Abstract
Background
Patient derived xenografts (PDXs) represent an essential tool in oncologic research, and we sought to further expand our repertoire of head and neck squamous cell carcinoma (HNSCC) while determining potential boundaries for this system.
Methods
We consented new patients for PDX development and determined if a 24-hour time delay from tumor excision to xenograft implantation affected PDX establishment. We developed a tissue microarray (TMA) from formalin fixed, paraffin embedded PDXs and their subsequent passages and carried out quantitative immunohistochemistry for EGFR, pEGFR, pAkt, pERK and ERCC1. First and last passaged PDXs were compared via a paired t-test to examine for the stability of protein expression across passages. We performed a similar comparison of the mutational profile of the patient tumor and resulting xenografts using a targeted sequencing approach.
Results
No patient/tumor characteristics influenced PDX take rate and the 24-hour time delay from tumor excision to xenograft implantation did not affect PDX establishment, growth or histology. There was no significant difference in biomarker expression between the first and last passaged PDXs for EGFR, pEGFR, pAkt, and ERCC1. For pERK there was a significant difference (p=0.002), but further analysis demonstrated this only arose in three of 15 PDXs. Targeted sequencing revealed striking stability of passenger and likely driver mutations from patient to xenograft.
Conclusions
The stability of protein expression across PDX passages will hopefully allow greater investigation of predictive biomarkers in order to identify ones for further pre-clinical and clinical investigation.
Keywords: Head and neck cancer, HNSCC, patient derived xenograft, PDX, mouse model
Introduction
Patient derived xenografts (PDXs), which are generated by directly implanting patient tumor tissue from a surgical resection or clinic biopsy either orthotopically or heterotopically into immunodeficient mice[1], have become a widely used model system for oncologic research. PDXs are hypothesized to more closely resemble a patient’s primary tumor than cell lines, and their histologic[2, 3] and molecular features[1, 2, 4, 5] mirror those of the primary cancer. PDXs have demonstrated establishment rates between 30 and 80% of the time across a range of tumor types.[1, 2, 4, 6] Following successful growth in the initial cohort of mice, tumors are excised and passaged into a new round of mice. In this manner, PDX tissue can be amplified and implanted into numerous mice to carry out therapeutic studies.[6, 7] Additionally, molecular analyses can be performed on pre-treatment tumors to identify biomarkers related to therapeutic response (predictive biomarkers). This represents an important facet and usage for PDXs, especially for cancers such as head and neck squamous cell carcinoma (HNSCC) where no currently validated predictive biomarkers exist.[8]
PDXs have become an essential tool in the preclinical development of novel therapeutics, with large cohort studies able to analogize a phase II clinical trial in terms of number of unique tumors studied.[9] While the largest cohorts of PDXs exist for breast, lung, colorectal, and pancreatic tumor types, a relatively small but growing number of HNSCC groups have been established. While characterization still differs from group to group, links between mutational profile and therapeutic response have now been investigated in several cases[4, 10, 11], and stability of proteomic markers from human to xenograft was evaluated in a single cohort[12].
Previously, our group established HNSCC PDXs from patients with both human papillomavirus (HPV) positive and HPV-negative cancers.[1] This initial work examined the stability in tumor histology and p16 expression across passages. Furthermore, we evaluated p53 and retinoblastoma expression of the PDXs to assess if this is related to HPV status and carried out initial chemoradiation experiments on a subset of PDXs. We subsequently determined that the time to re-implantation or storage solution used to house the tumor during the time delay did not have any impact on the maintenance of previously established PDXs.[13]
In this work, we sought to further define our population of HNSCC PDXs, determine potential boundaries of this model and expand the future utility of this system. First, we continued to consent patients and expand our repertoire of PDXs and determine whether disease or demographic factors impacted PDX establishment rates. Next, we assessed whether the time (up to 24 hours) from tumor excision in the operating room to implantation in the immunodeficient mice impacted initial PDX establishment, growth potential and histology. Finally, we evaluated the stability of both mutational and protein expression markers across PDX passaging. Using a targeted cancer mutation panel, we investigated the stability of mutations from the primary patient sample to multiple generations of PDX. Using quantitative IHC we determined whether any significant changes existed in the expression of putative predictive protein biomarkers across passaged PDXs. This work has important implications for the field of head and neck cancer research as it relates to the ongoing struggle to identify suitable predictive biomarkers to aid in the treatment of patients with HNSCC.
Materials and Methods
Mice, PDX propagation, and tumor harvesting
Six to eight week old female NOD-SCID gamma (NSG, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (Jackson Laboratories) were used for PDX establishment and amplification. UW-SCC1—36 PDXs were previously established and propogated in the lab.[1] New PDXs were generated and passaged in a similar manner; detailed methods are available in the Supplemental information.
New PDX establishment
Continued approval by the University of Wisconsin Institutional Review Board was obtained to discuss tissue donation with patients presenting to the clinic with newly diagnosed or recurrent HNSCCs. Consenting patients completed a form regarding tobacco/alcohol use, gender and age. At the time of surgery a small section of their tumor was obtained for PDX establishment in NSG mice as described previously[1]. Briefly, after receipt from the operating surgeon, tissue was mixed with a 1:1 mixture of media (Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2.5 μg/mL amphotericin B) and matrigel (catalog #354230, BD Biosciences, Inc) and minced into less than 1 mm3 pieces. 100–200 μl of the mixture was injected subcutaneously into two flanks of two to four NSG mice through an 18-gauge needle such that all sites received roughly equivalent amounts for tumor tissue. We previously demonstrated that neither time nor storage solution impacted tumor growth potential nor histology in a subsequent passage for previously established PDXs.[13] We expanded on these initial findings by evaluating whether a 24 hour time delay from initial excision in the operating room to ultimate implantation in the NSG mice impacted tumor take rate, growth potential, or histology for two new PDXs (UW-SCC63 and 64). After up to six months mice were sacrificed, any tumors harvested, weighed, and histological characteristics were evaluated by a board certified pathologist (C.Z.L). Additional details provided in supplemental methods.
Tissue microarray development and immunohistochemistry
A 196 core tissue microarray (TMA) was developed utilizing tissue from formalin fixed paraffin embedded (FFPE) tumors from each passaged PDX. All tumors were represented on the microarray by duplicate cores. The TMAs were sectioned (5 μm) and H&E stains were carried out on the thirtieth section. Remaining sections were stained by immunohistochemistry (IHC) for the expression of EGFR, pEGFR, pAkt, pERK and ERCC1 by standard IHC techniques[14], and detected with DAB. Additional details available in supplemental methods.
Tissue microarray analysis
The TMA was scanned by the UW TRIP lab’s Vectra System and analyzed by inForm Software v1.4.0 (PerkinElmer, Waltham, MA). We followed a standardized approach to quantitatively evaluate the expression of the TMA cores for each biomarker.[14] The inForm Software outputs the DAB mean optical density (MOD) as the measure of expression of each marker as a continuous value from 0 to 1. To evaluate the stability of protein expression, the expression of each biomarker in the first and last passaged PDXs were compared. The DAB MOD values for all the PDXs were compared between the first and last passages using a paired t-test. All tests were two sided, and SAS/STAT software (version 9.4) was used to perform these analyses. Additional analyses were carried out for passaged PDXs with respect to pERK expression. For PDXs with more than two passages, all passages were compared via a one-way analysis of variance (ANOVA) test while for PDXs with only two passages a two sample t-test with equal standard deviations was used (Graphpad Prism v6.0d). For all statistical analyses a p-value less than 0.05 was considered statistically significant. Additional details in supplemental information.
Hotspot mutational analysis
To investigate the stability of mutations from the original patient tumor to the initial PDX and subsequent passages through mice we employed an amplicon based next generation sequencing cancer panel. Total genomic DNA was isolated from FFPE tissue and sequenced using the Illumina TruSeq Cancer Amplicon panel run on a MiSeq2000. The DNA sequencing reads were adapter and quality(Q20) trimmed and aligned to the reference genome, GRCh37. Variants were called using MuTect[15] version 1.4 followed by annotation with SnpEff.[16] Variants were further filtered by minimum allele frequency and annotated by comparing to published studies searchable on cBioPortal for Cancer Genomics http://www.cbioportal.org/ [17, 18]. Additional method details in supplemental information.
Results
Patient and tumor characteristics do not predict PDX establishment
An additional 28 patients have been consented for PDX establishment, for a total of 65 patient consents to date.[1] We have obtained specimens from 34 of these patients, and their tissue was implanted in the NSG mice as described in the Materials and Methods. These tumors derived from numerous anatomic sites of the head and neck including the floor of mouth (n=11), base of tongue (n=7), tonsil (n=4), buccal mucosa (n=4) and oral tongue (n=3) as well as one tumor each from the retromolar trigone, supraglottis, nasopharynx, alveolar ridge and hypopharynx. In addition to the HNSCC tumor type we were able to generate a single adenoid cystic carcinoma PDX (UWSCC-60). The other 31 patients did not have tissue collected due to insignificant tumor quantity (tumor could not be spared for our work without impacting patient care), alterations in the surgical schedule or logistical complications. Overall, 27 of the 34 implanted tumors have established themselves as PDXs, which amounts to a take rate of 79.4%.
Specific characteristics of each patient/tumor that we collected tissue from is detailed in Supplemental Table S2. PDX take rate was compared to these characteristics to determine if any were related to the successful establishment of a PDX (Table 1). There was no statistically significant difference in PDX take rate based on age, gender, HPV status (p16 staining), alcohol use, tobacco use, tumor stage, nodal status, tumor differentiation or anatomic site. We confirmed that we had sufficient number of samples to support this finding with the following power calculation assuming a roughly ~75% overall engraftment rate. The null hypothesis is that the probability of PDX did not develop given a patient characteristic (ie nodal status) is no more than 0.6 i.e., H0: p≤0.6, and the alternative hypothesis that PDX established is H1: p≥0.80. A sample size of 34 patients and a one-sided alpha significance level of 0.05 gives approximately 82% power to test this hypothesis.
Table 1.
Patient and tumor characteristics and their influence on PDX establishment.
| Characteristica | Category | PDX did not develop | PDX established | P-valueb |
|---|---|---|---|---|
| Age at diagnosis (yrs) | <60 | 2 (15.4%) | 11 (84.6%) | 0.682 |
| >60 | 5 (23.8%) | 16 (76.2%) | ||
| Gender | Female | 2 (20.0%) | 8 (80.0%) | 1.000 |
| Male | 5 (20.8%) | 19 (79.2%) | ||
| Alcohol use (drinks/week) | Occasional (0–6) | 2 (13.3%) | 13 (86.7%) | 0.366 |
| Moderate (7–20) | 1 (12.5%) | 7 (87.5%) | ||
| Heavy (>20) | 4 (36.4%) | 7 (63.6%) | ||
| Tobacco use (pack-years) | 0–19 | 1 (7.1%) | 13 (92.9%) | 0.364 |
| 20 or greater | 5 (25.0%) | 15 (75.0%) | ||
| T Stage | T1 | 2 (25.0%) | 6 (75.0%) | 0.735 |
| T2 | 3 (30.0%) | 7 (70.0%) | ||
| T3 | 0 (0.0%) | 4 (100.0%) | ||
| T4 | 2 (16.7%) | 10 (83.3%) | ||
| Nodal status | Negative | 3 (23.1%) | 10 (76.9%) | 1.000 |
| Positive | 4 (19.0%) | 17 (81.0%) | ||
| Differentiation | Well | 3 (21.4%) | 11 (78.6%) | 1.000 |
| Moderate | 3 (20.0%) | 12 (80.0%) | ||
| Poor | 1 (20.0%) | 4 (80.0%) | ||
| HPV status (p16 IHC) | Negative | 5 (20.8%) | 19 (79.2%) | >0.999 |
| Positive | 2 (20.0%) | 8 (80.0%) | ||
| Anatomic site | Non-oropharynx | 4 (17.4%) | 19 (82.6%) | 0.656 |
| Oropharynx | 3 (27.3%) | 8 (72.7%) |
N (%) for categorical variables (all other characteristics).
P-values obtained via Fisher’s Exact test.
Time to implantation does not influence PDX establishment
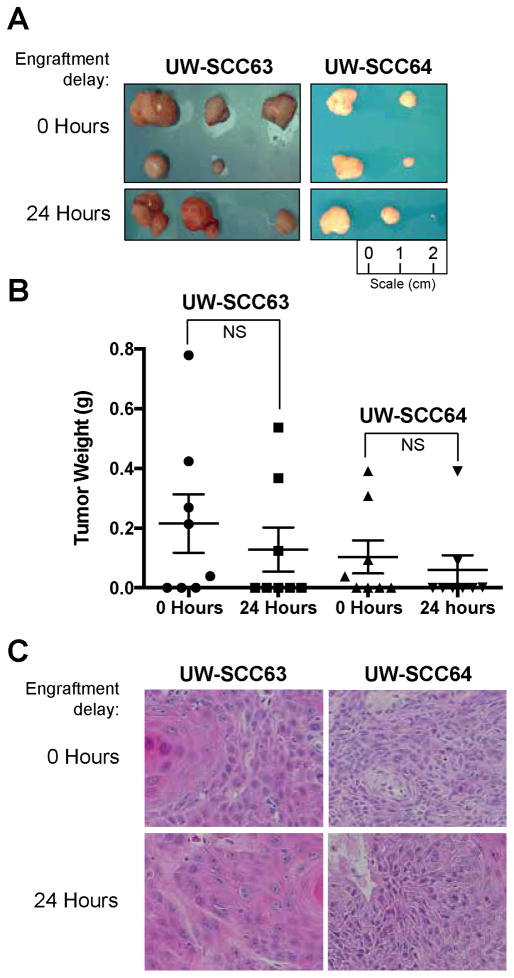
We determined whether a 24 hour delay between excision and engraftment in the NSG mice had any impact on PDX establishment or histological features using two recently consented patients with newly diagnosed head and neck cancers (Figure 1). As described previously, pre-implantation tumor weights were approximately equal for the time 0 and 24 hour groups within each PDX.[13]
Figure 1. Engraftment delay of 0 and 24 hour groups for UW-SCC63 and UW-SCC64 PDXs.
(A) At time of harvest (UW-SCC63-12 weeks post-implantation, UW-SCC64 – 9 weeks post implantation) tumors were excised from the mice and images taken of all available tumors. (B) Box plots comparing the weight of each tumor at time of harvest. A weight of 0 grams was used for implantation sites that did not produce any tumors (NS: Not significant). (C) Representative images from the H&E stained slides for both groups from UW-SCC63 and UW-SCC64 PDXs.
Tumors were harvested when larger masses for a given PDX reached approximately 1cm3. The NSG mice with the UW-SCC63 tumors were euthanized 12 weeks after initial implantation. Five of eight tumors grew in the time 0 group and three of eight were present in the 24 hour group. More importantly, there was no significant difference in mean tumor weight between these two groups (p=0.489). The mean tumor weight was 0.216 grams (g) (standard deviation (SD) 0.276 g) for the time 0 group and 0.129 g (SD 0.209 g) for the 24 hour tumors. Furthermore, tumors from both groups had similar histological features including moderate differentiation, 30–35% keratinization, 5% necrosis/cystic change and the presence of infiltrative features. Tumors from both the time 0 and 24 hour groups have been successfully passaged two times.
Nine weeks after implantation, mice bearing the UW-SCC64 PDXs were euthanized, and all tumors were resected from each mouse. Four of eight tumor sites grew in the time 0 group and three of eight developed in the 24 hour cohort. There was no statistically significant difference between the mean tumor weight for mice in the 0 versus 24 hour groups (p=0.564), and the average tumor weight for mice in the 0 hour group was 0.104 g (SD 0.156 g) and at 24 hours the average was 0.060 g (SD 0.137 g). Additionally, all tumors had similar histological attributes that included moderate differentiation, no keratinization, 30–60% necrosis/cystic change and an infiltrative phenotype. Similar to the UW-SCC63 PDX, tumors from both the time 0 and 24 hour cohorts have already been successfully passaged three times.
Putative protein biomarkers remain stable across passaged PDXs
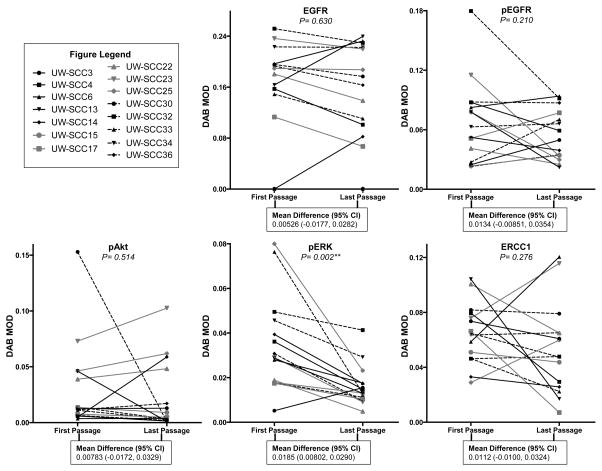
From the 22 PDXs that had been previously established and passaged in NSG mice,[1] 15 had high quality tissue available from multiple passages to be included in the TMA and biomarker analyses. To evaluate potential changes in previously described biomarker expression across passages, we aggregated protein expression data (DAB MOD values) from the first and last passages for each PDX and determined if there were any significant changes in biomarker expression for the 15 PDX cohort (Figure 2). For EGFR the mean difference (95% confidence interval (CI)) in DAB MOD values was 0.00526 (−0.0177, 0.0282) between the first and last passages, which was not significantly different (p=0.630). There was also no significant difference between the first and last passages for pEGFR (p=0.210), pAkt (p=0.514) and ERCC1 (p=0.276). With a mean difference of 0.0185 (0.00802, 0.0290), pERK represented the only biomarker where there existed a statistical difference between the first and last passages (p=0.002). Supplemental Figure 1 A and B contains representative IHC images for each biomarker from the first and last passages of every PDX.
Figure 2. Comparing biomarker expression between the first and last passages of each PDX.
Depiction of the protein expression (DAB MOD value) of EGFR, pEGFR, pAkt, pERK and ERCC1 for the first and last passages of each PDX (DAB MOD: 3,3′-diaminobenzidine mean optical density). The mean difference and 95% confidence intervals (CI) are presented for the comparisons between the first and last passages of each PDX by biomarker (**P<0.01). Tissue data was used for all biomarkers except ERCC1, where the nuclear fraction was solely utilized (nuclear DAB MOD values).
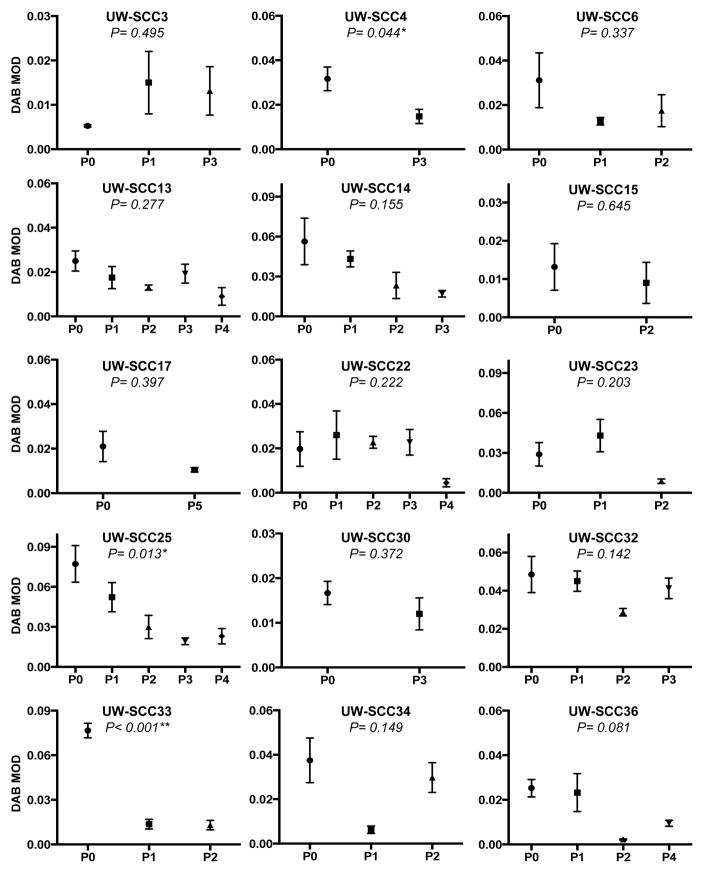
Owing to the statistically significant difference demonstrated between the first and last passages for pERK, we carried out further analyses for this biomarker (Figure 3). The expression for pERK was compared among each passage within individual PDXs. There was a statistically significant difference across passages for only three of the 15 PDXs: UW-SCC4 (p=0.044), UW-SCC25 (p=0.013) and UW-SCC33 (p<0.001).
Figure 3. Passaged PDXs and pERK expression.
Depiction of the mean with standard error of the mean for the pERK expression (DAB MOD value) for each IHC image analyzed for every passage within individual PDXs on the TMA (*P<0.05, **P<0.01; P0: passage 0, P1: passage 1, etc).
Cancer hotspot mutation analysis reveals stability of genetic alterations from patient to PDX
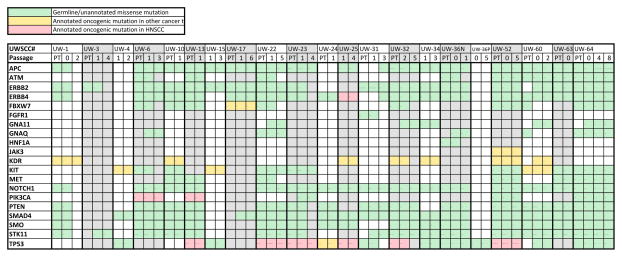
Using archived FFPE tissue from both patient clinical pathology samples and resulting PDXs we used an amplicon based next generation sequencing (NGS) cancer panel to identify mutations in our cohort and evaluate their stability from patient to xenograft and across PDX passages. Of the 34 patient samples that successfully formed PDXs, we were able to generate sequence data of sufficient quality at multiple passages for 21. Non-synonymous mutations are summarized in Figure 4, and were annotated based on published studies searchable on cBioPortal. We identified a high frequency of previously unannotated mutations that are potentially germline alterations, mutations in KDR, cKIT, FBXW7, and JAK3 that have been annotated as oncogenic in other tumor types, as well as known HNSCC oncogenic mutations in PIK3CA and TP53. The PIK3CA SNPs were the well-described helical domain activating mutations E545K and E542K. Consistent with TCGA and other published reports, the TP53 mutations were primarily loss of function mutations clustered in the HPV-negative tumors. 0 of 6 HPV positive tumors and 7 of 15 HPV negative tumors contained TP53 mutations (p<0.045 Chi-squared test). Specific variant calls are listed in Supplemental Figure 2. Due to the archival nature of these samples we did not have matched normal tissue to confirm the somatic nature of any mutations. Strikingly, for both the oncogenic and unannotated mutations, the presence of the variant was generally conserved from the original patient sample to the xenograft and across PDX passage where multiple passages were available. Additionally, the allele frequency of a given mutation was also generally stable from generation to generation (data not shown). These results indicate that both passenger and likely driver mutations in HNSCC PDXs reflect the mutational profile of the original patient and are stable across multiple PDX passages, increasing the utility of the model system.
Figure 4. Mutational profile of patient and xenograft samples.
Summary of non-synonymous variants identified in cancer targeted sequencing panel. UWSCC numbers correspond to patients in Table S2. PT column indicates patient primary tumor sample, other passage columns indicate first and last xenograft passages with usable sequence data for a given tumor. Green boxes- Germline/unannotated missense mutation. Yellow boxes – Annotated oncogenic mutation in other cancer type. Red boxes – Annotated oncogenic mutation in HNSCC.
Discussion
PDXs represent a critical model system for oncologic research, and they have been developed for many carcinomas including lung[3, 19], pancreatic[20, 21], breast[2], renal[6] and head and neck.[1, 4] In this work we sought to better characterize our cohort of HNSCC PDXs in order to maximize the applicability of this model for advancing oncologic research and discovery.
First, we determined whether any patient or tumor characteristics were related to successful PDX establishment. Interestingly, although our earlier work demonstrated a significant relationship between nodal status and PDX take rate (p=0.020),[1] this relationship did not hold true with the addition of the new patients (p=1.000). This included HPV status, as determined by p16 staining, where we were able to generate 8 HPV+ PDXs with a similar engraftment rate as HPV-tumors. The effect of HPV status on engraftment has been inconsistent in published reports[10, 12]; our results demonstrate that PDXs can be reliably generated for this tumor type and provide a critical tool for studying this disease sub-type. Overall, we have shown that PDXs can be established from a wide range of tumor or patient characteristics, highlighting that this model system captures the variety of the head and neck tumors.
Prior to undertaking this work, our group and others believed strongly that a short time interval from tumor excision to implantation was critical for xenograft establishment[3, 6, 22, 23]. In our prior work, we solely showed that tumors could develop at both time 0 and 24 hours[13], but we did not examine the histology nor the capability for these tumors to be passaged into subsequent rounds of mice. Here we demonstrated that for two PDXs similar numbers of tumors developed in the time 0 and 24 hour. Moreover, the tumors from these PDXs had similar histological features at both time points, and tumors could be successfully passaged into subsequent rounds of NSG mice. While additional tumors will need to be analyzed to confirm this effect, these initial cases combined with our previous work demonstrating the ability to successfully passage tumors up to 48 hours post-excision in established PDXs[13] suggest that handling time is not critical for successful HNSCC PDX establishment, further expanding the utility of this model system.
Most importantly, we believe the PDX model could play a pivotal role in identifying predictive biomarkers for cancers such as HNSCC for which there are no clinically validated biomarkers to aid in therapeutic decision-making[24, 25]. Current investigations have been underway to identify predictive biomarkers for standard HNSCC therapies including cisplatin, cetuximab and radiation. Lower levels of the nucleotide excision repair pathway member ERCC1 are correlated with cisplatin sensitivity[14, 26, 27]. Investigators have studied total EGFR levels in relation to cetuximab response[28, 29], while others evaluated the association between total EGFR and ERCC1 with respect to radiation sensitivity.[30, 31] Our work with cell line xenografts extended these analyses to include related EGFR-signaling members including pEGFR, pAkt and pERK in relation to both cetuximab and radiation response[14].
Since therapeutic studies are often carried out on PDXs at different passages, we must be confident that putative predictive biomarkers remain stable across PDX passages. Given the potential importance of EGFR, pEGFR, pAkt, pERK and ERCC1 expression, we evaluated if these proteins were stable across passaged PDXs. Importantly, our results demonstrated that for the 15 PDXs analyzed, these biomarkers were stable between the first and last passages, except for pERK. However, when the pERK data was analyzed in greater detail, we demonstrated that only three of the 15 PDXs had significant changes across passages. Consistent with other investigations[12] these results indicate that while protein expression is typically stable across passage, this is not uniformly true and care must be taken when utilizing such markers.
Similar to the protein expression data, we also found reliable stability of the mutational profile from the original patient tumor to multiple passages of PDX. While this cancer focused panel did not enable a genome wide analysis, it nonetheless revealed that the majority of both passenger and likely driver variants were present in all instances of the tumor and xenograft. For those where the variant call was not consistent, in many cases it was identified but was slightly below the 5% allele frequency threshold we set to report a variant. Our results are consistent with another HNSCC PDX cohort assayed with the same targeted sequencing cancer panel[10] which also demonstrated reliable mutation stability across passages. This stability greatly expands the utility of the PDX model as it allows for the use of later generation tissue and greater amplification of tissue to perform larger studies. This knowledge discovered for our cohort enables the use of targeted therapeutics tailored to a specific mutational profile such as PIK3CA activated or TP53-wt, and we are undertaking studies to exploit these insights.
This work carries important limitations that must be taken into consideration. First, it would have been important to compare the protein biomarker expression in the passaged PDXs to the primary patient tissue; however, we did not have access to this tissue when creating the TMAs. Second, variability in each PDX’s growth rate, the time to perform multiple passages and the appropriate use of tumor resulted in different last passages for these PDXs. We conducted further analyses (data not shown) where we compared P0 to the last available passage (P3 or P5) separately for each protein and the results were consistent with the overall comparison between the first and last passages. While there are numerous putative biomarkers currently under investigation, we elected to focus on these five as our lab and others have investigated these in relation to standard of care therapies. Although the targeted sequencing panel highlights frequently mutated genes in a range of cancer types, it certainly is not exhaustive and detects only SNPs and small INDELs not major truncations or inversions, and we have not yet assayed our cohort for gene copy gain or loss. As mentioned previously, it would be important to evaluate other elements of the initial PDX establishment procedure aside from solely time (up to 24 hours), but owing to the limited amount of primary patient tissue we received from the operating room we could only generate two groups per PDX for this experiment. In a challenge broadly affecting all groups working with PDX models, the immunodeficient nature of the murine hosts removes an essential component of anticancer defenses and therapeutic response. We recognize this limitation and have begun to address it by generating mouse models with humanized immune systems through collaborators at our institution. A humanized PDX approach will enable the investigation of the role of the immune response in anticancer defense across wide array of characterized human tumors in contrast to genetic mouse models that typically rely on a single alteration to drive oncogenesis.
In this work we continued to expand our cohort of our head and neck cancer PDXs while searching for potential boundaries in this model system. We determined that PDXs can be successfully established even if there is a 24 hour time delay between the initial tumor excision in the operating room and implantation in the NSG mice allowing more flexibility in obtaining fresh patient tumor tissue and increasing the number of PDXs generated for oncologic research. Moreover, we revealed that putative predictive biomarkers maintain stability across passaged PDXs. This is important information as we attempt to utilize predictive biomarker identification in the PDXs and translate this information into therapeutic pre-clinical and clinical studies[32].
Supplementary Material
Highlights.
Head and neck cancer PDXs can be generated for a wide range of tumor characteristics.
A 24-hr delay post surgical resection does not impact engraftment rate.
Protein expression biomarkers are stable across PDX passages
Mutational profile of PDXs are stable from patient tumor to multiple PDX generations.
Acknowledgments
Financial support and disclosures: Supported by a R00 CA160639 and Department of Human Oncology Seed Grant (RJK), UWCCC and UWCCC/MIR/WID institutional grants (PFL), and a UW ICTR-Shapiro fellowship (APS). IO is supported by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520 and the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR000427.
We acknowledge Paul M. Harari, Kwang P. Nickel, Hao-Shun Huang, Alexandra D. Torres, Robert Z. Yang, and Eric A. Armstrong for assistance with initial xenograft implantation. We also would like to acknowledge the University of Wisconsin Translational Research Initiatives in Pathology laboratory, in part, supported by the UW Department of Pathology and Laboratory Medicine and UWCCC grant P30 CA014520, for use of its facilities and services. The author(s) thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing the DNA sequencing facilities and services.
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kimple RJ, Harari PM, Torres AD, Yang RZ, Soriano BJ, Yu M, et al. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:855–64. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature medicine. 2011;17:1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong X, Guan J, English JC, Flint J, Yee J, Evans K, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:1442–51. doi: 10.1158/1078-0432.CCR-09-2878. [DOI] [PubMed] [Google Scholar]
- 4.Keysar SB, Astling DP, Anderson RT, Vogler BW, Bowles DW, Morton JJ, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol. 2013;7:776–90. doi: 10.1016/j.molonc.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng S, Creighton CJ, Zhang Y, Sen B, Mazumdar T, Myers JN, et al. Tumor grafts derived from patients with head and neck squamous carcinoma authentically maintain the molecular and histologic characteristics of human cancers. J Transl Med. 2013;11:198. doi: 10.1186/1479-5876-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivanand S, Pena-Llopis S, Zhao H, Kucejova B, Spence P, Pavia-Jimenez A, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Science translational medicine. 2012;4:137ra75. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jimeno A, Feldmann G, Suarez-Gauthier A, Rasheed Z, Solomon A, Zou GM, et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Molecular cancer therapeutics. 2009;8:310–4. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucs AV, Saltman B, Chung CH, Steinberg BM, Schwartz DL. Opportunities and challenges facing biomarker development for personalized head and neck cancer treatment. Head & neck. 2013;35:294–306. doi: 10.1002/hed.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 10.Klinghammer K, Raguse JD, Plath T, Albers AE, Joehrens K, Zakarneh A, et al. A comprehensively characterized large panel of head and neck cancer patient-derived xenografts identifies the mTOR inhibitor everolimus as potential new treatment option. Int J Cancer. 2015;136:2940–8. doi: 10.1002/ijc.29344. [DOI] [PubMed] [Google Scholar]
- 11.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–9. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Wheeler S, Park Y, Ju Z, Thomas SM, Fichera M, et al. Proteomic Characterization of Head and Neck Cancer Patient-Derived Xenografts. Mol Cancer Res. 2016;14:278–86. doi: 10.1158/1541-7786.MCR-15-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein AP, Saha S, Liu CZ, Hartig GK, Lambert PF, Kimple RJ. Influence of handling conditions on the establishment and propagation of head and neck cancer patient derived xenografts. PloS one. 2014;9:e100995. doi: 10.1371/journal.pone.0100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein AP, Swick AD, Smith MA, Blitzer GC, Yang RZ, Saha S, et al. Xenograft assessment of predictive biomarkers for standard head and neck cancer therapies. [Accepted November 18, 2014];Cancer medicine. doi: 10.1002/cam4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani P, Platts A, Wang lL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer research. 2009;69:3364–73. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrido-Laguna I, Uson M, Rajeshkumar NV, Tan AC, de Oliveira E, Karikari C, et al. Tumor engraftment in nude mice and enrichment in stroma-related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:5793–800. doi: 10.1158/1078-0432.CCR-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4652–61. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 22.Wennerberg J, Trope C, Biorklund A. Heterotransplantation of human head and neck tumours into nude mice. Acta oto-laryngologica. 1983;95:183–90. doi: 10.3109/00016488309130933. [DOI] [PubMed] [Google Scholar]
- 23.Rygaard J, Povlsen CO. Heterotransplantation of a human malignant tumour to “Nude” mice. Acta pathologica et microbiologica Scandinavica. 1969;77:758–60. doi: 10.1111/j.1699-0463.1969.tb04520.x. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira MB, De Souza JA, Cohen EE. Role of molecular markers in the management of head and neck cancers. Current opinion in oncology. 2011;23:259–64. doi: 10.1097/CCO.0b013e328344f53a. [DOI] [PubMed] [Google Scholar]
- 25.Langer CJ. Exploring biomarkers in head and neck cancer. Cancer. 2012;118:3882–92. doi: 10.1002/cncr.26718. [DOI] [PubMed] [Google Scholar]
- 26.Handra-Luca A, Hernandez J, Mountzios G, Taranchon E, Lacau-St-Guily J, Soria JC, et al. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3855–9. doi: 10.1158/1078-0432.CCR-07-0252. [DOI] [PubMed] [Google Scholar]
- 27.Jun HJ, Ahn MJ, Kim HS, Yi SY, Han J, Lee SK, et al. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. British journal of cancer. 2008;99:167–72. doi: 10.1038/sj.bjc.6604464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licitra L, Storkel S, Kerr KM, Van Cutsem E, Pirker R, Hirsch FR, et al. Predictive value of epidermal growth factor receptor expression for first-line chemotherapy plus cetuximab in patients with head and neck and colorectal cancer: analysis of data from the EXTREME and CRYSTAL studies. European journal of cancer. 2013;49:1161–8. doi: 10.1016/j.ejca.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. The New England journal of medicine. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 30.Mehra R, Zhu F, Yang DH, Cai KQ, Weaver J, Singh MK, et al. Quantification of excision repair cross-complementing group 1 and survival in p16-negative squamous cell head and neck cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:6633–43. doi: 10.1158/1078-0432.CCR-13-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang K, Ang KK, Milas L, Hunter N, Fan Z. The epidermal growth factor receptor mediates radioresistance. International journal of radiation oncology, biology, physics. 2003;57:246–54. doi: 10.1016/s0360-3016(03)00511-x. [DOI] [PubMed] [Google Scholar]
- 32.Stein AP, Swick AD, Smith MA, Blitzer GC, Yang RZ, Saha S, et al. Xenograft assessment of predictive biomarkers for standard head and neck cancer therapies. Cancer Med. 2015 doi: 10.1002/cam4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.