. 1996. Ph.D thesis. University of Illinois, Urbana.
- 23.Ingham, D. J., and S. G. Lazarowitz. 1993. A single missense mutation in the BR1 movement protein alters the host range of the squash leaf curl geminivirus. Virology 196:694-772. [DOI] [PubMed] [Google Scholar]
- 24.Ingham, D. J., E. Pascal, and S. G. Lazarowitz. 1995. Both geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology 207:191-204. [DOI] [PubMed] [Google Scholar]
- 25.Koncz, C., and J. Schell. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 203:383-396. [Google Scholar]
- 26.Kragler, F., M. Curin, K. Trutnyeva, A. Gansch, and E. Waigmann. 2003. MPB2C, a microtubule-associated plant protein, binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol. 132:1870-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for investigating intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leanna, C. A., and M. Hannink. 1996. The reverse two-hybrid system: a genetic scheme for selection against specific protein-protein interactions. Nucleic Acids Res. 24:3341-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, B., and L. A. Heaton. 2001. An Arabidopsis thaliana protein interacts with a movement protein of turnip crinkle virus in yeast cells and in vitro. J. Gen. Virol. 82:1245-1251. [DOI] [PubMed] [Google Scholar]
- 30.Mariano, A. C., M. O. Andrade, A. A. Santos, S. M. B. Carolino, M. L. Oliveira, M. C. Baracat-Pereira, S. H. Brommonshenkel, and E. P. B. Fontes. 2004. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 318:24-31. [DOI] [PubMed] [Google Scholar]
- 31.Mas, P., and R. N. Beachy. 1998. Distribution of TMV movement protein in single living protoplasts immobilized in agarose. Plant J. 15:835-842. [DOI] [PubMed] [Google Scholar]
- 32.McGarry, R. C., Y. D. Barron, M. F. Carvalho, J. E. Hill, D. Gold, E. Cheung, W. L. Kraus, and S. G. Lazarowitz. 2003. A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell 15:1605-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean, B. G., J. Zupan, and P. C. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7:2101-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montano, M. 2001. Qualitative and quantitative assessment of interactions. Methods Mol. Biol. 177:99-106. [DOI] [PubMed] [Google Scholar]
- 35.Mouchon, A., M.-H. Delmotte, P. Formstecher, and P. Lefebvre. 1999. Allosteric regulation of the discriminative responsiveness of retinoic acid receptor to natural and synthetic ligands by retinoid X receptor and DNA. Mol. Cell. Biol. 19:3073-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noueiry, A. O., W. J. Lucas, and R. L. Gilbertson. 1994. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmatal transport. Cell 76:925-932. [DOI] [PubMed] [Google Scholar]
- 37.Oparka, K. J. 2004. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 9:33-41. [DOI] [PubMed] [Google Scholar]
- 38.Pascal, E., P. E. Goodlo.ve, L. C. Wu, and S. G. Lazarowitz. 1993. Transgenic tobacco plants expressing the geminivirus BL1 protein exhibit symptoms of viral disease. Plant Cell 5:795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascal, E., A. A. Sanderfoot, B. M. Ward, R. Medville, R. Turgeon, and S. G. Lazarowitz. 1994. The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell 6:995-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pooma, W., W. K. Gillette, J. L. Jeffrey, and I. T. D. Petty. 1996. Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology 218:264-268. [DOI] [PubMed] [Google Scholar]
- 41.Qin, S., B. M. Ward, and S. G. Lazarowitz. 1998. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72:9247-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichel, C., J. Mathur, P. Eckes, K. Langenkemper, C. Koncz, J. Schell, B. Reiss, and C. Maas. 1996. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc. Natl. Acad. Sci. USA 93:5888-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rojas, M. R., A. O. Noueiry, W. J. Lucas, and R. L. Gilbertson. 1998. Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form- and size-specific manner. Cell 95:105-113. [DOI] [PubMed] [Google Scholar]
- 44.Samach, A., J. E. Klenz, S. E. Kohalmi, E. Risseeuw, G. W. Haughn, and W. L. Crosby. 1999. The unusual floral organs gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20:433-445. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sanderfoot, A. A., D. J. Ingham, and S. G. Lazarowitz. 1996. A viral movement protein as a nuclear shuttle: the geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 110:23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderfoot, A. A., and S. G. Lazarowitz. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soellick, T., J. F. Uhrig, G. L. Bucher, J. W. Kellmann, and P. H. Schreier. 2000. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad. Sci. USA 97:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutz, F., E. Izaurralde, I. Mattaj, and M. Rosbash. 1996. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol. Cell. Biol. 16:7144-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzfira, T., Y. Rhee, M. H. Chen, T. Kunik, and V. Citovsky. 2000. Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu. Rev. Microbiol. 54:187-219. [DOI] [PubMed] [Google Scholar]
- 51.Vidal, M. 1997. The reverse two hybrid system, p. 109-147. In P. L. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 52.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93:10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal, M., P. Braun, C. Elbert, J. D. Boeke, and E. Harlow. 1996. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc. Natl. Acad. Sci. USA 93:10321-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H., L. Hao, C. Y. Shung, G. Sunter, and D. M. Bisaro. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward, B. M., and S. G. Lazarowitz. 1999. Nuclear export in plants: use of geminivirus movement proteins for an in vivo cell based export assay. Plant Cell 11:1267-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward, B. M., R. Medville, S. G. Lazarowitz, and R. Turgeon. 1997. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weigel, D., and J. Glazebrook (ed.). 2002. Arabidopsis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Xiang, C., P. Han, I. Lutziger, K. Wang, and D. Oliver. 1999. A mini binary vector series for plant transformation. Plant Mol. Biol. 40:711-717. [DOI] [PubMed] [Google Scholar]