Abstract
Lysine acetylation is a reversible post-translational modification (PTM) of cellular proteins and represents an important regulatory switch in signal transduction. Lysine acetylation, in combination with other PTMs, directs the outcomes as well as the activation levels of important signal transduction pathways such as the nuclear factor (NF)-κB pathway. Small molecule modulators of the ‘writers’ (HATs) and ‘erasers’ (HDACs) can regulate the NF-κB pathway in a specific manner. This review focuses on the effects of frequently used HAT and HDAC inhibitors on the NF-κB signal transduction pathway and inflammatory responses, and their potential as novel therapeutics.
Introduction
Lysine acetylation is a reversible post-translational modification (PTM) of cellular proteins and represents an important regulatory switch in signal transduction cascades [1,2]. An increasing number of studies highlight the importance of lysine acetylation as a key PTM, directing the outcomes as well as the activation levels of important signal transduction pathways such as the nuclear factor (NF)-κB pathway. For example, acetylation of NF-κB transcription factors p65 and p50 plays an important part in their nuclear localization and transcriptional activity [3]. Similar phenomena have been observed for other pathways [4]. Next to this, acetylation of histones connected to specific genes has an important role in gene-specific transcription in the NF-κB pathway [3]. Furthermore, an increasing number of reports describe significant levels of crosstalk between lysine acetylation and other PTMs, such as ubiquitinylation, methylation and phosphorylation, in the NF-κB pathway. For example, competition between acetylation and ubiquitinylation on the same lysine residues is observed for transcription factor p65 [5]. This highlights the fact that acetylation is not a sole determining factor but, rather, is a regulator working in concert with other PTMs at multiple levels in signaling cascades.
Lysine acetylations are generally regulated by ‘writers’ and ‘erasers’, which are denoted as histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively, owing to their original discovery as histone-modifying enzymes. An important future challenge is to identify and quantify distinct HAT and HDAC activities in distinct signaling pathways such as the NF-κB pathway, as well as their aberrations in disease (models). Considering the importance of lysine acetylation in the NF-κB pathway (Fig. 1), small molecule modulators of HATs and HDACs have great potential to regulate this signaling cascade specifically, which is an important aim in drug discovery.
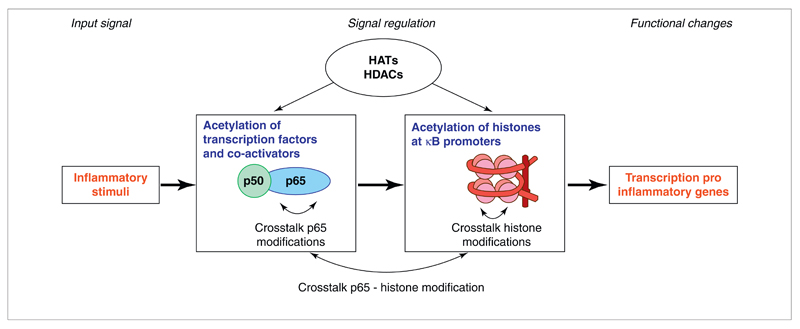
Figure 1.
Schematic representation of the diverse roles of lysine acetylation in the activation of the nuclear factor (NF)-κB pathway. Lysine acetylations of the transcription factors as well as their co-activators play an important part in the duration of the response and the signaling output. Lysine acetylation status of the histones works in concert with acetylation status of the transcription factors to enable or disable transcription of specific genes. Crosstalk of acetylation with other PTMs is an important component in the NF-κB pathway. Abbreviations: HATs, histone acetyltransferases; HDACs, histone deacetylases.
Focusing on the NF-κB pathway, here we summarize the effects of lysine acetylation of the p65 transcription factor as well as histones. In addition, we highlight the role of crosstalk between lysine acetylation and other PTMs such as methylation and phosphorylation. Furthermore, we discuss the effects of frequently used small molecule HAT and HDAC inhibitors on the NF-κB signal transduction pathway and inflammatory responses in vitro and in vivo.
Lysine acetylation as a regulator of the NF-κB pathway
In 2001, it was discovered that acetylation of p65 inhibits binding to the inhibitory complex IκBα, and thus stimulates gene transcription; whereas deacetylation promotes IκBα binding and nuclear export [6]. This study triggered intense interest in lysine acetylations of the seven lysine residues (122, 123, 218, 221, 310, 314, 315) of p65 that are subject to this PTM. These acetylations have specific roles in activation of the NF-κB pathway and have been previously reviewed [7,8]. Importantly: acetylation of lysines 122 and 123 decreases DNA binding [9]; acetylation at lysines 218 and 221 increases binding to κB enhancers; and acetylation at lysine 310 is essential for full transcriptional activity [10]. In addition, acetylations of specific lysine residues in histone H3 and H4 play an important part in NF-κB-mediated gene transcription as reviewed [3].
Lysine acetylation does not act alone: crosstalk between lysine acetylation and other PTMs
The acetylation of lysine residues in the NF-κB transcription factor and in histones (and numerous other cellular targets) can be dramatically affected by the PTM state of other constituent amino acids. These so-called ‘crosstalk’ mechanisms act, presumably, via increasing or decreasing the affinity of the substrate protein for the respective HAT or HDAC complexes involved in their acetylation. A recent review nicely illustrates the importance of crosstalk between PTMs on the NF-κB transcription factor [8]. In addition, previous reviews illustrate the importance of crosstalk between lysine acetylation and other PTMs in the histones [11–14]. Here, we highlight some specific examples that demonstrate the crucial involvement of crosstalk in NF-κB activation as well as in histones implicated in inflammation. The examples described below are limited to known cases of crosstalk within the same protein (cis crosstalk). In addition, a growing number of examples make it clear that similar mechanisms also operate in modulating protein–protein interactions including those between the peptides tails of different histones (trans crosstalk).
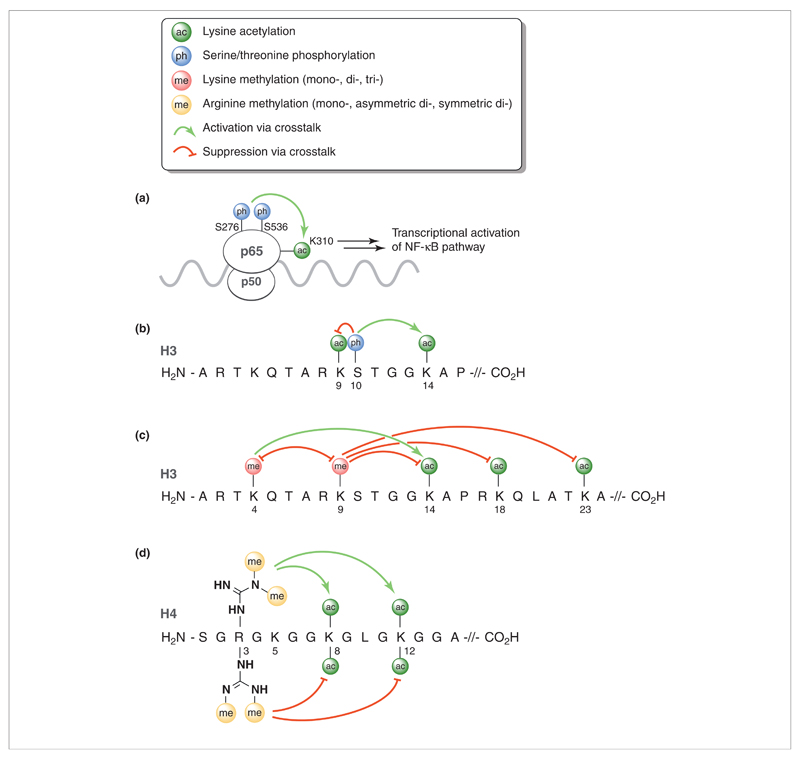
A specific example of crosstalk in the NF-κB pathway involves the phosphorylations of p65 at serines 276 and 536, which serves to enhance the p300-mediated acetylation of lysine 310. This, in turn, leads to an overall transcriptional activation of the NF-κB pathway (Fig. 2a) [15]. In addition, it has been found that phosphorylation of serine 276 is required for binding of p65 to the coactivator CREB-binding protein (CBP), which promotes proinflammatory gene transcription.
Figure 2.
Examples of various post-translational modifications (PTMs) and their crosstalk interactions with lysine acetylation in the p65 transcription factor and histone proteins. Abbreviation: NF-κB, nuclear factor κB.
Phosphorylation also has a major role in the crosstalk observed within histone proteins. One of the earliest reported and best-studied examples of crosstalk in histones involves the phosphorylation of serine 10 in histone 3 (H3S10) and its effect on lysine acetylation (Fig. 2b). Several kinases are known to phosphorylate H3S10. These include AuroraB and other members of the Aurora/Ipl 1 kinase family, as well as kinases implicated in transcriptional regulation such as the yeast non-specific serine/threonine protein kinase 1 (Snf1) and mammalian Proto-oncogene serine/threonine protein kinase 1 (Pim1), Ribosomal s6 kinase (Rsk), Mitogen and stress activated kinase 1 (Msk1), and Mitogen and stress activated kinase 2 (Msk2) kinases [16–18]. Phosphorylation of H3S10 leads to the stimulation of acetylation at lysine 14 (H3K14) with the prototypical histone acetyltransferase general control nonderepressible (Gcn)5 displaying an up to tenfold preference for acetylation of H3K14 when H3S10 is phosphorylated [19,20]. In contrast to the enhancement of acetylation at H3K14, phosphorylation of H3S10 completely blocks acetylation at the directly adjacent lysine 9 (H3K9) residue [21], clearly illustrating the varying crosstalk effects that the same PTM can impart.
In addition to serine phosphorylation, the methylation of lysine and arginine side chains is another common PTM known to impact lysine acetylation via crosstalk. The side chain of lysine can be mono-, di- or tri-methylated, often with varying crosstalk effects (Fig. 2c). For example, lysine methylation at lysine 4 in histone H3 (H3K4) leads to increased acetylation of H3K14 and other lysine residues in histone H3 by p300 and other acetyltransferases [22]. Further evidence suggests that the degree to which H3K4 is methylated directly influences the associated extent of H3 acetylation [23]. As a result, the trimethylation of H3K4 is generally seen as a marker for transcriptional activation given its direct role in promoting acetylation. Conversely, trimethylation of lysine 9 in histone 3 (H3K9) is linked to transcriptional repression and is associated with reduced histone H3 acetylation. In vitro studies have revealed that trimethylation of H3K9 leads to inhibition of H3 acetylation at lysines 14, 18 and 23 [23]. Given their opposing effects, it is perhaps not surprising that methylations of H3H4 and H3K9 appear to be mutually exclusive [22,24,25].
Likewise, the arginine side chain can be either mono-methylated or, more commonly, di-methylated to yield either asymmetric or symmetric dimethyl arginine. Interestingly, numerous examples of crosstalk between arginine methylation and lysine acetylation are known. A specific example involves the asymmetric dimethylation of arginine 3 in histone 4 (H4R3) by protein arginine N-methyltransferase (PRMT)1, which leads to increased histone 4 acetylation by p300, with the largest enhancement observed for acetylation at lysine 8 and 12 (Fig. 2d) [26]. Given its role in promoting H4 acetylation, the asymmetric dimethylation of H4R3 by PRMT1 is generally associated with the activation of gene transcription. In a striking example of the subtleties at play in histone crosstalk, it has also been found that symmetric dimethylation at H4R3, as performed by PRMT5, antagonizes the crosstalk effects induced by PRMT1 (Fig. 2d). Counter to asymmetric dimethylation at H4R3, the PRMT5-mediated symmetric dimethylation of the same arginine residue is associated with deacetylation of H4 and leads to repressed transcription [27,28]. In addition, a recent report by Zheng and colleagues has shown that the acetylation of lysine 5 in histone 4 differentially affects methylation at H4R3 [29]. Acetylation of H4K5 was found to deactivate the asymmetric dimethylation of H4R3 by PRMT1 while enhancing symmetric dimethylation at the same arginine by PRMT5. From this, it is clear that crosstalk mechanisms play a significant part in acetylation of the p65 transcription factor and histones. Exploiting such mechanisms could provide avenues for the development of novel therapeutics designed to affect acetylation.
HATs in the NF-κB pathway
On the basis of their primary structure homology, HATs have been divided into five families. Three families that have been studied extensively are the Gcn5-related N-acetyltransferase (GNAT) family, represented by p300/CBP-associated factor (PCAF) and Gcn5; the p300/CBP family, including CBP and p300; and the MYST (the acronym MYST derives from the four founding members of this HAT family: mammalian MOZ, yeast Y bf2/Sas3 and Sas2, and mammalian Tip60) family including males absent on the first (MOF) and TAT-interacting protein 60 (Tip60) [30]. The HATs p300 and PCAF acetylate lysines 122 and 123 of p65 [9] and p300 has been described to acetylate lysines 310, 314 and 315 [31]. The role of HATs in acetylation of the NF-κB transcription factor as well as acetylation of the histones connected to κB promoters has been reviewed [3]. Interestingly, a recent study demonstrates that Tip60 is a co-activator of several NF-κB target genes and exerts its action via protein–protein interactions with p65. It appears that Tip60 binds earlier to the κB promoters than p65 and simultaneously promotes histone acetylation, which indicates that Tip60 could serve as a platform to promote NF-κB-mediated gene transcription [32].
HAT inhibitors mainly lead to inhibition of the NF-κB pathway
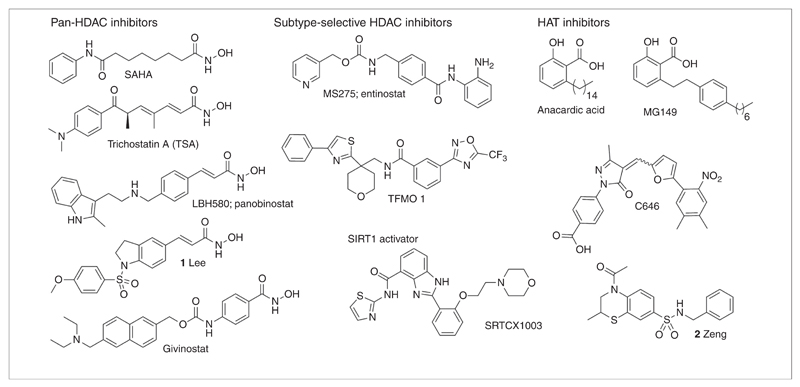
Several inhibitors of histone acetyltransferases are known and have been previously reviewed [3,33]. The natural product anacardic acid (Fig. 3, Table 1) is a small molecule inhibitor of HATs such as p300 and PCAF [34] and inhibits NF-κB-mediated gene transcription [35]. This inhibitor has been used as a starting point for the development of novel inhibitors such as the alkylidene malonates [36]. The novel anacardic acid derivative MG149 (Fig. 3, Table 1) demonstrates selectivity toward the MYST type of HATs: Tip60 and MOF [37], and this molecule effectively suppresses suberanilohydroxamic acid (SAHA)-induced hyperacetylation [38]. In addition, DNA microarrays demonstrated that MG149 inhibits the p53 and the NF-κB pathways, as well as a very limited number of other pathways [38].
Figure 3.
Structures of small molecules that interact with writers (histone acetyltransferases; HATs) or erasers (histone deacetylase; HDACs) of lysine acetylations.
Table 1. Small molecules that interfere with writers (HATs) or erasers (HDACs) of lysine acetylations, and their selectivity and effects on disease models for NF-κB-mediated inflammation.
| Compound | Selectivity | Effects in disease models | Refs |
|---|---|---|---|
| HAT inhibitors | |||
| Anacardic acid | p300 and PCAF inhibitor | Inhibits the NF-κB pathway | [35] |
| MG149 | Tip60 and MOF selective inhibitor | Inhibits SAHA-induced hyperacetylation | [37,38] |
| Inhibits the NF-κB and p53 signaling pathways | |||
| C646 | p300 selective inhibitor (Ki 0.4 μm) | Increases apoptosis by inhibition of the androgen receptor and NF-κB pathway | [44,46,47] |
| Repression of gene expression | |||
| Inhibition of COX-2 expression | |||
| Compound 2 (Fig. 3) | p300 HAT inhibitor (IC50 3.4 μm) | – | [45] |
| HDAC inhibitors or activators | |||
| SAHA | Pan-HDAC inhibitor | Suppression of LPS-induced cytokine release in vitro and in vivo | [50,62] |
| Trichostatin A (TSA) | Pan-HDAC inhibitor | Inhibition of TNFα in vitro | [49,62] |
| LBH580 (panobinostat) | Pan-HDAC inhibitor | Enhances NF-κB activation | [61,62] |
| ITF2357 (givinostat) | Pan-HDAC inhibitor | Therapeutic benefit in patients suffering from juvenile idiopathic arthritis | [56,62] |
| Compound 1 (Fig. 3) | Unknown | Inhibition of IL6, PGE2, TNFα and NO in RAW 264.7 macrophages | [55] |
| Anti-inflammatory in paw edema mouse model | |||
| MS275 (Entinostat) | HDAC1-3 selective inhibitor | Enhances NF-κB activation | [58,62] |
| TFMO 1 | Class IIa selective inhibitor | – | [63] |
| SRTCX1003 | SIRT1 activator | Inhibits NF-κB activation | [67] |
COX, cyclooxygenase; HDAC, histone deacetylase; IL, interleukin; LPS, lipopolysaccharide; MOF, males absent on the first; NF-κB, nuclear factor κB; NO, nitric oxide; PCAF, p300/CBP associated factor; PGE, prostaglandin E; RAW, Abelson murine leukemia virus transformed; SAHA, suberanilohydroxamic acid; SIRT, Sirtuin; Tip60, TAT-interacting protein 60; TNF, tumor necrosis factor.
Next to this, HTS identified the isothiazolones as HAT inhibitors [39]. However, attempts to optimize this class failed to give inhibitors with higher potency and selectivity [40–42], which could mainly be attributed to the exceptionally high reactivity of isothiazolones for thiolates [43]. Fortuitously, virtual screening enabled the identification of C646 (Fig. 3, Table 1) as the first potent, selective and cell-permeable p300 HAT inhibitor (Ki 0.4 μm) [44]. In addition, another recent virtual screening study describes the identification of a novel cell-permeable inhibitor (compound 2, Fig. 3, Table 1) of p300, which is active at the same concentrations as C646 [45]. This demonstrates the strength of virtual screening as a strategy for identification of novel inhibitors for challenging targets such as HATs.
Regarding the use of C646 in cellular models, one study on prostate cancer cell lines demonstrated that siRNA-mediated and C646-mediated inhibition of p300 increase apoptosis, which is, among other things, caused by inhibition of the androgen receptor and the NF-κB pathway [46]. Another group demonstrated that p300 binds to the cyclooxygenase (COX)-2 promoter, and that inhibition of p300 activity by C646 diminished the p300 promoter binding and the expression of COX-2. Interestingly, this study was done in animal models in which C646 was administered via a lumbar intrathecal catheter, which demonstrates that HAT inhibitors can be administered locally in animal models [47]. Taken together these data demonstrate that C646 performs very well in cell-based studies and upon local administration in animal models. Inhibition of p300 by siRNA and the chemical inhibitor C646 leads to subsequent inhibition of, among others, the NF-κB pathway.
HDAC classes
The HDACs are a highly homologous group of enzymes that are classified based on their primary and secondary structure. The zinc-binding HDACs are denoted class I (HDAC 1, 2, 3 and 8), class IIa (HDAC 4, 5, 7 and 9), class IIb (HDAC 6 and 10) and class IV (HDAC 11). In addition, there is a group of NAD+-dependent HDACs, which are denoted class III (sirtuin 1–7) [48].
Inhibitors of zinc-dependent HDACs have ambiguous effects on the NF-κB pathway
Although HDAC inhibitors were initially discovered as anticancer agents, many studies indicate their ability to suppress inflammatory responses. In this respect, early evidence stems from a study demonstrating that phenylbutyrate and trichostatin A (Fig. 3, Table 1) inhibit tumor necrosis factor (TNF)α expression in inflamed tissues in a rheumatoid arthritis animal model [49]. Another early study demonstrates that HDAC inhibitor SAHA (Fig. 3, Table 1) inhibits lipopolysaccharide (LPS)-induced cytokine release in vitro and in vivo [50]. Interestingly, this anti-inflammatory effect was observed at much lower concentrations than tumor suppressive effects in vitro. The anti-inflammatory potency of HDAC inhibitors has been described in several reviews [51–53]. It was observed that HDAC inhibitors such as SAHA and Trichostatin A (TSA) delay and reduce NF-κB nuclear translocation and gene expression upon TNFa stimulation [54]. The potential of HDAC inhibitors to suppress inflammation is also nicely illustrated by compound 1, recently described by Lee et al. (Fig. 3, Table 1) [55]. This novel HDAC inhibitor exhibits nanomolar HDAC inhibitory potency and demonstrated a strong suppressive effect on inflammatory mediator expression in animal models. Most interestingly, a recent study applied the HDAC inhibitor givinostat (Fig. 3, Table 1) in a relatively small patient group suffering from systemic-onset juvenile idiopathic arthritis, and demonstrated a clear therapeutic benefit and excellent safety profile [56].
In contrast to inhibiting inflammation, other studies have also demonstrated that certain HDAC inhibitors can also lead to the stimulation of proinflammatory gene transcription. Initial studies on NF-κB acetylation reported that HDACs 1–3 (class I) can deacetylate p65 and negatively regulate gene transcription [6,57]. In addition, HDAC activity leads to histone deacetylation, which is generally associated with inhibition of gene transcription. Both factors imply that HDAC inhibitors stimulate proinflammatory gene transcription, which is supported by several studies. Indeed, it was demonstrated that SAHA [58], TSA [59,60], MS275 [58,61,62] and LBH589 [61] enhance NF-κB activation (Fig. 3, Table 1).
Toward elucidation and selective modulation of specific HDACs in the NF-κB pathway
Such contradictory findings can be explained by applications of different cell types and the lack of selectivity of the employed HDAC inhibitors. Most frequently, applied HDAC inhibitors target all zinc-dependent HDACs, because most rely on the strongly zinc-coordinating hydroxamic acid functionality (carried by pan-HDAC inhibitors; Fig. 3, Table 1) [62]. This hampers elucidation of the relevance of HDAC activity in specific disease models. However, over the past few years, more selective inhibitors have become available. An example includes entinostat/MS275 (Fig. 3, Table 1), which is HDAC1–3 (class I) selective [62]. Also, the first HDAC class IIa (HDAC 4, 5, 7 and 9) selective inhibitors with a trifluoromethyloxadiazole scaffold (TFMO; Fig. 3, Table 1) were recently identified [63]. Interestingly, this novel class of inhibitors lacks the usual hallmark of class I and class IIb inhibition: cell death or apoptosis. This is beneficial for applications in non-oncology areas such as inflammation.
Several studies have already shed more light on the role of specific HDACs in the NF-κB pathway. A noteworthy recent study reports that HDAC3 deacetylates p65 on lysines 122, 123, 314 and 315, and that it is a positive regulator of inflammatory gene expression [64], which might seem contradictory to the previously mentioned initial studies [6,57] and studies using MS275 [58,61,62]. However, note that these concern not only HDAC3 but also HDAC1 and 2. This indicates that future studies should be aimed at developing more selective HDAC inhibitors that target, for example, HDAC3. Additionally, there have also been interesting reports on the role of SIRT type HDACs in the NF-κB pathway [65]. For example, HDAC SIRT1 deacetylates p65 at lysine 310 and thus inhibits the NF-κB transcriptional activity [66]. Interestingly, the small molecule SIRT1 activator SRTCX1003 (Fig. 3, Table 1) inhibits NF-κB activation in vitro and LPS-induced production of proinflammatory cytokines in BALB/c mice [67]. Also, SIRT2 interacts with p65, and deacetylates lysine 310, thereby inhibiting NF-κB transcriptional activity. Sirt2 knockdown results in increased expression of a subset of NF-κB target genes, indicating involvement of Sirt2 in the expression of a subset of NF-κB target genes [68]. This suggests that SIRT activators have a good potential for further development into molecules that suppress NF-κB activation in inflammation.
Concluding remarks
Lysine acetylation is a key regulator of the NF-κB pathway, which works in concert with other PTMs via complex crosstalk mechanisms to determine the signaling output. Importantly, small molecule modulators of its writers (HATs) or erasers (HDACs) have been demonstrated to regulate NF-κB signaling, suggesting that these are potential drugs for inflammatory diseases. An ongoing challenge toward regulation of the NF-κB pathway is the development of highly potent molecules that selectively target specific HATs or HDACs. Fortuitously, this field has seen remarkable progress over the past few years. Several inhibitors now demonstrate specific effects in distinct disease models in cellular systems and animal studies, which is very promising for drug discovery. A noteworthy example is HAT inhibitor C646. Owing to its high selectivity and potency for p300, this inhibitor mainly leads to inhibition of pathways that are connected to NF-κB, the androgen receptor and the glucocorticoid receptor in cellular and animal models. Another promising example is HAT inhibitor MG149, which mainly inhibits expression related to the NF-κB and p53 pathways.
Additionally, some currently described HDAC inhibitors show encouraging effects in animal models and even patients; pan-HDAC inhibitor givinostat displayed promising therapeutic benefits in patients suffering from juvenile idiopathic arthritis. An important consideration in the development of such agents is the capacity for HDAC inhibitors to show either activation or inhibition of inflammatory responses. We presume that a main cause of these ambiguities is that different reports employed different cell types. In addition, we speculate that they could also be explained by HDAC (non) selectivity of the inhibitors used. In the light of this, it is interesting that a recent study demonstrated that HDAC3 is a positive regulator of inflammatory gene expression, which deacetylates p65 on four specific lysines. This suggests that HDAC3-specific inhibitors, and perhaps more specific inhibitors for other HDACs, could unambiguously suppress inflammatory responses. Also, the role of studied SIRT type HDACs seems to be mainly inhibition of NF-κB gene transcription and, in line with this, SIRT1 activator SRTCX1003 encouragingly reduced inflammatory responses in mice. This indicates that activators of SIRT type HDACs could be also be effective in suppressing inflammatory responses. However, it often remains unclear which specific HDACs need to be targeted by inhibitors or activators. Thus, a challenge lies ahead in determining whether specific HDACs target specific lysine acetylations in specific signaling cascades, or if their effects are (much) broader (a challenge which also still applies to HATs). Finally, in addition to the current approaches of pursuing modulators of HDACs and HATs, agents capable of inhibiting (or enhancing) the various enzymes that install PTMs that crosstalk with acetylation could also find use as drugs for modulating histone (de)acetylation.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.drudis.2013.11.012.
Acknowledgments
We thank Massimo Ghizzoni from Axon Medchem for critically reading the manuscript.
References
- 1.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 3.Ghizzoni M, et al. Histone acetyltransferases are crucial regulators in NF-kappaB mediated inflammation. Drug Discov Today. 2011;16:504–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusio-Kobialka M, et al. Increased acetylation of lysine 317/320 of p53 caused by BCR-ABL protects from cytoplasmic translocation of p53 and mitochondria-dependent apoptosis in response to DNA damage. Apoptosis. 2012;17:950–963. doi: 10.1007/s10495-012-0739-9. [DOI] [PubMed] [Google Scholar]
- 5.Li H, et al. Regulation of NF-kappaB activity by competition between RelA acetylation and ubiquitination. Oncogene. 2012;31:611–623. doi: 10.1038/onc.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, et al. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 7.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, et al. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22:1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiernan R, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, et al. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JS, et al. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izzo A, Schneider R. Chatting histone modifications in mammals. Brief Funct Genomics. 2010;9:429–443. doi: 10.1093/bfgp/elq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 15.Chen LF, et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 17.Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- 18.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 20.Lo WS, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 21.Edmondson DG, et al. Site-specific loss of acetylation upon phosphorylation of histone H3. J Biol Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 23.Nightingale KP, et al. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 24.Vakoc CR, et al. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Nishioka K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 27.Fabbrizio E, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal S, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, et al. Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J Biol Chem. 2011;286:20323–20334. doi: 10.1074/jbc.M110.207258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buerki C, et al. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JW, et al. New molecular bridge between RelA/p65 and NF-kappaB target genes via histone acetyltransferase TIP60 cofactor. J Biol Chem. 2012;287:7780–7791. doi: 10.1074/jbc.M111.278465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Balasubramanyam K, et al. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 35.Sung B, et al. Anacardic acid (6-nonadecyl salicylic acid), an inhibitor of histone acetyltransferase, suppresses expression of nuclear factor-kappaB-regulated gene products involved in cell survival, proliferation, invasion, and inflammation through inhibition of the inhibitory subunit of nuclear factor-kappaBalpha kinase, leading to potentiation of apoptosis. Blood. 2008;111:4880–4891. doi: 10.1182/blood-2007-10-117994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sbardella G, et al. Identification of long alkylidenemalonates as novel small molecule modulators of histone acetyltransferases. Bioorg Med Chem Lett. 2008;18:2788–2792. doi: 10.1016/j.bmcl.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Ghizzoni M, et al. 6-Alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur J Med Chem. 2012;47:337–344. doi: 10.1016/j.ejmech.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legartova S, et al. Basic nuclear processes affected by histone acetyltransferases and histone deacetylase inhibitors. Epigenomics. 2013;5:379–396. doi: 10.2217/epi.13.38. [DOI] [PubMed] [Google Scholar]
- 39.Stimson L, et al. Isothiazolones as inhibitors of PCAF and p300 histone acetyltransferase activity. Mol Cancer Ther. 2005;4:1521–1532. doi: 10.1158/1535-7163.MCT-05-0135. [DOI] [PubMed] [Google Scholar]
- 40.Dekker FJ, et al. Inhibition of the PCAF histone acetyl transferase and cell proliferation by isothiazolones. Bioorg Med Chem. 2009;17:460–466. doi: 10.1016/j.bmc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Ghizzoni M, et al. Reactivity of isothiazolones and isothiazolone-1-oxides in the inhibition of the PCAF histone acetyltransferase. Eur J Med Chem. 2009;44:4855–4861. doi: 10.1016/j.ejmech.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Gorsuch S, et al. Synthesis of isothiazol-3-one derivatives as inhibitors of histone acetyltransferases (HATs) Bioorg Med Chem. 2009;17:467–474. doi: 10.1016/j.bmc.2008.11.079. [DOI] [PubMed] [Google Scholar]
- 43.Wisastra R, et al. Isothiazolones; thiol-reactive inhibitors of cysteine protease cathepsin B and histone acetyltransferase PCAF. Org Biomol Chem. 2011;9:1817–1822. doi: 10.1039/c0ob00464b. [DOI] [PubMed] [Google Scholar]
- 44.Bowers EM, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng FQ, et al. Structure-based identification of drug-like inhibitors of p300 histone acetyltransferase. Yao Xue Xue Bao. 2013;48:700–708. [PubMed] [Google Scholar]
- 46.Santer FR, et al. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10:1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- 47.Zhu XY, et al. p300 exerts an epigenetic role in chronic neuropathic pain through its acetyltransferase activity in rats following chronic constriction injury (CCI) Mol Pain. 2012;8:84. doi: 10.1186/1744-8069-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregoretti IV, et al. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Chung YL, et al. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8:707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 50.Leoni F, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci U S A. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villagra A, et al. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2009;29:157–173. doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 52.Grabiec AM, et al. Function of histone deacetylase inhibitors in inflammation. Crit Rev Immunol. 2011;31:233–263. doi: 10.1615/critrevimmunol.v31.i3.40. [DOI] [PubMed] [Google Scholar]
- 53.Dinarello CA, et al. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol Med. 2011;17:333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imre G, et al. Histone deacetylase inhibitors suppress the inducibility of nuclear factor-kappaB by tumor necrosis factor-alpha receptor-1 down-regulation. Cancer Res. 2006;66:5409–5418. doi: 10.1158/0008-5472.CAN-05-4225. [DOI] [PubMed] [Google Scholar]
- 55.Lee HY, et al. 1-Arylsulfonyl-5-(N-hydroxyacrylamide)indolines histone deacetylase inhibitors are potent cytokine release suppressors. ChemBioChem. 2013;14:1248–1254. doi: 10.1002/cbic.201300201. [DOI] [PubMed] [Google Scholar]
- 56.Vojinovic J, et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2011;63:1452–1458. doi: 10.1002/art.30238. [DOI] [PubMed] [Google Scholar]
- 57.Ashburner BP, et al. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai Y, et al. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol. 2005;25:5429–5444. doi: 10.1128/MCB.25.13.5429-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim YK, et al. Activation of NF-kappaB by HDAC inhibitor apicidin through Sp1-dependent de novo protein synthesis: its implication for resistance to apoptosis. Cell Death Differ. 2006;13:2033–2041. doi: 10.1038/sj.cdd.4401915. [DOI] [PubMed] [Google Scholar]
- 60.Sato T, et al. Enhancement of inflammatory protein expression and nuclear factor Kappab (NF-Kappab) activity by trichostatin A (TSA) in OP9 preadipocytes. PLoS ONE. 2013;8:e59702. doi: 10.1371/journal.pone.0059702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosato RR, et al. Histone deacetylase inhibitors activate NF-kappaB in human leukemia cells through an ATM/NEMO-related pathway. J Biol Chem. 2010;285:10064–10077. doi: 10.1074/jbc.M109.095208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Khan N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 63.Lobera M, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 64.Ziesche E, et al. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res. 2013;41:90–109. doi: 10.1093/nar/gks916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebastian C, et al. From sirtuin biology to human diseases: an update. J Biol Chem. 2012;287:42444–42452. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H, et al. SIRT1 activators suppress inflammatory responses through promotion of p65 deacetylation and inhibition of NF-kappaB activity. PLoS ONE. 2012;7:e46364. doi: 10.1371/journal.pone.0046364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rothgiesser KM, et al. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.