Abstract
Replication of viruses in species other than their natural hosts is frequently limited by entry and postentry barriers. The coronavirus that causes severe acute respiratory syndrome (SARS-CoV) utilizes the receptor angiotensin-converting enzyme 2 (ACE2) to infect cells. Here we compare human, mouse, and rat ACE2 molecules for their ability to serve as receptors for SARS-CoV. We found that, compared to human ACE2, murine ACE2 less efficiently bound the S1 domain of SARS-CoV and supported less-efficient S protein-mediated infection. Rat ACE2 was even less efficient, at near background levels for both activities. Murine 3T3 cells expressing human ACE2 supported SARS-CoV replication, whereas replication was less than 10% as efficient in the same cells expressing murine ACE2. These data imply that a mouse transgenically expressing human ACE2 may be a useful animal model of SARS.
The severe acute respiratory syndrome coronavirus (SARS-CoV) has been identified as the etiological agent of SARS, an acute pulmonary syndrome resulting in progressive respiratory failure and death in close to 10% of reported cases (2, 4, 10, 11). The SARS-CoV S protein, like that of other coronaviruses, mediates infection of receptor-expressing target cells (5, 8). It was recently demonstrated that angiotensin-converting enzyme 2 (ACE2) is a functional receptor for SARS-CoV and shown that a 193-amino-acid receptor-binding domain of the S protein is sufficient to bind ACE2 with high affinity (13, 20). A soluble form of ACE2, anti-ACE2 antibodies, and an antibody recognizing the ACE2-binding domain of the S protein each efficiently blocked SARS-CoV replication or infection by S protein-pseudotyped retrovirus (13, 18). Moreover, the tissue distribution of ACE2 in the lungs, kidney, and gastrointestinal tract (7, 9) is consistent with the isolation of virus from each of these tissues in infected humans and animals (2, 10-12). Collectively, these studies suggest that ACE2 is the primary physiologically relevant receptor for SARS-CoV.
SARS-CoV likely originated from one or more animal sources, and the virus can infect a number of species but does not appear to cause disease except in some primates, domestic cats, and ferrets (4, 6, 14, 17, 19). The virus has been shown to replicate in the respiratory tracts of mice challenged with SARS-CoV, but despite high challenge titers, the virus was cleared in all cases within 7 days (17). However, these mice raised neutralizing antibodies that were sufficient to prevent reinfection or infection of naïve mice following passive transfer of immune sera. These studies, as well as recent work demonstrating antibody-mediated protection in DNA-vaccinated mice (21), raise the possibility that a subunit vaccine may be sufficient for control of the virus. However, the lack of disease in mice makes them an imperfect model for evaluation of SARS vaccines and therapeutics.
Rodents are of particular interest in the study of SARS, not only because they can provide convenient animal models of human disease but also because both mice and rats have established roles in the dissemination of other viruses that infect humans. Rats in particular have been proposed to be vectors for SARS-CoV (15). Here we examine the ability of mouse and rat ACE2 to bind the SARS-CoV S protein and to mediate infection by an S protein-pseudotyped retrovirus. We also investigate the ability of SARS-CoV to replicate in murine cells. We show that mouse ACE2 is substantially less efficient than human ACE2 in supporting SARS-CoV infection but that murine cells otherwise allow for efficient replication of SARS-CoV. We also show that rat ACE2 is substantially less efficient than even the mouse receptor at binding SARS-CoV S protein or supporting S protein-mediated infection. These data imply that a mouse transgenically expressing human ACE2 could be useful in evaluating candidates for SARS vaccines and therapeutics and for studying the unusual severity of SARS.
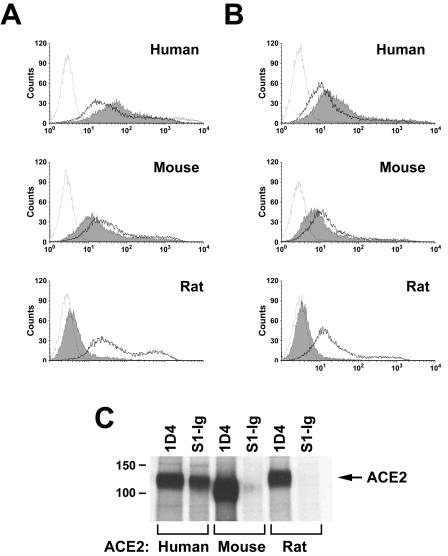
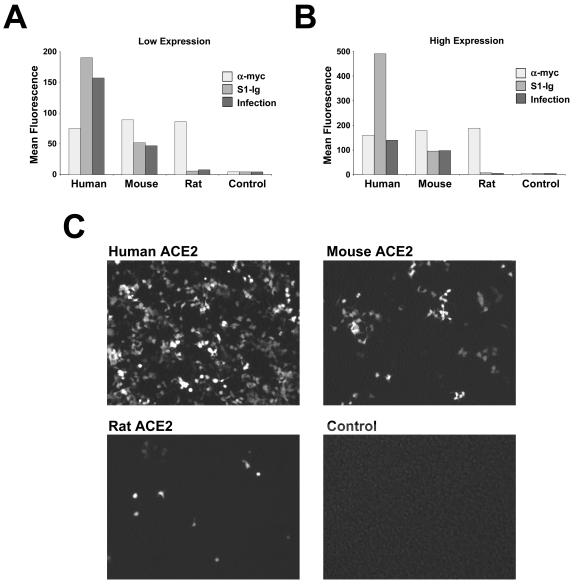
It was previously shown that the S1 region of the SARS-CoV S protein, expressed as a fusion protein with the Fc domain of human immunoglobulin G1 (IgG1) (S1-Ig), efficiently binds human ACE2 expressed on the surface of 293T cells (13, 20). In a similar assay, plasmids expressing human or rodent ACE2 molecules N-terminally tagged with 10 amino acids recognized by the anti-Myc antibody 9E10 were transfected into 293T cells. The cells were transfected with 10 (Fig. 1A) or 5 (Fig. 1B) μg of plasmid (in order to vary receptor expression for subsequent experiments) and analyzed by flow cytometry for their ability to bind 9E10 (a marker of receptor cell surface expression) or S1-Ig. As shown in Fig. 1A and B, human, mouse, and rat ACE2 were expressed efficiently as indicated by 9E10 binding, with mouse and rat ACE2 expression slightly but consistently higher than that of human receptor. Nonetheless, S1-Ig bound human ACE2 substantially more efficiently than it bound murine ACE2. Binding of S1-Ig to rat ACE2 was only slightly greater than binding of secondary antibody alone. No signal above that seen with secondary antibody alone was observed for mock-transfected 293T cells (data not shown).
FIG. 1.
SARS-CoV S1 domain binds rodent ACE2 less efficiently than human ACE2. (A) 293T cells transfected with 10 μg of plasmid encoding N-terminally Myc-tagged ACE2 of the indicated species were stained with the anti-Myc-tag antibody 9E10 (heavy line), with S1-Ig (a fusion of the SARS-CoV S1 domain with the Fc domain of human IgG1 [shaded area]), or with secondary antibody alone (light line), and analyzed by flow cytometry. (B) Same as for panel A, except that 5 μg of plasmid was used for transfection. (C) 293T cells transfected with plasmids encoding C-terminally tagged ACE2 of the indicated species were metabolically labeled with [35S]cysteine and [35S]methionine and lysed as previously described (20). Cell lysates were immunoprecipitated with either the antitag antibody 1D4 or S1-Ig and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
These initial observations were confirmed by immunoprecipitation of ACE2 from different species (Fig. 1C). 293T cells were transfected with plasmids expressing human, mouse, or rat ACE2, each expressing a nine-residue C-terminal tag recognized by the antirhodopsin antibody 1D4 (3). Cells were metabolically labeled with [35S]cysteine and [35S]methionine and subsequently lysed. Cell lysates were immunoprecipitated with protein A and either 1D4 or S1-Ig and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As shown in Fig. 1C, each ACE2 variant was readily precipitated by 1D4. As expected, human ACE2 was also precipitated by S1-Ig. However, only a faint band could be detected from lysates of mouse ACE2-transfected cells precipitated with S1-Ig. S1-Ig did not detectably bind rat ACE2. The amount of murine ACE2 precipitated by S1-Ig, which is relatively small compared with the amount recognized by S1-Ig on the surface of cells, is likely due to the more stringent wash conditions in the former assay. These results indicate that murine ACE2 binds less efficiently to the S1 domain of SARS-CoV than does human ACE2 and that rat ACE2 binds even less efficiently. Consistent with this observation, 293T cells expressing human receptor formed syncytia with S protein-expressing cells more efficiently than cells expressing mouse receptor do (Fig. 2). Few or no syncytia were observed in mixtures containing cells expressing rat ACE2, consistent with the substantially lower affinity of this receptor for S protein.
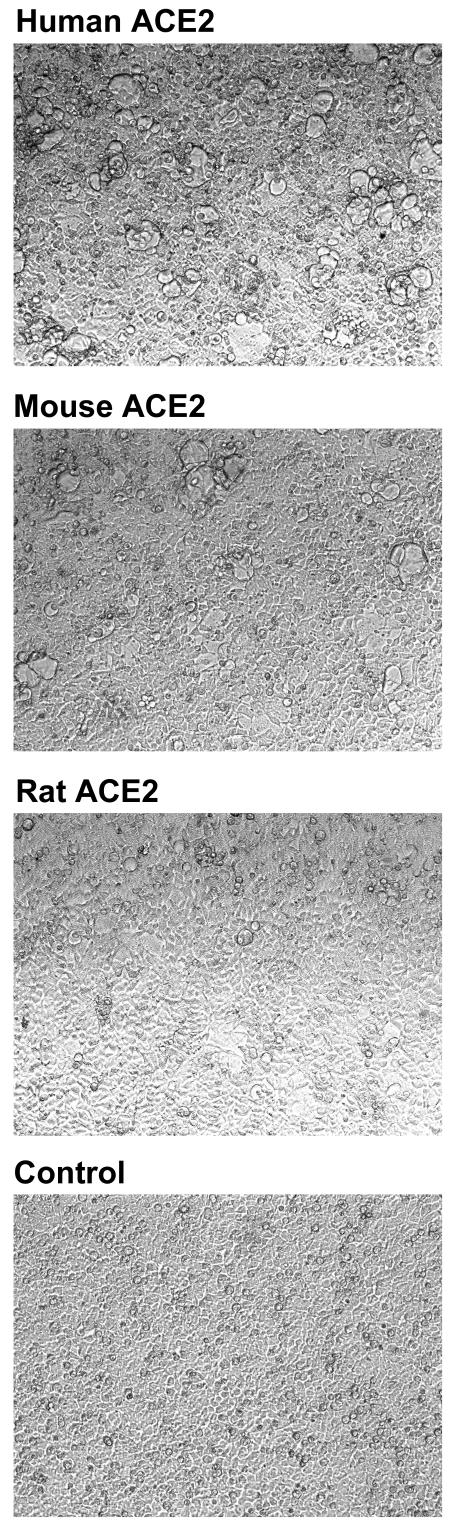
FIG. 2.
Rodent ACE2 mediates less-efficient syncytial formation with S protein-expressing cells than human ACE2. 293T cells transfected with plasmids encoding ACE2 of the indicated species or with vector alone (control) were mixed at a 1:1 ratio with 293T cells transfected with a plasmid encoding a codon-optimized form of a SARS-CoV S protein. Multinucleated syncytia were observed 48 h after the cells were mixed. The experiment shown is representative of three experiments with similar results.
Previous studies have shown that retroviruses pseudotyped with the S protein of SARS-CoV efficiently infect ACE2-expressing cells, including transfected 293T cells and Vero E6 cells (16, 20). We incubated an aliquot of the cells analyzed by flow cytometry (Fig. 1A and B) with a simian immunodeficiency virus (SIV) (1) lacking a functional env gene, expressing green fluorescent protein (GFP), and pseudotyped with SARS-CoV S protein. As shown in Fig. 3A to C, S protein-pseudotyped SIV-GFP efficiently infected 293T cells transfected with human ACE2 but less efficiently infected mouse ACE2-transfected cells, despite higher expression of the murine receptor. Some infection above the background level seen in mock-transfected cells was also observed in cells transfected with rat ACE2. It was observed that differences between the ability of mouse and human receptors to support S protein-mediated infection were consistently greater in cells expressing smaller amounts of receptor (compare Fig. 3A and B), a condition more reflective of expression on cells and tissues that naturally express ACE2 (13).
FIG. 3.
Infection with S protein-pseudotyped retrovirus is less efficient in cells expressing rodent ACE2 than in those expressing human ACE2. (A) An aliquot of cells analyzed as shown in Fig. 1B was incubated with S protein-pseudotyped SIV-GFP. After 48 h, the cells were harvested and analyzed by flow cytometry for GFP expression. GFP levels indicating infection (dark gray) are compared with anti-Myc antibody (light gray) and S1-Ig (medium gray) recognition in the same cells. (B) Same experiment as shown in panel A except that cells analyzed as shown in Fig. 1A were infected with SIV-GFP. (C) Typical image of GFP expression of cells transfected and infected as shown in panel B. The figure is representative of two similar experiments with similar results.
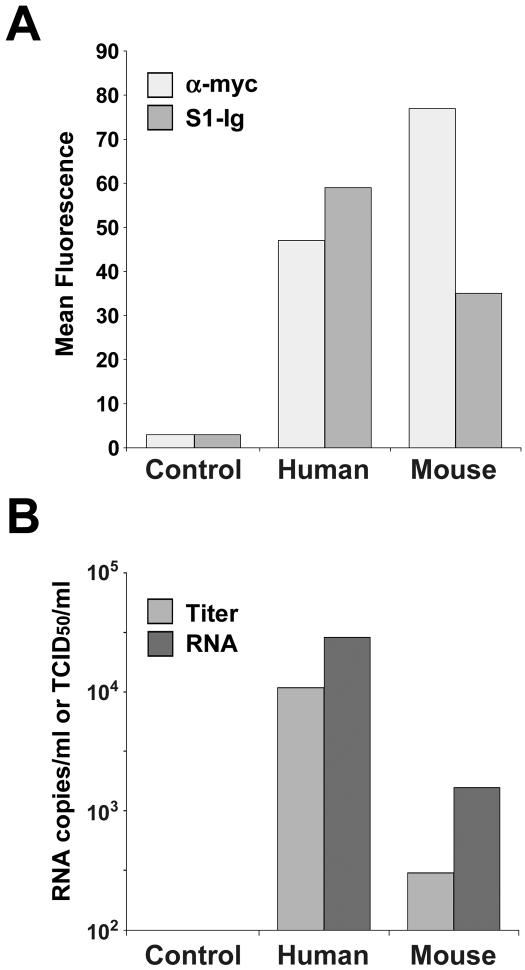
Finally, we examined the ability of SARS-CoV to replicate in murine cells transfected with human or murine ACE2. As shown in Fig. 4A, both murine and human ACE2 were efficiently expressed on murine 3T3 cells, as indicated by recognition by the antitag antibody 9E10. Transfected 3T3 cells were incubated with 1.3 × 104 50% tissue culture infective doses of SARS-CoV, and both viral titers and RNA copies were measured at peak levels (day 2 for virus, day 4 for RNA). Consistent with infection by S protein-pseudotyped retrovirus, SARS-CoV replication was substantially more efficient in human ACE2-expressing cells than in cells expressing murine receptor (Fig. 4B). Viral titers, determined based on the cytopathicity observed in Vero E6 cells in two separate experiments, were more than 10 times as high in human ACE2-expressing 3T3 cells as in cells expressing murine ACE2 at higher levels. Viral RNA levels, measured by semiquantitative PCR, were approximately 10-fold higher in human ACE2-expressing cells. Both viral titers and RNA levels were below the limits of detection of these assays in mock-transfected cells (control). These data suggest that transgenic expression of human ACE2 in the lungs and other ACE2-expressing tissues of mice will permit more efficient replication of SARS-CoV in these mice.
FIG. 4.
Replication of SARS-CoV in murine cells is enhanced by human receptor. (A) Murine 3T3 cells were transfected with plasmid encoding Myc-tagged variants of human or mouse ACE2 as indicated, or with vector alone (control), and analyzed by flow cytometry using the anti-Myc-tag antibody 9E10 (light gray bars) or S1-Ig (medium gray bars). (B) An aliquot of the cells analyzed as shown in panel A was incubated with SARS-CoV for 4 days. Peak viral titers (medium gray) were determined by measuring the cytopathicity in Vero E6 cells incubated with cell supernatants as previously described (13). Viral RNA levels (dark gray) were determined by semiquantitative reverse transcription-PCR of cell supernatants, as previously described (13). Viral titers and RNA levels of control cells were below 100 50% tissue culture infective doses (TCID50) per ml and 100 copies per ml, the limits of detection in both assays. The figure shows an experiment representative of two experiments with similar results.
ACE2 is a functional receptor for SARS-CoV, and many data are consistent with a physiological role for ACE2 in the replication of virus in the infected host (13). Here we show in several biochemical and functional assays that the ACE2 molecules of mice and rats are substantially less efficient than human ACE2 in binding the SARS-CoV S protein and in supporting infection with S protein-pseudotyped viruses and replication of SARS-CoV. These data are consistent with the rapid clearance of virus in challenged mice and the absence of reports of successful infection in rats (6, 17). We also demonstrate that SARS-CoV can replicate in murine 3T3 cells if these cells express human or mouse ACE2; again, the human receptor is markedly more efficient than the mouse form.
Although straightforward, these observations imply several conclusions useful for future studies of SARS-CoV. First, differences between rodent and human receptors will be useful in the mapping of the S protein-binding region of ACE2. In particular, chimeras and point mutation variants of rat and human receptors, which are 83% identical in their respective mature ectodomains, can readily be used to identify residues critical to the inability of the rat receptor to support efficient infection. Second, these studies imply that the low efficiency of the mouse receptor limits SARS-CoV replication efficiency and therefore the usefulness of wild-type mice in evaluating vaccines and therapies. Mice transgenically expressing human ACE2 will likely permit greater viral replication and may exhibit SARS-like symptoms. Finally, our data imply that significant adaptation of SARS-CoV to human receptor must have occurred if either mice or rats served as reservoirs or vectors for zoonotic transmission of the virus.
REFERENCES
- 1.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667-676. [DOI] [PubMed] [Google Scholar]
- 4.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 4:4. [DOI] [PubMed] [Google Scholar]
- 7.Harmer, D., M. Gilbert, R. Borman, and K. L. Clark. 2002. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 532:107-110. [DOI] [PubMed] [Google Scholar]
- 8.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu, T., Y. Suzuki, J. Imai, S. Sugano, M. Hida, A. Tanigami, S. Muroi, Y. Yamada, and K. Hanaoka. 2002. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2). DNA Sequence 13:217-220. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 11.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung, W. K., K. F. To, P. K. Chan, H. L. Chan, A. K. Wu, N. Lee, K. Y. Yuen, and J. J. Sung. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, C. Luzeriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng, S. K. 2003. Possible role of an animal vector in the SARS outbreak at Amoy Gardens. Lancet 362:570-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sui, J., W. Li, A. Murakami, A. Tamin, L. J. Matthews, S. K. Wong, M. J. Moore, A. St. Clair Tallarico, M. Olurinde, H. Choe, L. J. Anderson, W. J. Bellini, M. Farzan, and W. A. Marasco. 2004. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weingartl, H. M., J. Copps, M. A. Drebot, P. Marszal, G. Smith, J. Gren, M. Andova, J. Pasick, P. Kitching, and M. Czub. 2004. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 10:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2003. A 193-amino-acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, Z. Y., W. P. Kong, Y. Huang, A. Roberts, B. R. Murphy, K. Subbarao, and G. J. Nabel. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]