Abstract
Pleconaril is a broad-spectrum antirhinovirus and antienterovirus compound that binds into a hydrophobic pocket within viral protein 1, stabilizing the capsid and resulting in the inhibition of cell attachment and RNA uncoating. When crystals of human rhinovirus 16 (HRV16) and HRV14 are incubated with pleconaril, drug occupancy in the binding pocket is lower than when pleconaril is introduced during assembly prior to crystallization. This effect is far more marked in HRV16 than in HRV14 and is more marked with pleconaril than with other compounds. These observations are consistent with virus yield inhibition studies and radiolabeled drug binding studies showing that the antiviral effect of pleconaril against HRV16 is greater on the infectivity of progeny virions than the parent input viruses. These data suggest that drug integration into the binding pocket during assembly, or at some other late stage in virus replication, may contribute to the antiviral activity of capsid binding compounds.
The family of small, icosahedral RNA animal viruses known as Picornaviridae (45) has been subdivided into nine different genera (22), including the rhinoviruses (the leading causative agents of viral upper respiratory tract infections in humans) and the enteroviruses (responsible for respiratory tract infections, but often associated with more severe systemic disease). Picornavirus capsids consist of 60 copies of 4 viral proteins, VP1, VP2, VP3, and VP4. Whereas the core structures of VP1, VP2, and VP3 are eight-stranded, antiparallel β-barrels, the small protein VP4 is an extended polypeptide chain located on the internal surface of the capsid. Rhino- and enteroviruses have a surface depression (canyon) running around each fivefold vertex (42), which is the binding site of many immunoglobulin-like cell surface molecules that are often utilized by these viruses as receptors (43).
Most rhino- and enteroviruses that have been studied structurally were found to have a pocket factor bound into a hydrophobic pocket within the VP1 β-barrel, beneath the floor of the “canyon.” This ligand is probably a lipid-like molecule derived from the host cell (10, 21, 33, 34), although solvent molecules, such as polyethylene glycol used in crystallization (47), have also been found to bind. It has been hypothesized that pocket factor binding in this hydrophobic cavity plays a role in maintaining picornavirus capsid stability (24, 34, 41). Such an effect could be important to maintain capsid integrity during cell-to-cell transport of the virus or to ensure proper capsid function once the virus engages the cellular receptor or encounters the reduced pH environment of the endosome. Several classes of low-molecular-weight compounds have been identified that displace pocket factor, leading to the inhibition of functions associated with the virus capsid, including attachment to cellular receptors for some viruses (17, 38, 46) and RNA uncoating (1, 12, 29, 31, 32, 46).
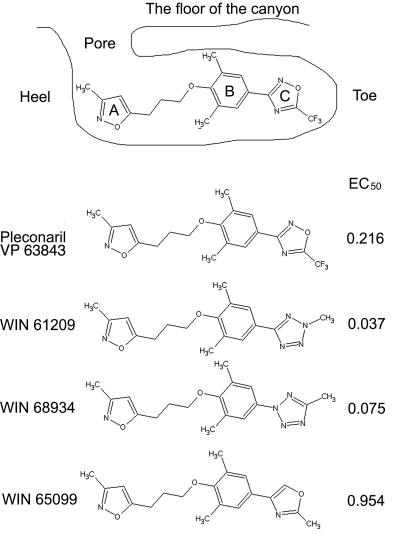
The cocrystal structures of several capsid-binding antiviral compounds complexed with human rhinovirus 14 (HRV14) (3, 6, 15, 48), HRV16 (14, 34), HRV1A (20), HRV3 (50), HRV2 (49), polioviruses (13, 19, 27), coxsackievirus A9 (18), and coxsackievirus B3 (33) are known. The hydrophobic binding pocket consists of a closed “toe” at one end and an open “pore” at the other. A well-characterized group of capsid binding compounds consists of a methylisoxazole ring (ring A), a substituted phenoxy group (ring B), and a five-member heteroatom ring (ring C) (Fig. 1). One such compound, pleconaril, has improved activity against a broad spectrum of HRV and enterovirus serotypes relative to its predecessors (26, 37, 40). This compound has been shown to be efficacious in treating picornavirus common colds in phase 3 human clinical testing (7, 16, 37, 40).
FIG. 1.
Schematic diagram of pleconaril bound into one of the pockets in the virion and the chemical structure of a selected set of antiviral compounds. The EC50 values, listed in the right column, are defined as the effective compound concentration (in μM) that gave 50% protection against virus-induced cytopathic effect to cells in a monolayer. A, methylisoxazole ring; B, substituted phenoxy group; C, five-member heteroatom ring.
We report here the crystal structures of HRV14 and HRV16 with bound pleconaril. Pleconaril was introduced either during virus assembly or by diffusion into the mature virus. When the viruses were grown in the presence of pleconaril, the occupancy was found to be higher than when the drug was introduced into the already-assembled viruses. Therefore, the antiviral compounds appear to affect viral replication during assembly.
MATERIALS AND METHODS
Crystal preparation.
Pleconaril was introduced into the virus by soaking crystals (soak_with), by cocrystallization (cryst_with), or by growing the virus in the presence of the compound (grown_with). HRVs, except for the grown_with sample, were propagated and purified as described previously (34, 42). When the virus was grown in the presence of pleconaril, HRV was adsorbed on HeLa cells for 2 h at room temperature, after which the infected cells were incubated at 33°C for 4 h. Pleconaril was then added to the medium at a final concentration of 2.85 μg/ml (the molecular mass of pleconaril is 381 Da), and the incubation was continued for an additional 6 h. The resultant virus was purified as described previously (34, 42). The hanging drop vapor diffusion method was used for crystallization, with the reservoir solution containing 0.25 M HEPES, 0.25 M NaCl, 0.1 M CaCl2, and 0.3% polyethylene glycol 8000. The crystallization conditions were the same for all methods of introducing the drug into the virus. For cryst_with, pleconaril was dissolved in 0.2% (vol/vol) dimethyl sulfoxide and introduced into the crystallization drop, giving a final concentration of 20 μg/ml. The crystallization solution for the grown_with crystals did not contain additional pleconaril. For soak_with HRVs, pleconaril was diffused into crystals by using the procedure described by Smith et al. (48). Crystals were soaked for a few minutes in a cryoprotectant solution containing 3% polyethylene glycol 8000 and 22% glycerol and were then flash frozen in a stream of nitrogen gas at 100 K.
TCID50 assay.
Virus titer was quantified in a 50% tissue culture infectious dose (TCID50) assay. HeLa Ohio cells were seeded into 96-well tissue culture cluster plates at 4 × 104 cells/well in Eagle's minimal essential medium (EMEM) supplemented with 5% fetal bovine serum, 5% Fetalclone (HyClone, Logan, Utah), glutamine, and antibiotics (vancomycin, amphotericin B, and gentamicin) (hereafter referred to as complete EMEM), followed by overnight incubation at 37°C. The growth medium was then removed, and serial 0.5 log10 dilutions of virus were added to the plates in duplicate. Subsequently, M199 medium supplemented with 5% fetal bovine serum and antibiotics was added after the virus had been absorbed. After 3 days of incubation at 33°C, the plates were fixed by the addition of 5% glutaraldehyde and stained with crystal violet (0.5% solution in water). After rinsing and drying steps, the optical density of residual stained cells was measured spectrophotometrically at 570 nm on a Molecular Devices Vmax tunable plate reader (Sunnyvale, Calif.). The serial dilution data were subjected to analysis by a four-parameter curve-fitting program, from which a TCID50 value was derived.
Pleconaril binding during virus assembly.
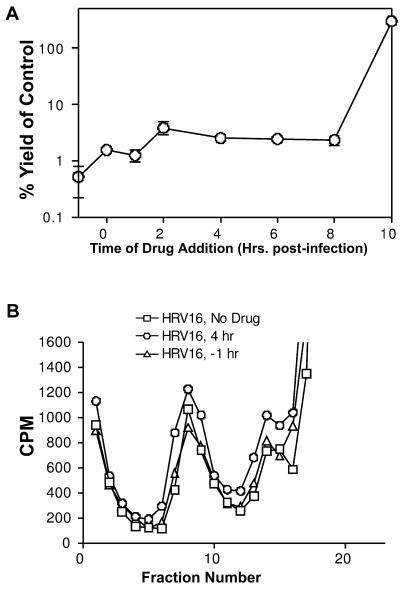
Time-of-drug-addition experiments were conducted to determine the stage at which pleconaril affected virus replication. Six-well tissue culture cluster plates were seeded with 1.6 × 105 HeLa Ohio cells in 4 ml of complete EMEM and incubated overnight at 37°C. The following day, the growth medium was removed, and the wells were infected with HRV14 or HRV16 at a multiplicity of infection of about 1 in duplicate for 1 h at 33°C. Pleconaril (7.5 μM) was added at t = −1 (during attachment), 0, 1, 2, 4, 6, 8, or 10 h postinfection. All wells were washed twice with 2 ml of Dulbecco's phosphate-buffered saline (DPBS) (Gibco) after the attachment phase of infection to remove inoculum virus. The plates were incubated for a total of 10 h (postattachment; a single cycle of HRV growth) and then frozen at −80°C. Samples were triple chloroform extracted prior to quantification by a TCID50 assay (Fig. 2).
FIG. 2.
Addition of pleconaril to HRV16 at differing times postinfection during a single cycle of growth. (A) The yield, expressed as a percentage with respect to virus growth when no drug was added, is shown as a function of the time the drug was added. (B) Particle production monitored by radiolabeling the virus with 35S methionine is plotted against the fraction number in a sucrose gradient. Fraction 1 was at the bottom of the gradient; fraction 18 was at the top.
In vitro radiolabeled pleconaril binding studies.
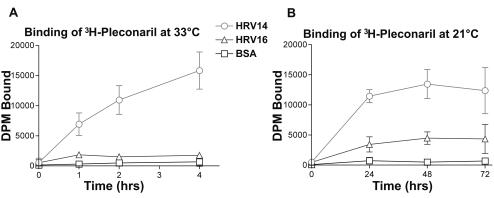
Binding of [3H]pleconaril to HRV14 and HRV16 (Fig. 3) was measured by the separation of free radiolabeled drug from drug bound to virus through G-50 Sephadex spin columns, a modification of a procedure used previously (12, 36). The Sephadex G-50 (fine) Quick Spin columns (Roche Diagnostics Corp., Indianapolis, Ind.) were equilibrated with 100 μl of 1× DPBS (without Ca and Mg), which was allowed to drip through the column directly prior to use. The 3H-labeled drug (with a specific activity of 15 Ci/mM) was mixed with 1 μg of purified HRV14 or HRV16 in 1× DPBS (without Ca and Mg) for a final concentration of 0.1 μM. In order to minimize nonspecific sticking of virus or drug, all binding reactions were performed in clear glass vials. Those reactions were incubated at 21°C and were measured over periods of 24, 48, and 72 h, by which time equilibrium binding was observed. Reactions incubated at 33°C were measured up to 4 h only, due to virus instability at later time points. At the end of the incubation period, 100 μl of the sample was added to the equilibrated G-50 Sephadex spin column and centrifuged at 250 × g for 4 min at room temperature in order to separate unbound drug from drug bound to virus. Total counts (before centrifugation) and bound counts (eluate) were determined by scintillation spectroscopy on a Wallac 1409 spectrophotometer. An equivalent amount of bovine serum albumin served as the control for nonspecific drug binding.
FIG. 3.
In vitro radiolabeled pleconaril binding studies at 33 and 21°C. One microgram of purified HRV14, HRV16, or BSA was incubated with 0.1 μM [3H]pleconaril for the times and at the temperatures indicated, followed by separation of bound and unbound drug on equilibrated G-50 Sephadex spin columns, as described in Materials and Methods. DPM, disintegrations per minute; BSA, bovine serum albumin.
X-ray diffraction data processing and density averaging.
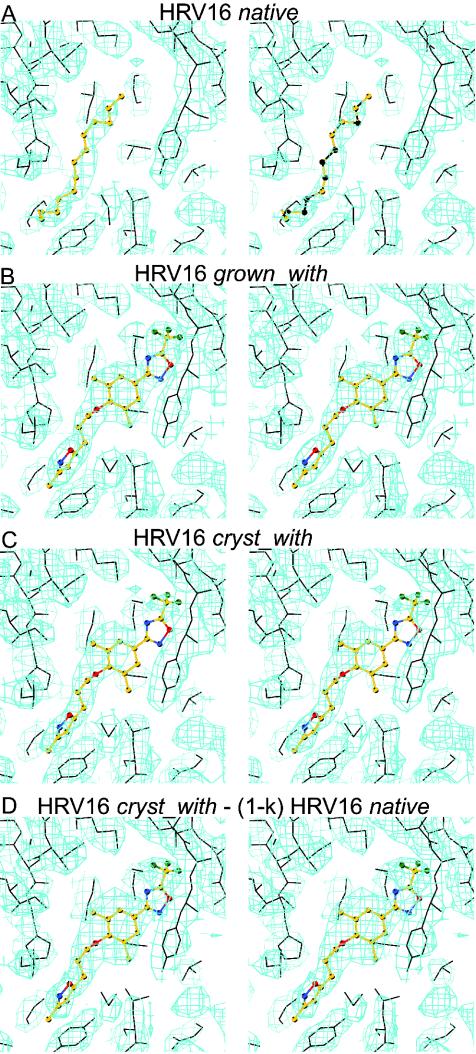
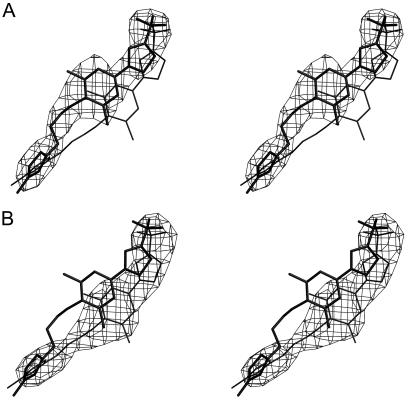
All diffraction data were collected at the Advanced Photon Source's 14BMC beam line (Table 1). Diffraction data sets were processed using the DENZO program (35) and scaled with the SNP (4) or SCALEPACK (35) program. The virus particle orientations and positions in the crystal unit cells had been determined previously (2, 14, 34) for room temperature data. The previous structures were used to calculate a phasing start for the low-temperature data sets. Phases were refined and extended by density averaging and solvent flattening (Table 1). The noncrystallographic symmetry parameters were refined by using the “climb” procedure in the ENVELOPE program (44), which resulted in movement of less than 0.15 Å of the viral center. The averaged maps (Fig. 4 and 5) showed that the pleconaril in the grown_with HRV16, grown_with HRV14, and soak_with HRV14 crystals was well defined, indicating that there was a drug molecule in nearly every pocket, whereas there was only partial occupancy in the cryst_with HRV16 crystals. The occupancy of pleconaril was determined by crystallographic refinement in every case except for the cryst_with HRV16 structure. In that case, the possible presence of pocket factor in pockets not occupied by pleconaril had to be considered. The effect of pocket factor on the estimation of pleconaril occupancy for the grown_with HRV16 data was negligible as the occupancy was nearly 100%. The pleconaril occupancy (k) can be determined by calculating vector difference maps, with Fourier coefficients FD. If it is assumed that FCOMP is composed of a drug-complexed (FD) component and an uncomplexed (FN) component, then FCOMP = kFD + (1 − k)FN. Here, FCOMP and FN represent the structure factors of the observed, pleconaril-associated, virus complex crystals and the native, pocket factor-complexed virus crystals, respectively. The occupancy of pleconaril was determined by adjusting the k value such that the electron density of the drug had about the same magnitude as the protein surrounding the binding pocket in difference maps computed with FD = [FCOMP − (1 − k) × FN]/k coefficients (Fig. 4D).
TABLE 1.
Data collection, density averaging, and refinement statistics
| Parameter | HRV16
|
HRV14
|
|||
|---|---|---|---|---|---|
| Native | Cryst_with | Grown_with | Soak_with | Grown_with | |
| Data collection and processing | |||||
| Space group | P22121 | P22121 | P22121 | P213 | P213 |
| Unit cell parameters (Å) | |||||
| a | 360.3 | 360.3 | 359.5 | 438.7 | 437.2 |
| b | 343.3 | 343.3 | 343.7 | ||
| c | 332.6 | 332.6 | 332.2 | ||
| Data collection temp (K) | 100 | 100 | 100 | 100 | 100 |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| No. of images | 300 | 454 | 61 | 150 | 60 |
| No. of crystals | 1 | 1 | 1 | 1 | 1 |
| Oscillation range/image (degrees) | 0.25 | 0.25 | 0.25 | 0.20 | 0.20 |
| Resolution (Å) | 20-2.5 | 20-2.8 | 20-2.7 | 20-2.5 | 20-3.3 |
| No. of observations | 1,294,215 | 1,224,998 | 1,107,533 | 1,675,367 | 545,390 |
| No. of unique reflections (σ cutoff [F]) | 710,094 (≥0σ) | 731,062 (≥3σ) | 452,981 (≥0σ) | 671,838 (≥0σ) | 320,952 (≥0σ) |
| % Overall data completenessa | 52.3 (20.9) | 95.1 (62.6) | 40.9 (34.9) | 70.3 (26.3) | 77.8 (68.2) |
| Rsyma,b | 0.038 (0.081) | 0.082 (0.191) | 0.133 (0.395) | 0.077 (0.456) | 0.164 (0.477) |
| Density averaging statistics | |||||
| Initial resolution (Å) | 2.8 | 2.8 | 2.8 | 2.8 | 3.3 |
| Final extended resolution (Å) | 2.5 | 2.8 | 2.7 | 2.5 | 3.3 |
| Overall R factora,c | 0.160 (0.370) | 0.163 (0.269) | 0.219 (0.339) | 0.111 (0.307) | 0.145 (0.266) |
| Overall correlation coefficienta,d | 0.92 (0.46) | 0.9 (0.77) | 0.87 (0.59) | 0.97 (0.68) | 0.94 (0.74) |
| CNS refinement statistics | |||||
| R factor (Rfree)c | 0.202 (0.206) | 0.247 (0.243) | 0.249 (0.248) | 0.217 (0.216) | 0.222 (0.228) |
| Rms bond length (Å) | 0.0060 | 0.0065 | 0.0061 | 0.0078 | 0.0096 |
| Rms bond angles (degrees) | 1.36 | 1.34 | 1.37 | 1.54 | 1.54 |
| Average B values (Å2) | 16.2 | 30.4 | 19.7 | 30.9 | 38.2 |
| Occupancy of pleconaril or pocket factore | 0.8 Pocket factor | 0.4 Pleconaril | 0.9 Pleconaril | 0.8 Pleconaril | 0.9 Pleconaril |
| PDB accession no. | 1ND2 | 1ND3 | 1NCR | 1NCQ | 1NA1 |
Parentheses indicate highest resolution.
Rsym = ΣhΣi|Ii(h) − <I(h)>|ΣhΣi Ii(h), where Ii(h) is the ith observation and <I(h)> is the mean of all measurements of I(h).
R factor = Σ|Fo−Fc|/ΣFo, where Fo and Fc are the observed and calculated amplitudes of the structure factor.
Correlation coefficient = Σ[(<Fo> − Fo)(<Fc> − Fc)]/Σ[(<Fo> − Fo)2 Σ (<Fc> − Fc)2]1/2, where Fo and Fc are defined as above.
For cryst_with HRV16 data, the occupancy of pleconaril was calculated from a vector difference map, while the occupancies for the other data sets were estimated by grouped occupancy refinement.
FIG. 4.
Electron densities and atomic models for (A) native HRV16, (B) grown_with HRV16, (C) cryst_with HRV16, and (D) a vector difference map for [cryst_with HRV16 − (1 − k) native HRV16] with a k value of 0.35 (see Materials and Methods). The atomic models of the pocket factor (A) and pleconaril (B, C, D) are shown in ball-and-stick representations. The protein model is shown in black bonds.
FIG. 5.
Electron densities of pleconaril in (A) grown_with HRV16 and (B) grown_with HRV14. The structures of pleconaril as it occurs in HRV16 (thick lines) and HRV14 (thin lines) are superimposed.
Atomic refinement of the pleconaril complexes.
Starting coordinates were based on the previously determined HRV14 and HRV16 structures. Pleconaril was built into the averaged electron density maps. Restrained individual positional and temperature factor (B) refinements were performed with the program CNS, by using constraints to define the noncrystallographic symmetry relationships (5) (Table 1). The maximum likelihood target function was used for refinement. The structural parameters for pleconaril used in the refinement were based on the charge distribution and geometry calculated for WIN52084 (28) and structures of trifluorinated ligands in the Protein Data Bank (PDB). The geometry and charge distribution for the putative pocket factor of HRV16 was calculated based on the coordinates of pocket factor in the 2.16-Å resolution native HRV16 structure (PDB accession number 1AYM) with the program XPLO2D (23). The positions and temperature factors of water molecules were refined assuming full occupancies. Water molecules whose temperature factors exceeded 50 Å2 were excluded from further refinement. The occupancy of pleconaril in the cryst_with HRV16 was determined by means of vector difference maps (see above). The occupancies of pleconaril in the soak_with HRV14, grown_with HRV16, and grown_with HRV14 were determined by grouped and unrestrained occupancy refinement, assuming the B factor of pleconaril to be the average B value of residues lying within 4 Å of the drug molecule. The same method was used to calculate the occupancy of a putative pocket factor in native HRV16. The refined coordinates of these data sets were then superimposed and compared with the coordinates of HRV16 complexed with WIN61209, WIN68934, and WIN65099 (Fig. 1).
The coordinates for the various structures have been deposited with the PDB (accession numbers are given in Table 1).
RESULTS AND DISCUSSION
Differences between HRVs at room temperature and 100 K.
Unlike previous studies of rhinovirus-antiviral complexes, the present study reports the first such structures identified at cryotemperatures. The freezing techniques (39) that have been introduced to macromolecular crystallography in the last decade or more have made it possible to collect complete three-dimensional data sets on a single crystal, thus greatly reducing the errors involved in data collection and eliminating errors due to the absence of complete data sets. However, the process of freezing might introduce some artifacts.
It was found that the structure of frozen native HRV16 is very similar to its structure at room temperature, with a root mean square (RMS) distance of 0.22 Å between equivalent Cα atoms. Similar results have also been reported for other rhinoviruses (8, 47). The largest difference between Cα atoms is 0.5 Å, where the temperature factors are greater than 40 Å2. Differences between side chain atoms are as big as 0.6 Å. A few larger differences can probably be attributed to model building errors in either the warm or cold structures. There is little if any difference in temperature factors between room temperature and frozen crystals for VP2 and VP3. However, the temperature factors for the frozen crystal are systematically about 5 Å2 lower in VP1 than in those of the room temperature crystal. The increased flexibility of VP1 at room temperature correlates with the increased sequence variability in VP1. No X-ray diffraction data were collected for frozen native HRV14 crystals. Hence, no exact structural comparison could be made between frozen and room temperature HRV14 crystals. Nevertheless, similar results were obtained when comparing warm native and frozen drug-complexed crystals of HRV14, showing that the freezing of these crystals produced only small structural changes outside the drug binding pocket.
Structures of virus-pleconaril complexes.
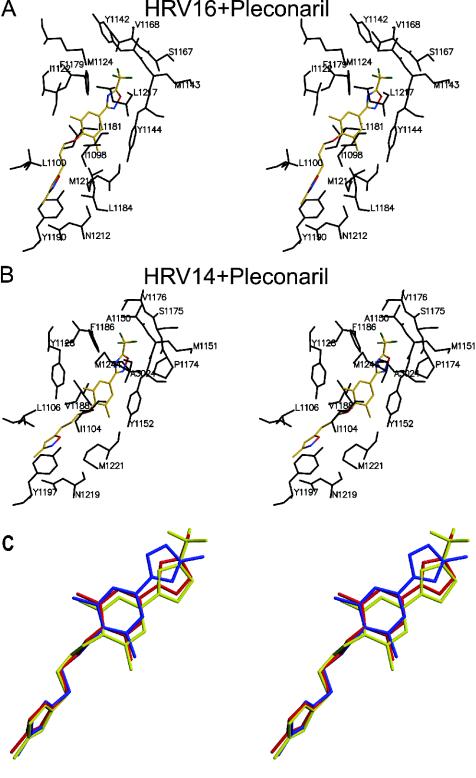
Pleconaril binds to HRV16 and HRV14 (Table 2 and Fig. 5 and 6), with ring A close to the entrance of the pocket and ring C at the toe end. This orientation is the same as in nearly all other related compounds, except for very long compounds, such as WIN52084 and WIN51711, that bind into the pocket in the opposite direction (3, 48). Rings B and C are roughly coplanar, whereas ring A is approximately perpendicular to the other two rings when pleconaril or related WIN compounds (Fig. 1) bind to either HRV16 or HRV14 (14). The primary difference in the binding of pleconaril to these two viruses is that rings B and C, which are in contact with Ile1122 (see Table 2 for an explanation of the numbering system) in HRV16 (Fig. 5), are displaced by the more bulky Tyr1128 in HRV14 (Fig. 5 and 6). Similar differences are observed for other compounds when bound to HRV14 (20) or HRV16 (34). The displacement of the pocket factor by pleconaril in HRV16 results in conformational changes of less than 0.8 Å for any virus atom. The largest of these changes is for the Tyr1144 that is displaced by ring B of pleconaril. More substantial changes occurred when pleconaril was introduced into empty HRV14 pockets, resulting in a displacement of the Cα atoms in residues 1213 to 1224 by as much as 4.3 Å, similar to displacements produced by other compounds (3, 48).
TABLE 2.
Binding sites of pleconaril in HRV14 and HRV16a
| HRV14
|
HRV16
|
|||
|---|---|---|---|---|
| Residue | Pleconaril | Residue | Pleconaril | |
| I1104 | Aliphatic chain and ring B | I1098.............................................. | Aliphatic chain and ring B | |
| L1106 | Ring A | L1100 | Ring A | |
| Y1128 | Aliphatic chain and ring B | I1122 | Ring B | |
| A1150 | Trifluoro group | M1124 | Trifluoro group | |
| M1151 | Trifluoro group | Y1142 | Ring C and trifluoro group | |
| Y1152 | Rings B and C and trifluoro group | M1143 | Trifluoro group | |
| P1174 | Ring C and trifluoro group | Y1144 | Rings B and C and trifluoro group | |
| S1175 | Trifluoro group | A1166 | Trifluoro group | |
| V1176 | Trifluoro group | S1167 | Trifluoro group | |
| F1186 | Ring C and trifluoro group | V1168 | Trifluoro group | |
| V1188 | Ring B | F1179 | Ring C and trifluoro group | |
| V1191 | Ring B | L1181 | Ring B | |
| Y1197 | Aliphatic chain and ring A | L1184 | Ring B | |
| N1219 | Ring A | Y1190 | Ring A | |
| M1221 | Ring A | Q1212 | Ring A | |
| M1224 | Ring B | M1214 | Rings A and B and aliphatic chain | |
| A3024 | Ring C | L1217 | Ring C and trifluoro group | |
Structurally aligned residues within 4 Å of pleconaril are listed on the same line. Residues are labeled with a four-digit notation. The first digit represents the viral protein, while the last three digits are the sequential amino acid number for that viral protein and that particular serotype.
FIG. 6.
Pleconaril in the environment of (A) HRV16 and (B) HRV14. Residues that have at least one atom lying within 4 Å of pleconaril are shown in black. Pleconaril is shown in color (C atom, yellow; O atom, red; N atom, blue; and F atom, green). (C) Positions and conformations of pleconaril (yellow), WIN61209 (blue), and WIN65099 (red) in HRV16.
Correlation between structure and EC50 values in HRV16.
The four compounds listed in Fig. 1 have similar binding sites on the virus, but ring C in pleconaril and WIN65099 is flipped by 180° compared with WIN61209 and WIN68934 (Fig. 6). Pleconaril and WIN65099 have higher 50% effective concentrations (EC50s) against virus-induced cytopathic effect to cells in a monolayer (see legend to Fig. 1) than WIN61209 and WIN68934, demonstrating that the flip in ring C correlates with the difference in their potency against HRV16. The flipped trifluoromethyloxadiazole in pleconaril and methyloxazoline in WIN65099 (rings C) are more polar and have a bigger volume than the corresponding methyltetrazole rings in WIN61209 and WIN68934 (Fig. 1) and probably have a less favorable interaction with the surrounding hydrophobic side chains, such as Tyr1144 and Phe1179. The EC50s in plaque reduction assays, for a selected set of similar compounds when bound to HRV14, correlate well with the binding dissociation constants (11). Thus, the EC50s of pleconaril and WIN65099 might be associated with high dissociation constants and, therefore, less favorable binding energy. The difference in EC50s between pleconaril and WIN65099 might result from the hydrophilic trifluoro (CF3) group in pleconaril or the additional nitrogen in ring C. Indeed, the environment of the CF3 group is somewhat hydrophilic as a consequence of the main chain atoms belonging to residues Tyr1142, Met1143, Tyr1144, and Ala1166. It is also possible that the CF3 group makes entry into the pocket more difficult.
Factors controlling the occupancy of pleconaril in HRVs.
Antiviral compounds, such as pleconaril, can be studied crystallographically when complexed with the virus by diffusion into grown crystals (soak_with), by cocrystallization (cryst_with), by incubation with the virus prior to crystallization, or by adding the drug to cells at the time of protein folding and virus assembly prior to purification and crystallization (grown_with). In all previous crystallographic examinations, the drug was added at a late stage after the virus had been fully assembled. Occupancy of pleconaril in HRV16 was surprisingly low (0.4) when cocrystallized (cryst_with) relative to its antiviral potency in cell culture (EC50 value of 0.2 μg/ml), considering the high concentration of the drug used during crystallization (20 μg/ml with a 1-month exposure). In contrast, the occupancy of pleconaril in HRV16 when added during viral propagation prior to virion assembly and subsequent crystallization was high (0.9), although there was no pleconaril in the mother liquor during crystallization, demonstrating that once pleconaril was introduced into the pockets of the virus, it could not diffuse out. The apparent irreversible binding of pleconaril to HRV16 and HRV14 when pleconaril was introduced during assembly is consistent with the inability to regain infectivity of the virus by attempts to extract pleconaril with organic solvents (see Materials and Methods). The in vitro radiolabeled drug binding studies at two different temperatures (Fig. 3) suggest that this effect probably cannot be attributed to the elevated temperature during the association of the drug with the virus in the grown_with experiments. It would seem more likely that pocket access constraints may be reduced during virus assembly, permitting higher occupancy of pockets than is observed in the soak_with or cryst_with experiments.
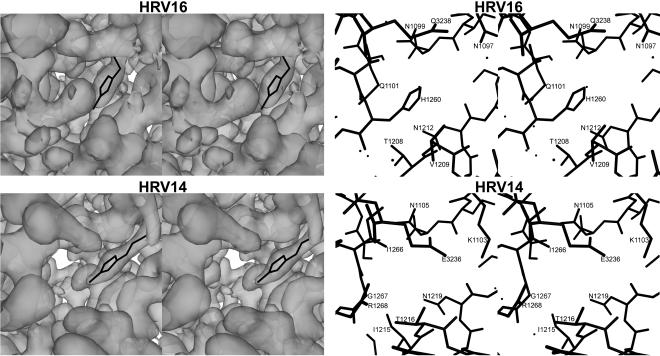
The pore leading to the drug-binding pocket of HRV16 is more restricted than that of HRV14 (Fig. 7). Specifically, His1260 is in the center of the pore of HRV16, whereas the equivalent residue in HRV14 is Gly1267. Although this residue is in a highly variable loop among different serotypes, it is a glycine in only HRV14 and HRV3, among known HRVs. This fact may relate to the absence of pocket factor when these viruses are purified because of the greater ease with which the pocket factor can enter and exit the pocket. Thus, the lower occupancy of pleconaril in cryst_with HRV16 (0.4), compared to that in soak_with HRV14 (0.8), may be due in part to the restriction at the entrance (pore) to the pocket. However, other compounds with similar potency as pleconaril have high occupancy (14) in soak_with crystals (Table 3), demonstrating that the occupancy is determined by both the nature of the compound and the residues lining the pocket and pore.
FIG. 7.
Left, stereographic views of the pore (the entry portal into the pocket that binds pocket factor or antiviral compounds), looking into the virus from the outside and showing the electron density surface. The electron densities were contoured at a 1.0 σ level. Pleconaril is shown in black. Right, atomic structure of the pore regions, shown in the same orientation as used on the left. The structure is slabbed to include only those atoms that are on the outside of the virus.
TABLE 3.
Virus-compound complex structural study of HRV16 and HRV14
| Druga | Serotype | Method of complex formation | Incubation time | Concn of the compound (μg/ml) | Occupancy | Reference |
|---|---|---|---|---|---|---|
| WIN61209 | HRV16 | Soak_with | 36 h | 100 | ∼1b | 14 |
| WIN68934 | HRV16 | Soak_with | 36 h | 100 | ∼1b | 14 |
| WIN65099 | HRV16 | Soak_with | 36 h | 100 | ∼1b | 14 |
| Pleconaril | HRV14 | Soak_with | 30 h | 2 | 0.8c | This paper |
| Pleconaril | HRV14 | Grown_with | 10 h | 2.85 | 0.9c | This paper |
| Pleconaril | HRV16 | Cryst_with | ∼1 mo | 20 | 0.4d | This paper |
| Pleconaril | HRV16 | Grown_with | 10 h | 2.85 | 0.9c | This paper |
The molecular mass of pleconaril is 381 Da, and the other WIN compounds have very similar molecular masses.
Occupancy was estimated by comparing the electron density height of the compound with that of the surrounding protein directly. The virus was almost fully occupied.
Occupancy was estimated by a refinement of the occupancy of the compound.
Occupancy was estimated by use of a vector difference map (3).
In time-of-drug-addition experiments, the addition of pleconaril at any time up to 8 h postinfection in a 10 h single cycle of growth resulted in a significant reduction of virus yield (Fig. 2A). Virus radiolabeling studies under conditions similar to those of the time-of-drug-addition experiments showed that the addition of pleconaril at any time in the single cycle of HRV16 growth did not significantly decrease particle production (Fig. 2B). This seemingly paradoxical finding (a profound antiviral effect in the absence of an effect on progeny virus production) can probably be attributed to the binding of pleconaril to progeny virions during assembly or at some point late in the virus growth cycle. The results are consistent with the observed occupancies of pleconaril in HRV16 crystals (low occupancy in previously assembled virus [0.4]; high occupancy when the drug is present during virus propagation [0.9]).
Several factors may contribute to the observed higher occupancy of pleconaril in HRV16 (and, to a lesser extent, in HRV14) when the drug is present during virus replication. It is likely that the drug-binding pocket is more readily accessed in virus intermediate particles, such as the 5S protomer, the 14S pentamer, or premature uncleaved virions. In addition or alternatively, intracellular concentrations of pleconaril, and in particular drug concentrations at the site of virus assembly, may be higher than those that can be achieved and maintained in solution in the extracellular medium or in the crystallization mother liquor. Whatever the reason for these findings, it is clear that in a single cycle of HRV16 growth, essentially all of the antiviral activity of pleconaril is exerted on progeny virions and not on input virus. If this antiviral activity is indeed due to more ready accessibility of the drug-binding pocket during virion assembly, it follows that it could represent an important mechanism for the antiviral activity of pleconaril (and other capsid binding compounds) against many HRV serotypes.
Acknowledgments
Figures 4, 5, and 6 were prepared by using MOLSCRIPT (25), Bobscript (9), and Raster3D (30). Figure 7 was prepared by using the programs DINO (http://cobra.mih.unibas.ch/dino) and POV-Ray (http://www.povray.org). We thank Suchetana Mukhopadhyay and Richard J. Kuhn for useful discussions, as well as Sharon S. Wilder and Cheryl A. Towell for help in the preparation of the manuscript. We are grateful to BIOCARS staff for help with data collection at the Advanced Photon Source beam line 14BMC.
This work was supported by an NIH grant (AI 11219) to M.G.R.
REFERENCES
- 1.Andries, K. 1995. Anti-picornaviral agents, p. 287-319. In D. J. Jeffries and E. DeCelercq (ed.), Antiviral chemotherapy. John Wiley and Sons, New York, N.Y.
- 2.Arnold, E., G. Vriend, M. Luo, J. P. Griffith, G. Kamer, J. W. Erickson, J. E. Johnson, and M. G. Rossmann. 1987. The structure determination of a common cold virus, human rhinovirus 14. Acta Crystallogr. Sect. A 43:346-361. [Google Scholar]
- 3.Badger, J., I. Minor, M. J. Kremer, M. A. Oliveira, T. J. Smith, J. P. Griffith, D. M. A. Guerin, S. Krishnaswamy, M. Luo, M. G. Rossmann, M. A. McKinlay, G. D. Diana, F. J. Dutko, M. Fancher, R. R. Rueckert, and B. A. Heinz. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotovsky, R., I. Steller, and M. G. Rossmann. 1998. The use of partial reflections for scaling and averaging X-ray area-detector data. J. Appl. Crystallogr. 31:708-717. [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, M. S., I. Minor, M. G. Rossmann, G. D. Diana, and K. Andries. 1991. Human rhinovirus 14 complexed with antiviral compound R 61837. J. Mol. Biol. 217:455-463. [DOI] [PubMed] [Google Scholar]
- 7.Diana, G. D., P. Rudewicz, D. C. Pevear, T. J. Nitz, S. C. Aldous, D. J. Aldous, D. T. Robinson, T. Draper, F. J. Dutko, C. Aldi, G. Gendron, R. C. Oglesby, D. L. Volkots, M. Reuman, T. R. Bailey, R. Czerniak, T. Block, R. Roland, and J. Oppermann. 1995. Picornavirus inhibitors: trifluoromethyl substitution provides a global protective effect against hepatic metabolism. J. Med. Chem. 38:1355-1371. [DOI] [PubMed] [Google Scholar]
- 8.Ding, J., A. D. Smith, S. C. Geisler, X. Ma, G. F. Arnold, and E. Arnold. 2002. Crystal structure of a human rhinovirus that displays part of the HIV-1 V3 loop and induces neutralizing antibodies against HIV-1. Structure 10:999-1011. [DOI] [PubMed] [Google Scholar]
- 9.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132-134. [DOI] [PubMed] [Google Scholar]
- 10.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, M. P., M. A. McKinlay, G. D. Diana, and F. J. Dutko. 1991. Binding affinities of structurally related human rhinovirus capsid-binding compounds are correlated to their activities against human rhinovirus type 14. Antimicrob. Agents Chemother. 35:1040-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, M. P., M. J. Otto, and M. A. McKinlay. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, R. A., C. N. Hiremath, D. J. Filman, R. Syed, K. Andries, and J. M. Hogle. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr. Biol. 4:784-797. [DOI] [PubMed] [Google Scholar]
- 14.Hadfield, A. T., G. D. Diana, and M. G. Rossmann. 1999. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc. Natl. Acad. Sci. USA 96:14730-14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadfield, A. T., M. A. Oliveira, K. H. Kim, I. Minor, M. J. Kremer, B. A. Heinz, D. Shepard, D. C. Pevear, R. R. Rueckert, and M. G. Rossmann. 1995. Structural studies on human rhinovirus 14 drug-resistant compensation mutants. J. Mol. Biol. 253:61-73. [DOI] [PubMed] [Google Scholar]
- 16.Hayden, F. G., D. T. Herrington, T. L. Coats, K. H. Kim, E. C. Cooper, S. A. Villano, S. Liu, S. Hudson, D. C. Pevear, M. Collett, and M. A. McKinlay. 2003. Efficacy and safety of oral pleconaril for treatment of picornavirus colds in adults: results of two double-blind, randomized, placebo-controlled trials. Clin. Infect. Dis. 36:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz, B. A., R. R. Rueckert, D. A. Shepard, F. J. Dutko, M. A. McKinlay, M. Fancher, M. G. Rossmann, J. Badger, and T. J. Smith. 1989. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 63:2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypiä, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 19.Hiremath, C. N. 1995. Binding of antiviral drug WIN51711 to the Sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14. Acta Crystallogr. Sect. D 51:473-489. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, F. J. Dutko, M. A. McKinlay, and D. C. Pevear. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-226. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Pevear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. Crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 22.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Family Picornaviridae, p. 657-678. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, San Diego, Calif.
- 23.Kleywegt, G. J. 1995. Dictionaries for heteros. CCP4/ESF-EACBM Newsl. Protein Crystallogr. 31:45-50. [Google Scholar]
- 24.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraulis, P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 26.Ledford, R. M., N. R. Patel, T. M. Demenczuk, A. Watanyar, T. Herbertz, M. Collett, and D. C. Pevear. 2004. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 78:3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lentz, K. N., A. D. Smith, S. C. Geisler, S. Cox, P. Buontempo, A. Skelton, J. DeMartino, E. Rozhon, J. Schwartz, V. Girijavallabhan, J. O'Connell, and E. Arnold. 1997. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of the three poliovirus serotypes. Structure 5:961-978. [DOI] [PubMed] [Google Scholar]
- 28.Lybrand, T. P. 1991. Molecular simulation and drug design. J. Pharm. Belg. 46:49-54. [PubMed] [Google Scholar]
- 29.McKinlay, M. A., F. J. Dutko, D. C. Pevear, M. G. Woods, G. D. Diana, and M. G. Rossmann. 1990. Rational design of antipicornavirus agents, p. 366-372. In M. A. Brinton and F. X. Heinz (ed.), New aspects of positive-strand RNA viruses. American Society for Microbiology, Washington, D.C.
- 30.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 31.Mosser, A. G., and R. R. Rueckert. 1996. Capsid-binding agents, p. 13-40. In D. D. Richman (ed.), Antiviral drug resistance. John Wiley and Sons, Inc., Chichester, England.
- 32.Mosser, A. G., and R. R. Rueckert. 1993. WIN 51711-dependent mutants of poliovirus type 3: evidence that virions decay after release from cells unless drug is present. J. Virol. 67:1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muckelbauer, J. K., M. Kremer, I. Minor, L. Tong, A. Zlotnick, J. E. Johnson, and M. G. Rossmann. 1995. Structure determination of coxsackievirus B3 to 3.5 Å resolution. Acta Crystallogr. Sect. D 51:871-887. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira, M. A., R. Zhao, W. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 36.Penefsky, H. S. 1977. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 252:2891-2899. [PubMed] [Google Scholar]
- 37.Pevear, D. C. 1999. Activity of pleconaril against enteroviruses. Adv. Exp. Med. Biol. 458:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pevear, D. C., M. J. Fancher, P. J. Felock, M. G. Rossmann, M. S. Miller, G. Diana, A. M. Treasurywala, M. A. McKinlay, and F. J. Dutko. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodgers, D. W. 2001. Cryocrystallography techniques and devices, p. 202-208. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography. Vol. F, crystallography of biological macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 40.Rogers, J. M., G. D. Diana, and M. A. McKinlay. 1999. Pleconaril. A broad spectrum antipicornaviral agent, p. 69-75. In J. Mills, P. A. Volberding, and L. Corey (ed.), Antiviral chemotherapy 5. New directions for clinical applications and research. Kluwer Academic/Plenum Publishers, New York, N.Y. [PubMed]
- 41.Rossmann, M. G. 1994. Viral cell recognition and entry. Protein Sci. 3:1712-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 317:145-153. [DOI] [PubMed] [Google Scholar]
- 43.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 44.Rossmann, M. G., R. McKenna, L. Tong, D. Xia, J. Dai, H. Wu, H. K. Choi, and R. E. Lynch. 1992. Molecular replacement real-space averaging. J. Appl. Crystallogr. 25:166-180. [Google Scholar]
- 45.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 46.Shepard, D. A., B. A. Heinz, and R. R. Rueckert. 1993. WIN compounds inhibit both attachment and eclipse of human rhinovirus 14. J. Virol. 67:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature (London) 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 49.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1181-1196. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0 Å resolution. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]