Abstract
Prokaryotic and eukaryotic genomic DNA is comprised of the four building blocks A, G, C and T. We have begun to explore the consequences of replacing a large fraction or all of a nucleoside in genomic DNA with a modified nucleoside. As a first step we have investigated the possibility of replacement of T by 2′-deoxy-5-(hydroxymethyl)uridine (5hmU) in the genomic DNA of E. coli. Metabolic engineering with phage genes followed by random mutagenesis enabled us to achieve approximately 75% replacement of T by 5hmU in the E. coli genome and in plasmids.
Graphical Abstract
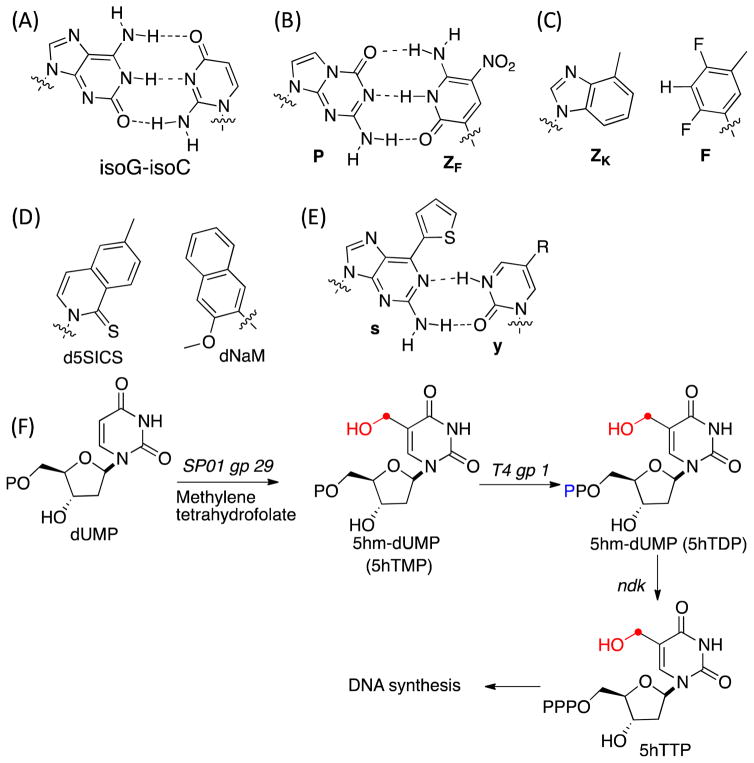
Prokaryotic and eukaryotic genomic DNA is comprised of the four building blocks A, G, C and T. The A-T and G-C hydrogen bonding and base-pair stacking interactions are important stabilizing non-covalent interactions in double helical DNA and play a central role in template-dependent replication and transcription. Modified bases have been observed in genomic DNA, including N6-methyladenine, 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxycytosine and 2′-deoxy-5-(hydroxymethyl)uridine (5hmU). 1,2 Of these 5mC and 5hmC have been established as stable epigenetic modifications in eukaryotes, including mammals, and play a key role in regulating transcription and organization of chromatin structure.3 In bacteria, 5mC also plays a role in restriction modification systems.4 The roles of all of these DNA modifications are an active area of research. In addition to studies of natural nucleoside modifications, a number of synthetic efforts have been made to introduce unnatural base pairs (UBPs) into a template DNA and utilize them for in vitro and in vivo replication, transcription and translation reactions. Leonard and coworkers developed several extended purines to investigate enzyme recognition and base pairing.5–7 Benner and coworkers have generated an isoG-isoC base pair (Figure 1A) and examined its in vitro replication, transcription and translation reactions.8–10 They further developed an unnatural PB-ZB base pair (Figure 1B) that functions in PCR with high efficiency.11–13 Kool and coworkers have developed a ZK-F base pair based on hydrophobic interactions rather than hydrogen bonding interactions (Figure 1C), and shown it to be selective during the replication by the Klenow fragment of E. coli DNA polymerase I.14–16 They also developed an xDNA based system for plasmid based expression system.17 Hirao and coworkers have developed an unnatural base pair between s–y (Figure 1E) that can be transcribed by T7 polymerase, 18,19 and Romesberg, Schultz and coworkers developed a series of selective, stable hydrophobic and metallo-base pairs.20,21 Moreover, Romesberg and co-workers demonstrated that the E. coli replication machinery could accurately replicate plasmids containing a d5SICS-dNaM base pair (Figure 1D). 22–25
Figure 1.
(A) – (E) Examples of unnatural base pairs; (F) Metabolic pathway to biosynthesize and incorporate 5hmU into genomic templates.
We have begun to explore the consequences of replacing a large fraction or even all of a nucleoside in genomic DNA with a modified nucleoside. As a first step we have investigated the possibility of replacement of T by 2′-deoxy-5-(hydroxymethyl)uridine (5hmU) in the genomic DNA of E. coli. Studies have detected 5hmU in mammalian DNA and its role as an epigenetic regulator is currently an active area of investigation.2 Thymidine replacement by the unnatural thymidine analog, 5-bromouridine, has been previously studied by Hanawalt and coworkers26 and more recently, Mutzel and coworkers studied thymidine replacement by 5-chlorouridine.27 Here we focused on 5hmU incorporation since a complete replacement of thymidine with 5hmU is observed in the SPO1 bacillus phage genome.28 In contrast to the previous studies, our goal is to engineer an autonomous E. coli strain that both biosynthesizes the modified base and efficiently incorporates it into the genome without addition of exogenous nucleoside analogs. This system also allows us to explore in an easily manipulatable system (E. coli) how 5hmU affects replication, transcription and repair.
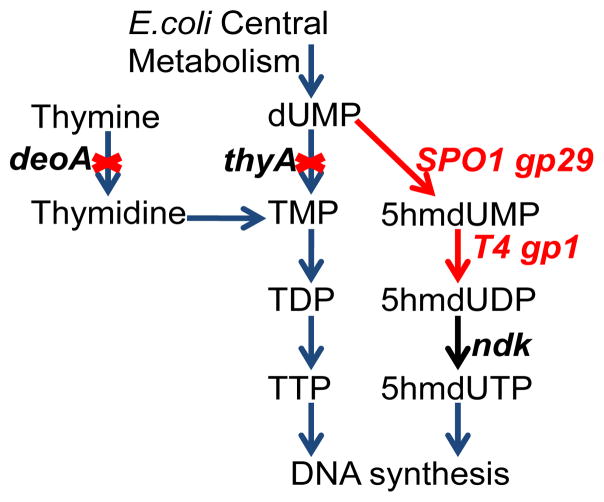
We used a similar metabolic approach to that used by SPO1 bacillus phage to engineer the E. coli pyrimidine triphosphate biosynthetic pathway to incorporate 5hmU into the E. coli genome. (Figure 2). 29 Briefly, deoxyuridine monophosphate (dUMP), the intermediate for thymidine triphosphate (TTP) biosynthesis in E. coli, was converted to 2′-deoxy-5-(hydroxymethyl)uridine triphosphate (5hmdUTP) by a series of reactions catalyzed by a hydroxymethylase, a mono-nucleotide kinase, and a nucleotide diphosphate kinase, which are encoded by SPO1 bacillus phage gp29, T4 bacteriophage gp1, and native E. coli ndk genes, respectively. The resulting 5hmdUTP is then incorporated into DNA by the endogenous DNA polymerase machinery. Since TTP competes with 5hmdUTP for base pairing in DNA synthesis, the genes thyA, deoA and trmA of E. coli were disrupted to lower the intra-cellular concentration of TTP.
Figure 2.
Metabolic engineering of the E. coli pyrimidine triphosphate biosynthetic pathway and incorporation of 5hmU into E. coli DNA.
The genes SPO1 gp29 and T4 gp1 were codon optimized and introduced as a part of a synthetic operon under the pBAD promoter with tetracycline resistance (pAM22). This operon together with native E. coli genes was expected to drive the biosynthesis of 5hmU (Figure 2). pAM22 was transformed into 3KO (BW25113 ΔthyA::specR ΔdeoA::FRT ΔtrmA::FRT-KanR) to give E. coli hT-1 and grown in the presence of thymidine supplementation. Individual colonies were picked, and an overnight culture (5%) was inoculated into LB medium containing tetracycline, thymidine (0.5 mg/L) and arabinose. Cells were harvested, and genomic DNA was isolated and digested to single nucleosides with phosphodiesterase I, calf intestinal phosphatase and benzonase.
The resulting pool was then analyzed by high performance liquid chromatography (HPLC, 254 nm). Along with the standard nucleosides in the genomic DNA (2′-deoxycytidine, 2′-deoxyadenosine, 2′-deoxy-guanosine and thymidine (T)), 5hmU was detected. We calculated 5hmU levels by HPLC peak area at 254nm with standards. Under the above growth conditions, we observed that approximately half of genomic thymidine was replaced by 5hmU [%5hmU/(5hmU+T) ~ 48%]. Moreover, plasmid DNA isolated from these strains showed 5hmU incorporation at somewhat higher levels than the genomic templates (%5hmU/(5hmU+T) ~ 62%, Figure S4).
Because 5hmU and TTP compete at the polymerase level for incorporation into genomic DNA, we investigated the effects of decreasing TTP levels by replacing the richer LB medium with defined M9 minimal media containing L-arabinose, tetracycline and defined levels of thymidine. No growth was observed for hT-1 strains in the absence of exogenous thymidine; and very poor growth was observed (OD600 ~ 0.2) for hT-1 strains grown in the presence of 0.5 mg/L thymidine. On the other hand, better growth was observed with 2 mg/L thymidine (OD600 ~ 0.6 to 0.8). However, genomic DNA analysis indicated that %5hmU/(5hmU+T) was ~ 52%, indicating no significant change in genomic 5hmU incorporation. To further lower the level of TTP, we tested the effect of expression of thymidylate 5′-phosphatase (TMPase, from Lactobacillus plantarum), which converts TMP to thymidine (Figure 4). Introduction of a construct containing TMPase (pAM26) into hT1 to afford hT3 strain (Table 1) resulted in %5hmU/(5hmU+T) of ~51%, again indicating no significant effect on the levels of 5hmU in the genomic templates.
Table 1.
Strain names and properties
| Strain | Properties/plasmid |
|---|---|
| 3KO | Thymidine auxotroph from E. coli BW25113 |
| hT-1 | 3KO transformed with pAM22 |
| hT-2 | 3KO transformed with pAM22 and pAM23 |
| hT-3 | 3KO transformed with pAM22 and pAM26 |
| hT-4 | 3KO transformed with pAM43 |
| 2hTC strains | NTG mutagenesis on hT-4 in the absence of exogenous thymidine |
We then attempted to increase the pool size of 5hmdUTP in order to increase 5hmU genomic content. During the biosynthesis of 5hmUTP, SPO1 gp29 converts dUMP to 5-hydroxythymidine-5′-mono-phosphate (5hmdUMP) using methylenetetrahydrofolate (MFH4), which is converted to tetrahydrofolate (FH4) in this process. E. coli glyA regenerates MFH4 from FH4 by converting serine to glycine. In order to maintain high levels of MFH4 to drive the formation of 5hmUTP, we tested the effect of glyA overexpression and serine supplementation. Serine supplementation of the overnight culture under induction conditions (hT-1 + serine) had little effect on the net 5hmU content [%5hmU/(5hmU+T) increased to ~53% from ~48%; Figure S6]. Overexpression of glyA was carried out by introducing plasmid pAM23 into hT-1, resulting in strain hT-2. The hT-2 cultures were grown as described above with supplementation by serine (10 mg/L) and thymidine (0.5mg/L); however, the %5hmU/(5hmU+T) was again ~48%, indicating no significant effect (Figure S5).
We next increased the copy numbers of the phage genes encoding the 5hmUTP biosynthetic pathway, which might also increase the 5hmUTP pool. A new construct was designed where SPO1 gp29 and T4 gp1 were introduced into a high copy number vector (Cdf ori and chloramphenicol resistance, pAM43) under the pBAD promoter. We transformed pAM43 into 3KO to generate hT-4, and this strain was grown as described above in LB medium in the presence of thymidine (0.5 mg/L), serine (10 mg/L) and L-arabinose. This modification increased the %5hmU/(5hmU+T) to around 62% as compared to 48% for hT-1. Finally, there was no significant effect when we incorporated the phage polymerase (SPO1 gp31) into our engineered metabolic network (%5hmU/(5hmU+T) ~ 50%, Figure S10).
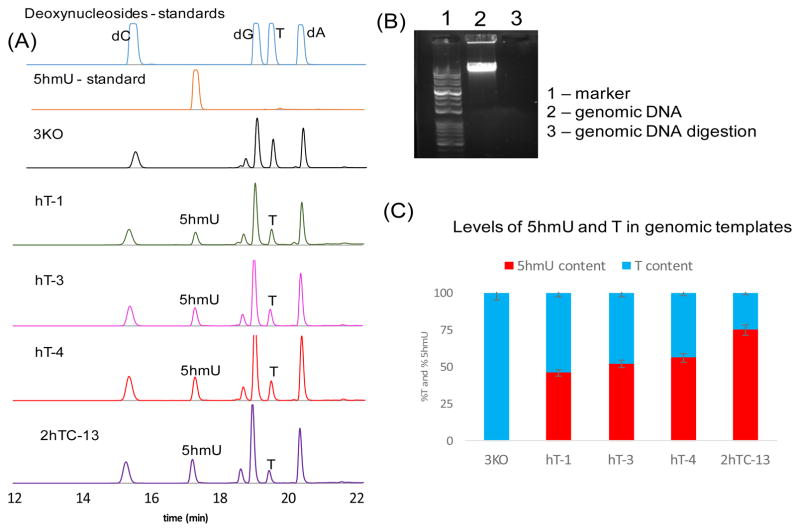
In all of the above experiments we observed that cells containing significant amounts of 5hmU in their genomes did not grow above OD600 ~ 0.7 (growth curve - Figure S11). These observations suggested that 5hmU incorporation into E. coli genomic DNA might limit growth by impairing processes such as replication, transcriptional, and repair, and as a consequence, alterations may be necessary in these processes to increase genomic 5hmU content. Because such changes may be difficult to predict and target by rational design, we decided to introduce genome-wide random mutations into hT-4 under selective pressure for 5hmU incorporation (thymidine starvation). The hT-4 strain was grown as described above in LB medium in the presence of serine (10 mg/L), tetracycline and L-arabinose. Cells were then subjected to methylnitronitrosoguanidine (NTG) treatment (see SI for mutagenesis procedure) and plated on LB-agar plates containing arabinose, tetracycline and serine with no exogenous thymidine. Of the 19 colonies isolated, 14 survived in liquid medium containing LB, serine, arabinose, chloramphenicol and no exogenous thymidine. All of these colonies showed higher 5hmU incorporation, and one of these strains (2hTC-13) showed %5hmU/(5hmU+T) = 73 ± 2 % (Figure 3). This phenotype as well as growth was stable for at least three generations (Figure S12–S14). Similar levels of 5hmU incorporation were also observed in plasmids (%5hmU/(5hmU+T) = 67 ± 4 %, Figure S8) isolated from these strains. In contrast to 3KO, 2hTC-13 strains took around 48hr to reach a maximum OD600 ~ 0.6 to 0.75 (growth curves – Figure S12–S14). We also tested their growth in a defined minimal medium in the absence or 0.05mg/L of thymidine, but no growth was observed in 48 hr. We next attempted to perform a second round of mutagenesis on 2hTC-13 to further increase 5hmU content. The recovered cells in this case were plated on M9 minimal medium containing casamino acids and thymidine (0.05 mg/L). Unfortunately, no colonies were observed. Thus, it appears under these experimental conditions, we seemed to have reached a maximum level of 5hmU incorporation in the genomic DNA of E. coli (%5hmU/(5hmU+T)) of ~ 75%.
Figure 3.
(A) HPLC traces (254 nm) of genomic DNA after digestion to single nucleosides. (B) Representative gel showing the complete digestion of the isolated genomic templates with phosphodiesterase I, calf intestinal phosphatase and benzonase. (C) Levels of 5hmU content in various strains. For strain details - see Table 1.
To summarize, we have used a metabolic engineering approach to incorporate 5hmU into the E. coli genome. We further performed random mutagenesis to increase 5hmU content ((%5hmU/(5hmU+T)) to ~ 75%. At present we do not know if 75% is the maximum possible replacement of T by 5hmU. Further mutagenesis studies and characterization of the mutations and mutant strains with increased 5hmU incorporation will likely provide insights into the key physiological regulators in E. coli that limit global genomic incorporation of modified nucleosides as well as the effect of 5hmU on replication, transcription and repair. At the same time we are investigating if the 5-hydroxylmethyl group of 5hmU can be modified enzymatically30 and whether other base or sugar modifications of the natural deoxynucleosides can be introduced into the genome at high levels, e.g., replacement of dC by 5hmdC, or dC by rC.
Supplementary Material
Acknowledgments
We would like to acknowledge Kristen Williams for her assistance in manuscript preparation. This work was supported by NIH R01 GM062159-14.
Footnotes
Supporting Information. HPLC data, experimental procedure and design of the constructs is described in the supporting information. The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Nabel CS, Manning SA, Kohli RM. ACS Chem Bio. 2012;7:20. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olinski R, Starczak M, Gackowski D. Mutat Res, Rev Mutat Res. 2016;767:59. doi: 10.1016/j.mrrev.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Münzel M, Globisch D, Carell T. Angew Chem Intl Ed. 2011;50:6460. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Romero MA, Cota I, Casadesus J. Curr Opin Microbiol. 2015;25:9. doi: 10.1016/j.mib.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Leonard NJ. Biopolymers. 1985;24:9. doi: 10.1002/bip.360240104. [DOI] [PubMed] [Google Scholar]
- 6.Leonard NJ, Hiremath SP. Tetrahedron. 1986;42:1917. [Google Scholar]
- 7.Lessor RA, Gibson KJ, Leonard NJ. Biochemistry. 1984;23:3868. doi: 10.1021/bi00312a012. [DOI] [PubMed] [Google Scholar]
- 8.Bain JD, Switzer C, Chamberlin AR, Benner SA. Nature (London) 1992;356:537. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 9.Horlacher J, Hottiger M, Podust VN, Huebscher U, Benner SA. Proc Natl Acad Sci U S A. 1995;92:6329. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Switzer C, Moroney SE, Benner SA. J Am Chem Soc. 1989;111:8322. [Google Scholar]
- 11.Kim HJ, Chen F, Benner SA. J Org Chem. 2012;77:3664. doi: 10.1021/jo300230z. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang ZY, Chen F, Alvarado JB, Benner SA. J Am Chem Soc. 2011;133:15105. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales JC, Kool ET. Nat Struct Biol. 1998;5:950. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 15.Moran S, Ren RXF, Kool ET. Proc Natl Acad Sci U S A. 1997;94:10506. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran S, Ren RXF, Rumney SIV, Kool ET. J Am Chem Soc. 1997;119:2056. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger AT, Peterson LW, Chelliserry J, Kleinbaum DJ, Kool ET. J Am Chem Soc. 2011;133:18447. doi: 10.1021/ja208025e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, Kimoto M, Sugiyama H, Hirao I, Yokoyama S. Bioorg Med Chem Lett. 2001;11:2221. doi: 10.1016/s0960-894x(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 19.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, Kodama K, Yokogawa T, Nishikawa K, Yokoyama S. Nat Biotechnol. 2002;20:177. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda S, Henry AA, Schultz PG, Romesberg FE. J Am Chem Soc. 2003;125:6134. doi: 10.1021/ja034099w. [DOI] [PubMed] [Google Scholar]
- 21.Atwell S, Meggers E, Spraggon G, Schultz PG. J Am Chem Soc. 2001;123:12364. doi: 10.1021/ja011822e. [DOI] [PubMed] [Google Scholar]
- 22.Henry AA, Romesberg FE. Curr Opin Chem Biol. 2003;7:727. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Degardin M, Lavergne T, Malyshev DA, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2014;136:826. doi: 10.1021/ja408814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Romesberg FE. Nature (London, U K) 2014;509:385. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa AK, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg FE. J Am Chem Soc. 2000;122:3274. [Google Scholar]
- 26.Hanawalt PC. Methods in Enzymology. Part A. Vol. 12. Academic Press; 1967. p. 702. [Google Scholar]
- 27.Marlière P, Patrouix J, Döring V, Herdewijn P, Tricot S, Cruveiller S, Bouzon M, Mutzel R. Angew Chem Intl Ed. 2011;50:7109. doi: 10.1002/anie.201100535. [DOI] [PubMed] [Google Scholar]
- 28.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML. J mol bio. 2009;388:48. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm K, Rueger W. Virology. 1992;189:640. doi: 10.1016/0042-6822(92)90587-f. [DOI] [PubMed] [Google Scholar]
- 30.Jolyon Terragni JB, Zheng Yu, Pradhan Sriharsa. Biochemistry. 2012;51:1009. doi: 10.1021/bi2014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.