Abstract
Three strikingly different alternative male mating morphs (aggressive “Independents”, semi-cooperative “Satellites” and female mimic “Faeders”) coexist as a balanced polymorphism in the ruff, Philomachus pugnax, a lek-breeding wading bird1,2,3. Major differences in body size, ornamentation, and aggressive and mating behaviour are inherited as an autosomal polymorphism4,5. We show that development into Satellites and Faeders is determined by a supergene6,7,8 consisting of divergent alternative, dominant, non-recombining haplotypes of an inversion on chromosome 11, which contains 125 predicted genes. Independents are homozygous for the ancestral sequence. One breakpoint of the inversion disrupts the essential Centromere protein N (CENP-N) gene, and pedigree analysis confirms lethality of inversion homozygotes. We describe novel behavioural, testes size, and steroid metabolic differences among morphs, and identify polymorphic genes within the inversion that are likely to contribute to the differences among morphs in reproductive traits.
Stable genetic polymorphisms for alternative reproductive strategies, involving differences in body size, structure, and behaviour, have been identified in a range of taxa9,10. These traits are clearly associated with fitness and putatively maintained by frequency-dependent selection. However, little is known about their genomic basis or mode of evolution, or why examples are relatively rare, in contrast to the widespread phenotypic plasticity in reproductive strategy in response to environmental variation10.
The ruff (Philomachus pugnax) is a Eurasian sandpiper with a highly polygynous, lek-based mating system involving competition and cooperation among three male morphs (Fig. 1), and extensive female mate choice and genetic polyandry1,2,11,12. Territorial “Independent” males, with hypervariable but predominantly dark ornamental plumage, defend small mating courts on leks. Non-territorial “Satellite” males, with predominantly white ornamental plumage, join Independents on courts, co-displaying to attract females, but competing for matings. Rare small female-mimicking “Faeder” males attend Independents’ courts and attempt rapid copulations when females solicit matings from ornamented displaying males3.
Figure 1.
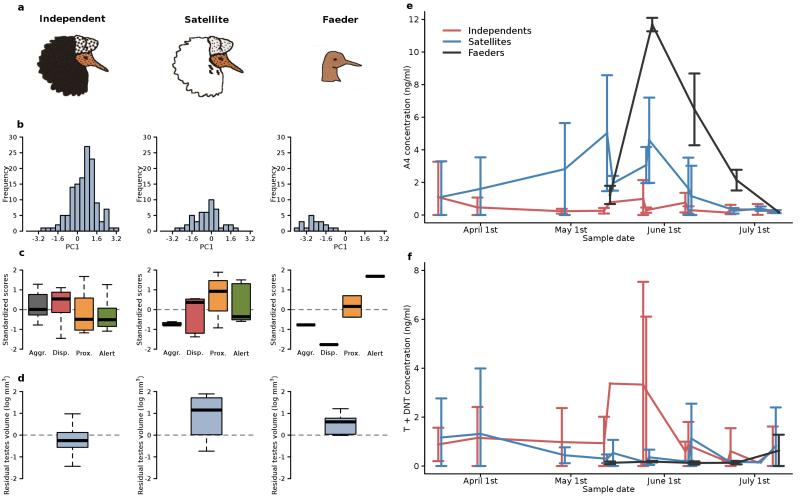
Comparison of of Independent, Satellite, and Faeder ruff males. a, Representative breeding plumages. b, Body size distributions5 (see Methods). c, Comparisons of morph behavioural profiles for Aggression (“Aggr.” – black), Display (“Disp.” – red), Proximity (“Prox.” – brown), and Alert stance (“Alert” – green). Morph-specific boxplots are plotted for each behaviour from the means of individual male ruffs, standardized across the sample population by subtracting the global mean and dividing by s.d.. Morphs differed in their distributions of behaviour (MANOVA on individual bird mean values; Wilks’s Lambda = 0.33, approximate F6,44 = 5.37, P < 0.0003). d, Residuals of the logarithm of testes volume index corrected for date differ among morphs (F2,37 = 6.31, P = 0.0045). Testes of Independents (n = 29) are substantially smaller than those of Satellites (n = 4, Tukey-adjusted P = 0.006) and Faeders (n = 6, P = 0.02). Boxplots indicate median (bold line), 25% and 75% quartiles (box) and ranges (whiskers). e, f, Seasonal patterns of circulating steroid concentrations in blood plasma of male ruffs: e, androstenedione (A4), f, testosterone plus dihydrotestosterone (T+dHT). Points are daily means±s.d. Independents (n = 11: red, jittered −1 day; Satellites (n = 9): blue; Faeders (n = 2): black, jittered +1 day. Independents have higher date-specific levels of T than Satellites (F1,97 = 10.01, P = 0.002), and Satellites higher levels of A4 than Independents (F1,97 = 17.03, P < 0.0001; Supplementary Table 1); Faeders appear similar to Satellites, but were not tested statistically due to n = 2.
Two autosomal Mendelian factors, Faeder (F) and Satellite (S), have been shown to be dominant to independent (i), but the precise genetic architecture of morph determination remains unclear4,5. We have maintained a pedigreed captive breeding population of several hundred ruffs since 1985. Adult males were phenotyped for mating behaviour (Independent, Satellite, Faeder), and the presence or absence of ornamental plumage (ornamented vs Faeder). Some adult females were also phenotyped, with small size indicating carriers of Faeder5, and aggressive behaviour following testosterone implantation permitting classification as Independents or Satellites13.
We explored the reproductive physiology of the morphs, with a particular focus on steroid metabolites known to affect these phenotypes13. The testes of Satellites, like those of Faeders3, were larger than those of Independents, despite their smaller body sizes (Fig. 1b, Supplementary Fig. 1), presumably to increase the efficiency of less frequent or effective copulations by producing larger numbers of sperm. Breeding Independents had higher circulating testosterone concentrations, while Satellites and Faeders had higher concentrations of androstenedione (Fig. 1e, f; Supplementary Table 1), suggesting fundamental morph differences in the regulation of the hypothalamus–pituitary–gonadal system that drives seasonal reproduction14,15.
To identify and characterise the loci controlling morph divergence we used a combination of genetics and genomics, based on both our captive population and wild ruffs. We generated a reference ruff genome from a single Independent male in the captive colony, using Illumina paired-end, 3-kb and 5-kb mate-pair data, and long-read PacBio data (Supplementary Fig. 2; Supplementary Table 2). The assembled genome has a span of ~1.17 Gb in 12,085 scaffolds longer than 1 kb, with 292 scaffolds longer than 877 kb (Supplementary Table 3). Genes were predicted using extensive new transcriptome data (Supplementary Table 4). Assessment of assembly quality using genomic read mapping, transcriptome read and assembly mapping, and comparison to the high-quality chicken (Gallus gallus) genome assembly suggest high completeness and contiguity (Supplementary Table 5; Supplementary Fig. 3). The draft genome was ordered and orientated using the chicken reference to yield a chromosome-level assembly. We identified and typed single nucleotide polymorphisms (SNPs) in the pedigree population using reduced representation, restriction-site associated DNA (RAD) sequencing16 at low density (i.e. based on SbfI restriction sites), identifying 1,068,556 SNPs. We mapped Faeder and Satellite to a genetic map based on 3,948 biallelic SNPs with minor allele frequency ≥ 0.1, which had been typed in 286 individuals with a success rate of ≥ 0.99.
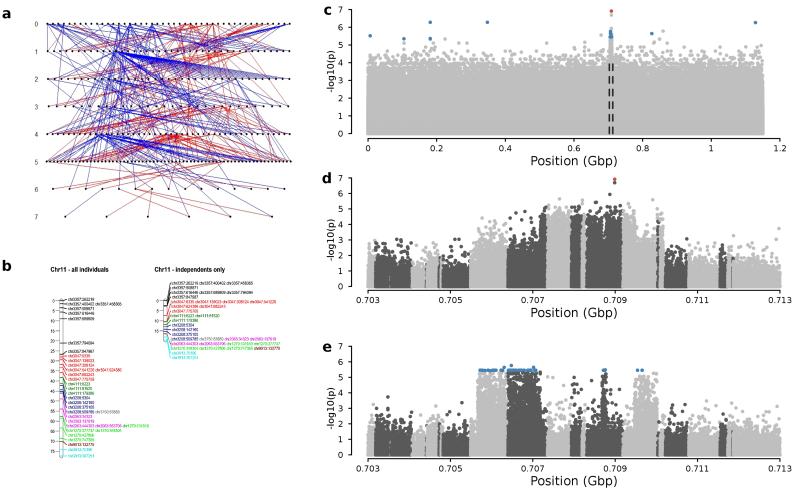
Faeder and Satellite each mapped significantly and uniquely (LOD >3), to the same region of chromosome 11 (Fig. 2; Supplementary Fig. 4), coincident with the region previously identified for Faeder using microsatellite markers17. The region of linkage encompasses about one fifth of the chromosome. Independent estimation of genetic maps for cohorts containing or excluding birds carrying Satellite or Faeder gave very different recombination lengths for this region (Fig. 2b), suggesting that it might contain a genomic rearrangement refractory to recombination between haplotypes.
Figure 2.
Genetic mapping and genome-wide association analysis identify the genomic region determining ruff reproductive morphs. a, The ruff linkage mapping pedigree. Paternal links are shown as red lines and maternal links as blue lines. b, Linkage maps (in centiMorgans) of the inversion region on ruff chromosome 11 support the presence of the inversion polymorphism, indicated by an ~70% shorter linkage map when only Independents are considered. SNPs on the same contig share the same colour. c, Association between markers and morphs based on 41 unrelated males (10 Faeder, 10 Satellite, 21 Independent). Alternating shades of grey indicate different contigs, ordered based on synteny with the chicken genome. The top panel shows −log10P-values of association across the entire ruff genome; middle panel (Faeder) and bottom (Satellite) show blow ups of the associated peak (0.703–0.713 Gbp, indicated by dashed lines in the top panel) for comparisons with independent. Red and blue dots indicate genome-wide significance (P < 0.05, 1000 permutations) for the Faeder and Satellite loci, respectively.
A separate genome-wide association study, using densely sampled SNPs (from RAD sequencing at PstI restriction sites) in a sample of 41 unrelated Faeder, Satellite and Independent birds, identified genome-wide significance of association with the mating morphs on contigs from the same section of chromosome 11 (Fig. 2c). The high resolution provided by the unrelated individuals revealed specific segments of this region uniquely associated with either Faeder or Satellite, indicating the presence of morph-specific haplotypes spanning several megabases of the chromosome.
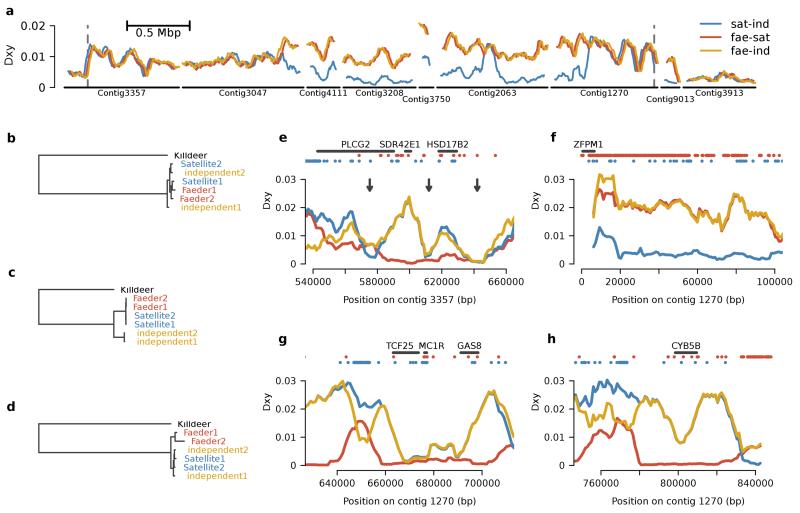
To map finely the variation in the region, we whole-genome sequenced an additional five males (one Independent, two Satellites and two Faeders) at 80x genome coverage (Supplementary Table 2). Nucleotide variation (measured by Watterson’s theta) was substantially greater in Faeder and Satellite individuals within the region of interest (Faeder = 0.011; Satellite = 0.011), compared to regions adjacent to the inversion (Faeder = 0.003; Satellite = 0.002). Divergence (measured by Dxy) between Faeder and the other morphs was high across the entire region (Fig. 3a), with morph-specific lineage sorting occurring within the region (Fig. 3c). These patterns are consistent with the presence of a large non-recombining inversion, and this was confirmed by the orientations of read pairs across the breakpoints (Supplementary Fig. 4).
Figure 3.
Sequence divergence among morph-determining haplotypes. a, Divergence (Dxy) among morphs across the inversion (4.4 Mbp) and flanking regions. Vertical dotted lines refer to the two inversion breakpoints. The gene CENP-N crosses the breakpoint located on Contig 3357.Each line is staggered by 15 kb to make each line visible. b, c, d, Evolutionary relationships among the three morphs. Maximum-likelihood trees showing the relationship among the sequences from two resequenced Satellites, two Faeders, and the non-inverted reference (Independent 1) and a resequenced Independent (Independent 2) for different regions within and around the inversion: b, adjacent to the inverted region (Contig 3913: 140,000–150,000), c, within regions of the inversion exhibiting high divergence from the reference (Contig 3357: 280,000–290,000), d, within regions of the inversion where Satellites show low divergence from Independents (Contig 3208). e, f, g, h, Divergence patterns between Faeder and Satellite across regions containing candidate loci involved in steroid hormone metabolism (PLCG2, SDR42E1, HSD17B2 and CYB5B), sperm motility (GAS8) and pigmentation (MC1R), plus also transcription factors expressed in the gonads (ZFPM1) and in proximity to MC1R showing Satellite-specific divergence (TCF25). Arrows indicate the presence of morph-specific deletions affecting the coding region of PLCG2 (see Supplementary Fig. 6) and deletions in noncoding regions surrounding HSD17B220. Dots indicate the presence of morph-specific nucleotide substitutions (Satellite – blue; Faeder – red).
One inversion breakpoint disrupts the Centromere protein N (CENP-N) gene between its fourth and fifth exons. CENP-N is essential for mitotic centromere assembly18. We therefore predicted that homozygosity for the inversion haplotypes would be lethal and our breeding data confirmed a complete absence of inversion homozygotes (Table 1). The other breakpoint appears to be in a non-coding repeat.
Table 1.
Segregation ratios demonstrating apparent lethality of inversion genotypes in matings between heterozygotes identified with diagnostic SNPs.
| Phenotype associated with SNP |
SNP* | Progeny | Genotypes | Deviation from 1:2:1 |
Deviation from 1:2† |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| χ 22 | P | χ 21 | P | |||||||
| Faeder | ++ | +I | II | |||||||
| A & B | All | Obs | 20 | 16 | 0 | 22.67 | 1.2 × 10−5 | 8.00 | 0.005 | |
| Exp | 9 | 18 | 9 | |||||||
| Males | Obs | 8 | 7 | 0 | 8.60 | 0.014 | 2.70 | 0.100 | ||
| Exp | 3.75 | 7.5 | 3.75 | |||||||
| Females | Obs | 12 | 9 | 0 | 14.14 | 8.5 × 10−4 | 5.36 | 0.021 | ||
| Exp | 5.25 | 10.5 | 5.25 | |||||||
| Satellite | ++ | +I | II | |||||||
| C | All | Obs | 20 | 18 | 0 | 21.16 | 2.5 × 10−5 | 6.37 | 0.012 | |
| Exp | 9.5 | 19 | 9.5 | |||||||
| Males | Obs | 9 | 9 | 0 | 9.00 | 0.011 | 2.25 | 0.134 | ||
| Exp | 4.5 | 9 | 4.5 | |||||||
| Females | Obs | 11 | 9 | 0 | 12.30 | 0.002 | 4.23 | 0.040 | ||
| Exp | 5 | 10 | 5 | |||||||
Data were obtained by pooling all offspring across reproductive events where both parents were heterozygous for diagnostic SNPs. I represents the inversion haplotype (either Faeder or Satellite associated), + is the ancestral-ordered haplotype (independent associated).
SNPs used: A, contig 1270:576,631; B, contig 3047:368,535; C, contig 1270:576,631. The sex ratios produced by these crosses show no suggestion of sex-specific lethality, i.e. no sex difference in the ratio of ++:+I (SNPs A, B and C Fisher Exact test, P = 1.00)
excluding inversion homozygote (II) class.
Since recombination is completely suppressed close to inversion breakpoints, regions in linkage disequilibrium with breakpoints are expected to have high divergence7,19. This expectation was supported by divergence analyses using our resequencing data. We identified 44,433 SNPs specific to one of the three haplotypes in the assembled reference (independent) inversion region. The inverted haplotypes showed several structural changes, i.e. regions with large (> 100 bp) deletions and duplications (Supplementary Fig. 5). Within some regions of the inversion, Satellites showed greater similarity to Independents (Fig. 3a, d), suggesting that Satellite originated through recombination or gene conversion between inverted Faeder and non-inverted independent alleles20.
We phased SNPs located in the inversion into longer haplotypic sequences and compared 100 gene sequences directly between the three morphs. Of these, 78% showed consistent morph-specific differences, including amino-acid substitutions, insertions and deletions (Supplementary Table 6). In addition, two of six genes adjacent to the inversion showed Faeder-specific protein sequence differences. Under neutral drift and in the absence of recombination we would expect consistent divergence across the inverted region. However, the genes in the inversion varied widely in their divergence among morphs, with several showing high divergence (Fig. 3e-h).
Several divergent protein-coding genes have predicted functions in hormonal systems, such as androsteroid homeostasis and plumage development relevant to ruff morph phenotypes (Fig. 3e-h and Supplementary Table 7). A key candidate is Estradiol 17-beta-dehydrogenase 2 (HSD17B2), encoding an enzyme that preferentially inactivates testosterone to androstenedione and estradiol to estrone21. Also present are Short-chain dehydrogenase reductase (SDR42E1), Palmitoyltransferase (ZDHHC7)22 and Cytochrome b5 (CYB5B) (Supplementary Table 7). The morph-specific alleles of these enzymes may alter steroid secretion levels and/or receptor responsiveness, driving morphological and neurological mechanisms responsible for contrasting anatomical, plumage and behavioural profiles (Fig. 1c). Genes involved in steroid metabolism have also been implicated in another polymorphic vertebrate – the white-throated sparrow Zonotrichia albicollis15 – in which the two morphs are similarly associated with an inversion. As in ruffs, the sparrow morphs differ in aggression and testosterone during the breeding season23. This suggests that convergent molecular pathways may contribute to the evolution of behavioural variation during reproduction. Another gene located in the ruff inversion, Melanocortin-1 receptor (MC1R), a locus that controls colour polymorphisms in other birds24, might account for the reduced melanin in Satellite display feathers (Fig. 1a). In Faeder (but not Satellite), the gene for 1-Phosphatidylinositol 4,5-bisphosphate phosphodiesterase γ-2 (PLCG2) has experienced complex deletions and rearrangement, including the loss of an exon encoding an SH3 protein-interaction domain, and is probably a loss-of-function allele (Supplementary Fig. 6). PLCG2 is a transmembrane signalling enzyme involved in cell receptor activation25 and interacts with epidermal growth factor receptor EGFR26. As EGFR is involved in the formation of feather arrays27, this locus is a candidate for the loss of secondary sexual expression of display feathers and behaviour in Faeders. Additional genes with roles in sperm motility and gonadal expression are also present (Supplementary Table 7).
As inversion homozygotes appear to be lethal, to maintain allelic frequencies, the fitness of individuals carrying the inversion, in one or both sexes, must exceed that of ancestral independent homozygotes. Heterozygous carriers of the inversion also have poor survival in crosses (Table 1). Higher reproductive success by Satellite and Faeder males is a likely explanation for how the survival disadvantage is offset, and their larger testis sizes suggest that they might be more successful in sperm competition, despite equal or lower mating rates2,11. Selection should also favour disassortative mating by individuals carrying the inversion, particularly by females. Since some ruff females mate with multiple morphs12, morph discrimination of mates, if it occurs, is not ubiquitous. Strong disassortative mating is a key feature of the white-throated sparrow system, although the causative inversion in this species is not lethal15.
Alternative reproductive morphs are predicted to evolve when strong reproductive skews provide low thresholds for invading forms9. The lek mating system of ancestral Independents would have satisfied this condition, but numerous other species that lack genetically polymorphic alternatives also do so10. The occurrence of genomic rearrangements that can provide viable substrates for differentiation must be rare, but by suppressing recombination and combining the fates of loci within the same genomic region, the inversion in the ruff enabled a phenotypically complex alternative strategy to evolve through the coevolution of genes affecting male behaviour, morphology, and fertility. As shown here and independently elsewhere20, the initial occurrence of one alternative can facilitate the evolution of multiple morphs,which has also occurred in other species9,10,28,29,30.
URLs
Assemblage gene prediction pipeline, https://github.com/sujaikumar/assemblage/blob/master/README-annotation.md; CLC Bio Assembler, http://www.clcbio.com/products/clc-assembly-cell/; Slide, https://github.com/mspopgen/slide; Evolib, https://github.com/mspopgen/Evolib; GWAS and population genetic pipelines, https://github.com/mspopgen/kuepper2015.
ONLINE METHODS
Samples
DNA samples were obtained from the Simon Fraser University colony, which was founded with 110 ruffs hatched from wild eggs collected in Finland prior to 19904,5 plus two Faeder males from the Netherlands in 2006 (under permits from the Canadian Food Inspection Agency, Canadian Wildlife Service, and Simon Fraser University Animal Care Committee). Additional blood samples were obtained from wild males in breeding plumage caught in the Netherlands (10 Faeders and 10 Satellites) between 2004–2008 or Belarus (one Independent and one Satellite) in 2014 (under permits from Dutch Ringing Centre, Animal Experimentation Committee of the University of Groningen and Belarus Bird Ringing Centre).
Male morph determination
Behavioural phenotypes of captive male ruffs were determined during the breeding season1,3,13,32 (Fig. 1c). Classification of ornamented males was based on ethological displays13, with some wild males assigned from plumage32,33,34,35. Faeders were definitively identified by small size, and lack of ornamental plumage, seasonal facial wattles, and epigamic display3,37.
Behavioural profiles
We quantified behaviour in captivity of 19 Independents, six Satellites and two wild-caught Faeders (Fig 1c). The Faeders were housed with 61 females. Two Independents and one Satellite were visible in an adjacent pen. These ornamented males were introduced to the Faeders and females during 1–2-hour morning observation periods. We scanned postures and relative positions of males at 2-minute intervals, and recorded all interactions, in 55 sessions over 50 days. Independents and Satellites were replaced at least every five or ten days, respectively.
Four behavioural variables summarized differences among morphs1,3,13. Aggressive behaviour included: total chases, bill points, bill thrusts, or fights per minute. Display was defined as the proportion of scans with squats, half-squats, and obliques. Proximity was the proportion of scans when positioned < 2 bird lengths from another displaying male. Alert stance was the proportion of scans standing on the lek with head up. Fig. 1c shows morph-specific means of rates calculated for each behaviour from the means of each male, standardized across the sample population.
Testes volume index
Lengths (L) and widths (W) of both right and left testes were measured by T.P. with callipers to the nearest 0.1 mm from pre-breeding and breeding ruffs that died during capture in the Netherlands during March–June in 1993–200538 (Supplementary Fig. 1a). A volume index including both testes, in mm3, was calculated assuming testes were cylindrical (L × W2 × 0.785). A non-breeding baseline index, measured during late winter, was defined as <120 ml3. Residuals from log(index) were calculated for males caught during 10 April–15 May each year showing gonadal recrudescence above baseline, using a quadratic regression controlling for date (F2,36 = 7.62, P = 0.0002, , Supplementary Fig. 1).
Steroid hormone measurement
Hormone levels were measured in blood plasma samples collected in 2003 and 200639. In 2003, we sampled 16 displaying males 3–9 years old held with four other males in two groups of ten (five Independents and five Satellites). Blood was sampled between 09:00–14:00 approximately every two weeks prior to, throughout, and after the breeding season (14 March–7 July) (n = 107) and plasma separated and stored at −20°C. In 2006, we sampled two groups of three Independents and two Satellites. Two Faeder males (see Behavioural profiles, above) were also sampled. Males had constant visual access to females, and physical access for 2–3 hours between 06:00–11:00, when lek attendance is highest in the wild40. Blood samples (n = 50) were collected between 10:00–12:00, immediately after males had access to females, approximately every two weeks between 14 May–2 July.
Plasma samples were analysed at the University of Alaska, Fairbanks in duplicates following established radio-immunoassay (RIA) procedures41,42. Thirty 2003 samples were extracted with HPLC-grade dichloromethane. Steroids were separated using diatomaceous earth/glycol chromatography (“column” RIA), such that testosterone (T), dihydrotestosterone (DHT), and androstenedione (A4) could be analysed from a single 100-μl plasma sample. T and DHT titres were strongly correlated (r230 = 0.95, P < 0.0001), thus the remaining 2003 samples were analysed in two assays without separation of steroids prior to RIAs (“direct” RIA), using a 100-μl sample for T+DHT (“total T”), and 50 μl for A4. All 2006 samples were assayed for total T and A4 using 100 μl plasma with steroids separated using diatomaceous earth/glycol chromatography (“short column RIA”). The antibody with the same cross-reactivity for T and DHT (T-3003 Research Diagnostics) was used for T, DHT, and total T RIAs, and the A4-specific antibody (A-1707 Wien Laboratories) was used for A4 RIAs. Mean ± s.d. percentage recoveries were: 55.5 ± 8.7, 45.8 ± 6.9, and 45.6 ± 7.3 for A4, T, and DHT, respectively, in the column assay; 69.1 ± 5.2 for total T and 67.7 ± 6.2 for A4 in direct assays; and 72.7 ± 7.0 for total T and 64.0 ± 10.0 A4 in short columns. Inter-assay CV was 21% for total T and 22% for AE, and intra-assay CVs were < 10%. Minimum detectability was 3.90 pg/sample for A4, and 1.95 pg/sample for T, DHT, and total T.
We tested for morph differences in levels and temporal patterns of circulating steroids (Figs. 1e, f) using generalized linear mixed models with log linked Poisson distribution. Date, its quadratic effect, and two-way interactions with morph were included as factors. Bird was a random factor, along with assay type and year (= social situation), which had no detectable effects (Supplementary Table 1).
Pedigree construction
Preliminary pedigree assignments from 1985–2013 were generated using 27 microsatellite markers43 in 756 ruffs. We assigned parentage including all possible candidate parents using Cervus44 and Colony45. Inconsistencies were resolved by manual inspection, incorporating housing information of the most likely candidates.
Genome sequencing
We sequenced the genome of an Independent male from the captive colony. Illumina HiSeq 2500 v4 150-bp paired reads were generated, using paired-end and mate-pair libraries with various insert sizes, resulting in 137x raw coverage (Supplementary Table 2, Supplementary Fig. 2). We used Pacific Biosciences RS II technology with P5–C3 chemistry to generate 8.8x coverage in long reads (mean length 5,713 bp).
Genome sequence assembly
The genome was de novo assembled using an integrated approach (Supplementary Fig. 2).
Cleaning and trimming of raw reads
The raw Illumina paired-end library reads were quality trimmed (> q30) and adapter sequences removed using fastqc-mcf (ea-utils.1.1.2-53746). Short reads (< 50 bases) were discarded. Mate-pair reads were quality trimmed (> q30) and adapter and linker sequences removed using CutAdapt47 1.3. We retained only read pairs containing Nextera linker sequence and remaining read length > 50 bases.
Removal of contaminant data
An initial assembly of raw paired-end reads was prepared using CLC Bio assembler (CLC bio 4.2.0, Aarhus, Denmark; see URLs). Contaminant data deriving from bacterial, parasite and viral genomes were identified using BLAST48 against the NCBI nr database, reporting the best hits with E-value < e−50. Contigs likely to be contaminant were extracted and reads mapping to these removed.
Kmer optimisation
The optimal kmer size for assembly was estimated with sga-preqc v0.10.1349 and kmergenie50 v1.5924 on one lane each for each paired-end library and a kmer sweep with ABySS51 v1.3.7) using all paired-end and mate-pair reads. Sga identified an optimum around k = 35, while kmergenie identified k = 30 and 38 as the optimal sizes and ABySS suggested k = 38. We used k = 38.
Genome assembly
Preliminary assembly of all paired-end and mate-pair reads using ABySS resulted in unitigs that were masked using RepeatMasker52 (version open-4.0.5), with mate-pair reads mapped using bwa-aln and bwa-sampe (v0.7.753). Reads where both of the pair mapped to the masked unitigs were retained. Filtered paired-end reads were then used to scaffold the unitigs using SSPACE54 (Basic 2.0). Further scaffolding was done with PacBio reads using PBJelly55 v14.1.14.
RNA sequencing and annotation
Previous RNA-Seq data in ruffs were available for genes expressed in feather follicles56. We generated new RNA-Seq data from egg, chick heart, lung and brain, female heart and brain, and male heart, brain and testes to obtain a wide variety of transcripts for gene annotation (Supplementary Table 2). Raw data were quality and adapter trimmed using CutAdapt 1.3), and assembled using Trinity57 (version r20140413). The initial transcripts were filtered for abundance (Trinity: align_and_estimate_abundance.pl, filter_fasta_by_rsem_values.pl; RSEM 1.2.758).
Gene prediction
We predicted genes using the Assemblage gene prediction pipeline (see URLs) up until the second round of Maker, using Maker 2.31.759, cegma 2.460, SNAP version 2006-07-2861 and GeneMark-ES 2.3e62. Highly-expressed, unique transcripts from the Trinity assembly and proteins from Uniprot/Swissprot were used as evidence. Predicted genes with an Annotation Edit Distance (AED) < 1 were selected from the Maker first-pass results and used as hints to train Augustus v3.0263 (Supplementary Table 4). Genes predicted by Augustus on the inversion-associated contigs were further annotated using blast2go64. Annotations were checked manually to identify incorrectly split or fused genes (n = 5 and 4, respectively). In total, we annotated 125 genes (101 with known homologues) within the inversion, with 89 (71%) supported by partial mRNA transcripts.
RAD sequencing
We chose 300 ruffs from the pedigree for low-density RAD sequencing and genetic mapping. For the initial GWAS we used high-density RAD sequencing of 41 unrelated males with established phenotypes originating from Finland (21 Independents) and the Netherlands (10 Faeders, 10 Satellites). For the pedigree analyses, we sampled to maximise pedigree completeness, morph representation and number of generations in our pedigree. Genomic DNA was digested using restriction enzymes SbfI (low-density) or PstI (high-density), following Baird et al.19. We pooled samples from different morphs during library preparation.
SNP calling
For calling SNPs in the pedigreed, low-density RAD sequences and in the resequenced individuals we used the GATK 3.2.2 variant-calling pipeline65. Reads were aligned to the reference genome with bwa-mem 0.7.10 and realigned using GATK RealignerTargetCreator and IndelRealigner to improve alignment quality before running GATK HaplotypeCaller. Genotypes were then called using GATK GenotypeGVCFs. For the analysis of high-density RAD sequences from unrelated individuals, demultiplexing and filtering were performed using Stacks v1.21. Mapping was performed using bwa-mem 0.7.10. Samtools v0.1.19-44428cd66 was used for filtering bam files. We only included properly mapped and paired reads, removing reads with non-primary, supplementary or terminal alignments, and with mapping quality > 30. SNP calling was performed using bcftools v0.1.19-44428cd67.
Confirming linkage to the Satellite- and Faeder-determining loci
We used twopoint linkage mapping to locate the Faeder and Satellite loci. Fourteen individuals lacking pedigree links with the remaining 286 RAD-sampled birds were excluded, resulting in an eight-generation pedigree with 286 paternal and maternal links, 186 maternal grandmaternal/grandpaternal links and 195 paternal grandmaternal/grandpaternal links (Fig. 2a). Phenotypes were known for 189 individuals (117 Independents, 38 Faeders and 34 Satellites); 85 additional individuals were known to not be Faeders. Twelve birds were unknowns.
Satellite and Faeder causal loci were both scored as segregating in the pedigree (Independents as 1/1 homozygotes, Satellites and Faeders as 1/2 at the Satellite and Faeder loci, respectively). We followed a two-stage procedure. First, 3,948 informative SNPs (3,901 had ≥ 50 informative meioses) were tested for linkage with Satellite and Faeder using CriMap v2.50368 (modified by Jill Maddox, Department of Veterinary Science, University of Melbourne, Australia). We split the pedigree into smaller families, using the crigen function of the Linkage Mapping Software (Xuelu Liu, Monsanto). We then performed twopoint linkage analysis. SNPs associated with Faeder or Satellite and LOD score > 3 were associated with Chromosome 11. In the second stage, all low-density RAD SNPs from contigs spanning the inversion (n = 3,810) were tested for linkage with the two morphs. Twopoint mapping usually finds relatively few markers co-segregating with a causal variant. However, numerous SNPs within the chromosome 11 contigs showed highly significant, perfect co-segregation (LOD score > 3 and recombination fraction of zero) with both Faeder and Satellite (Fig. 2b), suggesting an inversion polymorphism.
Confirming the inversion with linkage mapping
An inversion polymorphism produces alternative marker orders segregating within the mapping panel. Thus any marker order tested will contain erroneous recombination events that lead to larger estimates of the map length. We tested whether maps from the complete dataset (286 birds) and from a subset of the data (Independents only, n = 215 birds) differed in map distances. SNPs on contigs spanning the inversion, with at least 70 informative meioses and separated by ≥50 kbp, which caused no parent–offspring mismatches in the pedigree, were retained for analyses, leaving 35 markers. Because of the conserved synteny between the ruff and chicken genomes, linkage map lengths were initially tested using the contig and SNP order inferred from homology to the chicken genome. Alternative orders attempted with the FLIPS option showed lower likelihoods and longer maps than the initial marker order. Sex-averaged map distances, estimated using the Crimap command CHROMPIC, were 74.9 cM for the complete dataset and 20.7 cM (28%) for the reduced dataset (Fig. 2b). The latter measure is consistent with other avian maps of chromosome 1169,70,71,72, supporting the presence of an inversion polymorphism in this region.
Genome-wide association study
Tests of association between markers and morphs were performed using GenABEL73. Correction for population stratification was performed by first calculating identity-by-state from all SNPs in the dataset and adding these as cofactors in the model. Genome-wide significance was assessed by performing 1,000 permutations of the data.
Inversion mapping with paired-end reads
Following Corbett-Detig et al.74 , we searched for an inversion in the region of interest identified by linkage mapping and genome-wide association study. We identified two breakpoints exhibiting morph-specific clustering of reads mapping in parallel to the (independent) reference genome at position 185,694 on Contig 3357 and position 821,901 on Contig 1270 (Supplementary Fig. 7). Resequenced Faeders and Satellites had reads mapping in parallel orientation at these breakpoints and reduced coverage (~50%) of properly mapped reads (Supplementary Fig. 7). Using allele-specific primers, we confirmed the predicted sequence across one breakpoint (Contig 1270) in three Satellite males. The inverted sequence at one breakpoint showed an inserted repetitive motif in Faeder but absent in independent, mapping to a non-LTR retrotransposon. These two breakpoints coincided with sharp changes in between-morph divergence (Dxy; Supplementary Fig. 7) and increased heterozygosity in Faeder and Satellite individuals, indicating that the inversion is heterozygous in Faeders and Satellites.
Confirming that the inversion haplotype is a lethal recessive
The pedigree enables tests of lethality of the inversion haplotype(s) carrying the Faeder and Satellite alleles. Since Satellite and Faeder morphs are imperfectly identified phenotypically, we first identified SNPs co-segregating with Faeder or Satellite. Eighteen SNPs co-segregated with Faeder with zero recombination fraction and a LOD score > 15. For two of these (Contig3047:314,715; Contig3047:314,697), we identified > 30 progeny where both parents were heterozygous for the inversion haplotype. Under Mendelian segregation, we expected a 1:2:1 ratio of genotypes AA:AB:BB among the progeny, where A is the ancestral-ordered and B is the inversion-associated allele. At both SNPs the ratio of genotypes deviated significantly from expectation (Table 1), suggesting that inversion homozygotes are lethal. Furthermore, the observed ratio of AA:AB also exceeded Mendelian expectations (1:2), suggesting that heterozygotes also carry a viability cost (Table 1).
These results were supported by two further SNPs that perfectly co-segregated with Satellite alleles and where > 30 progeny of heterozygous × heterozygous matings were observed (Table 1). For both, the lack of inversion homozygotes and the deficit of heterozygotes were significant. The lethality of the inversion was not sex-specific and is probably not morph-specific, although this is difficult to confirm due to some unknown female phenotypes. In both sexes, the absence of inversion homozygotes was statistically significant and we produced more non-inversion homozygotes than heterozygotes, although the departure from the expected 1:2 ratio was only significant in females.
We did not specify family structure in Table 1, as fertilizations in birds are independent events. A more conservative test estimated a G-test statistic for each of 5 paternal half-sibships with 4 or more offspring (27 offspring across 5 families), and compared the summed the G-test statistics against a chi-square distribution. The complete absence of inversion homozygotes remained statistically significant (Chi-square = 12.2, df = 5, P = 0.03).
Population genomics and phylogenomics
Divergence (Dxy) and heterozygosity were obtained using Evolib (see URLs), with sliding windows, nucleotide diversity and Tajima’s D calculated using Slide (see URLs). Maximum-likelihood reconstruction of morph phylogenies was performed using RAxML75 under the generalised time-reversible substitution model with a gamma model of rate heterogeneity (GTRGAMMA), with maximum-likelihood searches performed on 50 randomized stepwise addition parsimony trees. Orthologous regions of the outgroup species, killdeer (Charadrius vociferus)76, were obtained through blasting and performing multiple alignment with the phased haplotypes.
Haplotype calling and analysis of morph-specific amino-acid differences
We established haplotypes of alleles differing from the reference for five 80x resequenced wild ruffs for all inversion contigs. We used read-backed phasing, implemented in GATK65 (version 3.3.0), phasing SNPs co-occurring on the same (or paired) sequence reads into the same haplotype. We refrained from haplotype calling in the region within 3 kbp of the breakpoints. Inversion haplotypes were, on average, longest for Faeders (14.1 kbp), then Satellites (3.4 kbp), then Independents (1.0 kbp). Deletions longer than 100 bp were identified, based on a morph-specific drop in read coverage (read depth reduced to approximately 50% in inversion carriers, Supplementary Fig. 5).
After phasing, we used GATK’s FastaAlternateReferenceMaker tool to generate haplotype-specific fasta files for each contig. Using these haplotypic sequences, we predicted genes and established the amino-acid sequences using Augustus (see ‘Annotation’ above). One hundred (94 with known homologues) of the 125 predicted genes in the inversion were identified by the trained Augustus algorithm established for gene prediction in the reference (Supplementary Table 6). We then aligned amino-acid sequences of all six haplotypes for these genes using the CLUSTALW algorithm (MEGA677 #6140220) and identified consistent morph-specific amino acid changes in 78 genes. Candidate gene predictions (Supplementary Table 7) were verified by comparing mRNA and BLAST evidence to available gene models suggested by Maker and Augustus. Where predictions conflicted, we chose the best supported model. We resolved the complex copy number variation, rearrangement and deletion in the Faeder haplotype of the PLCG2 locus by combining coverage maps with rearrangement-spanning read pairs.
Supplementary Material
Acknowledgments
This work was supported by a grant to T.B., J.S. & M.B. from the UK BBSRC (BB/J0189371), grants to D.B.L. from NSERC, The H.F. Guggenheim Foundation, National Geographic Society and NSF, and an EU Marie–Curie fellowship to C.K.. The Simon Fraser University Work–Study program facilitated the maintenance of the captive population since 1993. Hormone work was supported by the NSF EPSCoR program at the University of Alaska Fairbanks. We thank the laboratory and bioinformatics staff at Edinburgh Genomics at the University of Edinburgh for Illumina sequencing and the Lausanne Genomic Technology Facility for PacBio sequencing.
Author Contributions
T.B., D.B.L., J.S., M.B. and C.K. designed the study. M.S. and J.E.R. analysed the genomic and transcriptomic data and assembled the genome. N.d.R., L.L.F, S.B.M. and C.K. undertook the laboratory work. D.B.L. bred, raised, and measured captive birds, and obtained tissue samples for genetic and hormone analyses. S.B.M and D.B.L. made behavioural observations. T.C.M., A.S.K. and J.C.W. measured hormones. T.P. and Y.I.V. obtained Faeder samples, donated Faeder founders to the captive population and obtained testis measurements. D.B.L., T.P., Y.I.V, N.K., P.P. and C.K. obtained tissue samples from wild birds. M.B. supervised the sequencing and genome assembly. C.K. and J.S. undertook the linkage analysis. C.K., L.L.F., N.d.R, S.B.M. and D.B.L. constructed the pedigree using microsatellite and SNP data. M.S., C.K., J.S., K.Z. and T.B. analysed the SNP data and performed molecular evolutionary analyses. T.B., D.B.L., C.K., M.S., J.E.R., M.B. and J.S. wrote the paper with input from the other authors.
Footnotes
Accession codes. The sequences reported in this paper have been deposited in the European Nucleotide Archive (ENA) database (Parent project: PRJEB11172, WGS PacBio: PRJEB11127, RNAseq: PRJEB10873, low-density RAD: PRJEB10868, high-density RAD: PRJEB10855, WGS Illumina: PRJEB10770, whole genome re-sequencing: PRJEB10677).
Note: Supplementary Information is available in the online version of the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.van Rhijn JG. The Ruff. T. & A.D. Poyser; London: 1991. [Google Scholar]
- 2.Widemo F. Alternative reproductive strategies in the ruff, Philomachus pugnax: a mixed ESS? Anim. Behav. 1998;56:329–336. doi: 10.1006/anbe.1998.0792. [DOI] [PubMed] [Google Scholar]
- 3.Jukema J, Piersma T. Permanent female mimics in a lekking shorebird. Biol. Lett. 2006;2:161–164. doi: 10.1098/rsbl.2005.0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lank DB, Smith CM, Hanotte O, Burke T, Cooke F. Genetic polymorphism for alternative mating behaviour in lekking male ruff, Philomachus pugnax. Nature. 1995;378:59–62. [Google Scholar]
- 5.Lank DB, Farrell LL, Burke T, Piersma T, McRae SB. A dominant allele controls development into female mimic male and diminutive female ruffs. Biol. Lett. 2013;9:20130653. doi: 10.1098/rsbl.2013.0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann A, Rieseberg L. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 9.Shuster SM, Wade MJ. Mating Systems and Strategies. Princeton University Press; Princeton, NJ: 2003. [Google Scholar]
- 10.Oliveira RF, Taborsky M, Brockman J, editors. Alternative Reproductive Tactics: An Integrative Approach. Cambridge University Press; Cambridge: 2008. [Google Scholar]
- 11.Hugie DM, Lank DB. The resident's dilemma: a female-choice model for the evolution of alternative male reproductive strategies in lekking male ruffs (Philomachus pugnax) Behav. Ecol. 1997;8:218–225. [Google Scholar]
- 12.Lank DB, et al. High frequency of polyandry in a lek mating system. Behav. Ecol. 2002;13:209–215. [Google Scholar]
- 13.Lank DB, Coupe M, Wynne-Edwards KE. Testosterone-induced male traits in female ruffs (Philomachus pugnax): autosomal inheritance and gender differentiation. Proc. R. Soc. B. 1999;266:2323–2330. [Google Scholar]
- 14.Knapp R, Neff BD. Steroid hormones in bluegill, a species with male alternative reproductive tactics including female mimicry. Biol. Lett. 2007;3:628–631. doi: 10.1098/rsbl.2007.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton BM, et al. Estrogen receptor alpha polymorphism in a species with alternative behavioral phenotypes. Proc. Natl Acad. Sci. USA. 2014;111:1443–1448. doi: 10.1073/pnas.1317165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baird NA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell LL, Burke T, Slate J, McRae SB, Lank DB. Genetic mapping of the female mimic morph locus in the ruff. BMC Genetics. 2013;14:109. doi: 10.1186/1471-2156-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8:e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamichhaney S, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nat. Genet. 2015;XX:xxx–xxx. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 21.Mindnich R, Möller G, Adamski J. The role of 17 beta-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 2004;218:7–20. doi: 10.1016/j.mce.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Pedram A, Razandi M, Deschenes RJ, Levina ER. DHHC-7 and -21 are palmitoylacyltransferases for sex steroid receptors. Mol. Biol. Cell. 2012;23:188–199. doi: 10.1091/mbc.E11-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maney D. Endocrine and genomic architecture of life history trade-offs in an avian model of social behavior. Gen. Comp. Endocr. 2008;157:275–282. doi: 10.1016/j.ygcen.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Mundy NI. window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. B. 2005;272:1633–1640. doi: 10.1098/rspb.2005.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunney TD, Katan M. PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem. Sci. 2011;36:88–96. doi: 10.1016/j.tibs.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 27.Atit R, Ronald AC, Niswander L. EGF signaling patterns the feather array by promoting the interbud fate. Dev. Cell. 2003;4:231–240. doi: 10.1016/s1534-5807(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 28.Joron M, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuster SM, Wade MJ. Equal mating success among male reproductive strategies in a marine isopod. Nature. 1991;350:608–610. [Google Scholar]
- 30.Sinervo B, Lively CM. The rock–paper–scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. [Google Scholar]
- 31.Bruford MW, Hanotte O, Brookfield JFY, Burke T. Multilocus and single-locus DNA fingerprinting. In: Hoelzel AR, editor. Molecular Genetic Analysis of Populations: A Practical Approach. 2nd edition IRL Press; Oxford: 1998. pp. 287–336. [Google Scholar]
- 32.Hogan-Warburg AJ. Social behaviour of the ruff, Philomachus pugnax (L.) Ardea. 1966;54:109–229. [Google Scholar]
- 33.Lank DB, Dale J. Visual signals for individual identification: the silent “song” of ruffs. Auk. 2001;118:759–765. [Google Scholar]
- 34.Dale J, Lank DB, Reeve HK. Signaling individual identity versus quality: a model and case studies with ruffs, queleas, and house finches. Am. Nat. 2001;158:75–86. doi: 10.1086/320861. [DOI] [PubMed] [Google Scholar]
- 35.van Rhijn J, Jukema J, Piersma T. Diversity of nuptial plumages in male ruffs Philomachus pugnax. Ardea. 2014;102:5–20. [Google Scholar]
- 36.Höglund J, Lundberg A. Sexual selection in a monomorphic lek-breeding bird: correlates of male mating success in the great snipe Gallinago media. Behav. Ecol. Sociobiol. 1987;21:211–216. [Google Scholar]
- 37.Stonor CR. On a case of a male ruff (Philomachus pugnax) in the plumage of an adult female. Proc. Zool. Soc. Lond. A. 1937;107:85–88. [Google Scholar]
- 38.Piersma T, Rogers KG, Boyd H, Bunskoeke EJ, Jukema J. Demography of Eurasian golden plovers Pluvialis apricaria staging in The Netherlands, 1949–2000. Ardea. 2005;93:49–64. [Google Scholar]
- 39.Morgan T. M.S. thesis. University of Alaska; Fairbanks: 2006. Hormonal regulation of alternative reproductive strategies. [Google Scholar]
- 40.Lank DB, Smith CM. Conditional lekking in ruff (Philomachus pugnax) Behav. Ecol. Sociobiol. 1987;20:137–145. [Google Scholar]
- 41.Goymann W, Wingfield JC. Competing females and caring males. Sex steroids in African black coucals, Centropus grillii. Anim. Behav. 2004;68:733–740. [Google Scholar]
- 42.Wingfield JC, Farner DS. The determination of five steroids in avian plasma by radioimmunoassay and competitive protein-binding. Steroids. 1975;26:311–327. doi: 10.1016/0039-128x(75)90077-x. [DOI] [PubMed] [Google Scholar]
- 43.Farrell LL, Dawson DA, Horsburgh GJ, Burke T, Lank DB. Isolation, characterization and predicted genome locations of ruff (Philomachus pugnax, AVES) microsatellite loci. Cons. Genet. Resour. 2012;4:763–771. [Google Scholar]
- 44.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang J. An improvement on the maximum likelihood reconstruction of pedigrees from marker data. Heredity. 2013;111:165–174. doi: 10.1038/hdy.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aronesty E. Expression Analysis. Durham, NC: 2011. ea-utils: Command-line tools for processing biological sequencing data. http://code.google.com/p/ea-utils. [Google Scholar]
- 47.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Simpson JT. Exploring genome characteristics and sequence quality without a reference. 2013. arXiv:1307.8026v1 [q-bio.GN] [DOI] [PMC free article] [PubMed]
- 50.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 51.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015 http://www.repeatmasker.org.
- 53.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] 2013.
- 54.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 55.English AC, et al. Mind the Gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ekblom R, Farrell LL, Lank DB, Burke T. Gene expression divergence and nucleotide differentiation between males of different colour morphs and mating strategies in the ruff. Ecol. Evol. 2012;2:2485–2505. doi: 10.1002/ece3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell MS, Holt C, Moore B, Yandell M. Genome Annotation and Curation Using MAKER and MAKER-P. Current Protocols in Bioinformatics. 2002;48:4.11.1–4.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 61.Korf I. Gene finding in novel Genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ter-Hovhannisyan V, Lomsadze A, Chernoff Y, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 64.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green P, Falls K, Crooks S. Documentation for CRIMAP. Version 2.4. Washington University School of Medicine; St Louis, MO, USA: 1990. http://www.animalgenome.org/tools/share/crimap/ [Google Scholar]
- 69.Groenen MAM, et al. A consensus linkage map of the chicken genome. Genome Res. 2000;10:137–147. doi: 10.1101/gr.10.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aslam ML, et al. A SNP based linkage map of the turkey genome reveals multiple intrachromosomal rearrangements between the turkey and chicken genomes. BMC Genomics. 2010;11:647. doi: 10.1186/1471-2164-11-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawakami T, et al. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol. Ecol. 2014;23:4035–4058. doi: 10.1111/mec.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Oers K, et al. Replicated high-density genetic maps of two great tit populations reveal fine-scale genomic departures from sex-equal recombination rates. Heredity. 2014;12:307–316. doi: 10.1038/hdy.2013.107. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aulchenko YS, Ripke S, Isaacs A, van Duijin CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 74.Corbett-Detig RB, Cardeno C, Langley CH. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics. 2012;192:131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.