Abstract
Human immunodeficiency virus type 1 (HIV-1) exists as a complex population of multiple genotypic variants in persons with chronic infection. However, acute HIV-1 infection via sexual transmission is a low-probability event in which there is thought to be low genetic complexity in the initial inoculum. In order to assess the viral complexity present during primary HIV-1 infection, the V1/V2 and V3 variable regions of the env gene were examined by using a heteroduplex tracking assay (HTA) capable of resolving these genotypic variants. Blood plasma samples from 26 primary HIV-1-infected subjects were analyzed for their level of diversity. Half of the subjects had more than one V1/V2 viral variant during primary infection, indicating the frequent transmission of multiple variants. This observation is inconsistent with the idea of infrequent transmission based on a small transmitting inoculum of cell-free virus. In chronically infected subjects, the complexity of the viral populations was even greater in both the V1/V2 and the V3 regions than in acutely infected subjects, indicating that in spite of the presence of multiple variants in acute infection, the virus does pass through a genetic bottleneck during transmission. We also examined how well the infecting virus penetrated different anatomical compartments by using the HTA. Viral variants detected in blood plasma were compared to those detected in seminal plasma and/or cerebral spinal fluid of six individuals. The virus in each of these compartments was to a large extent identical to virus in blood plasma, a finding consistent with rapid penetration of the infecting variant(s). The low-probability transmission of multiple variants could be the result of transient periods of hyperinfectiousness or hypersusceptibility. Alternatively, the inefficient transfer of a multiply infected cell could account for both the low probability of transmission and the transfer of multiple variants.
Characterization of the earliest stage of human immunodeficiency virus type 1 (HIV-1) infection, primary infection, is critical for gaining a better understanding of HIV-1 pathogenesis. Determining what variants are transmitted and what factors, if any, affect transmission will greatly aid the development of vaccines and other prophylactic therapies (such as microbicides). Primary infection studies may also lead to a better understanding of the role of innate host defense.
Primary infection can be roughly subdivided into two phases: acute and early. Peak viremia is reached and resolved during the acute phase while the virus replicates initially in the absence of immune response. Primary infection symptoms begin, on average, 2 weeks after infection (3, 31, 45, 54), most likely the result of the initial immune response. During early primary infection, the immune response matures leading to seroconversion and a steady state of HIV-1 replication (4, 6, 8, 23, 28, 38, 63, 66, 67). Primary infection ends with seroconversion and stabilization of the Western blot pattern (7, 31). Identifying subjects during primary infection is difficult but critical to its study. The absence of HIV-1 antibodies as measured by enzyme-linked immunosorbent assay (ELISA; seronegative) defines the acute primary infection stage. The detection of evolving antibody specificity, measured by Western blotting, is an indicator of early primary infection. Western blot antibody specificities typically become diagnostic within 1 to 2 weeks of ELISA positivity (19).
Transmission probabilities associated with the most common HIV-1 exposures are relatively low (18, 22, 58), which a priori would be expected to create a genetic bottleneck that would result in the infecting variant being genetically less complex than the swarm of donor virus strains. Transmission can occur from either cell-free virus (e.g., infection via plasma-derived blood products) (34) or from cell-associated virus (e.g., infected cells in semen) (68). However, in most cases it is not clear which of these forms of virus is transmitted.
Studies of viral diversity during primary infection have generated variable results. The presence of homogeneous (1, 11, 20, 27, 37, 39, 43, 44, 55, 56, 63, 64, 67) or heterogeneous (10, 14, 16, 25, 29, 30, 32, 36, 42, 48, 53, 57, 60, 65, 68) virus populations have been documented. Evidence for differences in viral heterogeneity during primary infection based on sex has also been reported (33, 47) but not seen consistently (36). These discrepancies are in part explained by the different regions of the viral genome that were compared, the different modes of transmission among populations, the different definitions of primary infection that were used, and different criteria by which viral populations were categorized as homogeneous versus heterogeneous.
We assessed viral diversity during primary infection by heteroduplex tracking assay (HTA), which resolves distinct genotypes in a gel-based assay (9, 12). Heterologous probes were used to enhance the detection of nucleotide differences of <2% between viral variants as described by Kitrinos et al. (26). The HTA is capable of accurately quantitating variants contributing as little as 3% to the total viral population (49). We applied this technology to reveal the presence of multiple coexisting variants of the V1/V2 and V3 variable regions of the viral env gene within infected people (26, 40, 41). Here we report studies to examine the complexity of the viruses present in the blood plasma of 26 subjects in a primary infection cohort. All subjects in the present study were infected through sexual contact. All but one subject was estimated to have acquired HIV-1 fewer than 90 days prior to sampling. Approximately 50% of the subjects harbored multiple viral variants, a result inconsistent with a low transmission probability from an inoculum consisting solely of cell-free virus. The initial viral populations present in the seminal plasma (SP) and/or cerebrospinal fluid (CSF) were also examined in a subset of subjects. Significant penetrance of the transmitted variants was observed in these compartments as early as 1 month after infection.
MATERIALS AND METHODS
Subjects.
The 26 subjects in the present study were part of a primary HIV-1 infection cohort enrolled at the University of North Carolina-Chapel Hill, Duke University, and Emory University. All but one of the subjects were identified after presentation with symptoms of an acute retroviral syndrome. The number of days postinfection was estimated by adding 2 weeks to the date of symptom onset; for one asymptomatic subject, a unique sexual exposure was reported, and transmission was confirmed by phylogenetic analysis. A negative HIV-1 antibody test within 30 days of diagnosis or an evolving Western blot pattern was necessary for inclusion, except for subjects Z04 and Z09, who were included based on interview and disease symptoms. Blood plasma samples were collected for all subjects, whereas CSF and/or seminal samples were examined where available. Subject characteristics are presented in Table 1. The average log10 viral load was 5.6, and the range was 3.7 to 7.3. The average CD4 count was 518 cells/μl (range, 128 to 1,161). All samples were collected with informed consent under an Institutional Review Board-approved protocol.
TABLE 1.
Primary infection subject characteristics
| Subject | Subject characteristicsa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WPI | Sex | Viral loadb
|
EIAc | WBd | Modee | |||
| Blood | CSF | Semen | ||||||
| Z01 | 7 | F | 4.5 | + | I | HT | ||
| Z02 | 4 | M | 5.5 | + | I | HT | ||
| Z03 | 5 | M | 5.3 | 4.3 | + | I | HM | |
| Z04 | 27 | M | 5.3 | + | + | HM | ||
| Z05 | 4 | M | 7 | NA | − | HT | ||
| Z06 | 6 | M | 5.7 | + | I | HM | ||
| Z07 | 10 | M | 3.7 | NA | I | HM | ||
| Z08 | 7 | M | 5.5 | + | I | HM | ||
| Z09 | 8 | M | 4.4 | 3.1 | + | NA | HM | |
| Z10 | 10 | M | 5.3 | 3.8 | + | I | HM | |
| Z11 | 10 | M | 6.1 | 3.1 | 5.9 | + | I | HM |
| Z12 | 8 | M | 5.1 | + | I | HM | ||
| Z13 | 10 | M | 4.7 | 3.1 | + | I | HT | |
| Z14 | 3 | M | 5.8 | − | − | HM | ||
| Z15 | 12 | F | 5.3 | + | I | HT | ||
| Z16 | 6 | M | 6.2 | NA | I | HM | ||
| Z17 | 5 | M | 6.4 | + | I | HM | ||
| Z18 | 5 | M | 6.5 | 3.3 | NA | − | HM | |
| Z20 | 4 | M | 7.1 | 3.8f | 4.3 | + | I | HM |
| Z23 | 6 | M | 5.3 | NA | NA | HM | ||
| Z24 | 5 | M | 7.3 | − | I | HT | ||
| Z25 | 10 | M | 4.7 | + | I | HM | ||
| Z26 | 6 | M | 6.1 | NA | − | HM | ||
| Z27 | 5 | M | 5.3 | 3.8 | NA | NA | HT | |
| Z29 | 5 | M | 5.7 | 3.8 | 3.7g | + | I | HM |
| Z30 | 5 | F | 6.2 | − | − | |||
All data refer to subject samples used in this study and not necessarily samples used to identify eligible study subjects. All compartment samples are matched for time unless otherwise noted. WPI, weeks post-infection; NA, not applicable; M, male; F, female.
That is, the log10 copies of viral RNA/milliliter of blood plasma, CSF, or seminal plasma.
Reactivity to sensitive HIV-1 ELISA (EIA).
Reactivity to HIV-1 Western blot (WB). −, Negative; I, incomplete; +, positive.
Mode of transmission: HT, heterosexual; HM, homosexual.
Sample from 1 week before blood plasma and seminal plasma samples were obtained.
Sample from 1 week after blood plasma and CSF samples were obtained.
The 17 subjects with established HIV-1 infection were part of a study at the University of North Carolina-Chapel Hill to assess the effects of antiretroviral treatment on the development of HIV-1-associated neurological dysfunction. The average log10 viral load was 4.9 (range, 4.2 to 6.4). The average CD4 count was 243 cells/μl (range, 10 to 500). All blood plasma samples were collected and analyzed in the absence of ongoing antiretroviral therapy treatment and with informed consent.
RNA extraction.
RNA was extracted from 140 μl of blood plasma, CSF, or SP by using the QIAamp viral RNA minikit as described by the supplier (Qiagen, Valencia, Calif.) with a final 60-μl elution volume. Samples with viral RNA loads of <10,000 copies/ml were concentrated by centrifugation of 1 ml of sample at 25,000 × g for 1.5 h at 4°C before RNA extraction. The virus pellet was resuspended in 140 μl of its supernatant, and RNA was extracted as described above.
RT-PCR.
The V1/V2 region of the HIV-1 env gene was reverse transcribed by using a modified Titan One-Step RT-PCR system (Roche Molecular Biochemicals). The Titan One-Step RT-PCR system protocol was as described in Kitrinos et al. (26). Briefly, the cDNA synthesis reaction consisted of 1× Titan RT-PCR buffer, 15 pmol of V2 primer, a 1 mM concentration of each deoxynucleoside triphosphate (Amersham Pharmacia), 5 mM dithiothreitol, 10 U of RNase inhibitor (Roche Molecular Biochemicals), 14 U of avian myeloblastosis virus (AMV) reverse transcriptase (RT) (Roche Molecular Biochemicals), and 5 to 10 μl of eluted viral RNA in a total volume of 20 μl. cDNA synthesis reactions were incubated at 42°C for 30 min and then at 95°C for 5 min to inactivate the AMV RT. A 30-μl PCR mix consisting of 1× Titan RT-PCR buffer, 15 pmol of V1 primer, 5 mM dithiothreitol, and 1.7 U of Titan or Expand enzyme was added to each 20-μl cDNA synthesis reaction for a final volume of 50 μl. PCR temperatures and cycle numbers were as described by Kitrinos et al. (26).
V3 RT-PCR products were generated by using the Expand One-Tube RT-PCR kit (Roche) modified as described by Nelson et al. (41). The Expand Kit reactions consisted of 5 to 10 μl of eluted RNA, 1× Expand HF Buffer, 2.5 mM MgCl2, 0.5 mM concentrations of each deoxynucleoside triphosphate, 15 pmol of V3R5 primer, and 12 U of AMV RT in a total volume of 20 μl. cDNA synthesis reverse transcription reactions were incubated at 42°C for 30 min, followed by 2 min at 99°C to inactivate the AMV RT. A 30-μl PCR consisting of 1× Expand HF buffer, 2.5 mM MgCl2, 15 pmol of V3L4 primer, and 2.6 U of Expand HF enzyme mix was then added to the 20-μl cDNA synthesis reaction for a final volume of 50 μl. The following PCR program was used to amplify the V3 products: 1 cycle of 95°C for 2 min and 45 s, 52°C for 45 s, and 72°C for 1 min, followed by three sets of 10 cycles of 95°C for 45 s, 52°C for 45 s, and 72°C for 1 min (with an additional minute for each set of 10 cycles). The last cycle had a 72°C extension for 7 min.
The gag fragment generated for vaccine analysis was obtained through RT-PCR of two separate primers jointly covering the p17 and p24 region (nucleotides 2546 to 3515, HXB2 numbering). The G1up (5′-GGGTGCGAGAGCGTCAGTATTAAG-3′) and G1dn (5′-TGAAAACATGGGTATTACTTCTGGGC-3′) primers amplify nucleotides 2545 to 3041. The G2up (5′-ACATCAGGCCATATCACCTAGAAC-3′) and G2dn (5′-TCCAATTTTTTACCTCCTGTGAAGC-3′) primers amplify nucleotides 2932 to 3515. The two env fragments were amplified with the E1 and E2 primer sets. The E1Bup (5′-CATACATTATTGTGCCCCGGCTGG-3′) and E1dn (5′-CGTTACAATTTCTGGGTCCCCTCC-3′) primers span the C2, V3, and C3 regions (nucleotides 6815 to 7346, HXB2 numbering). The E2Bup (5′-AAACATGTGGCAGGAAGTAGGAAAAGC-3′) and E2dn (5′-GTGCAAATGAGTTTTCCAGAGCAACC-3′) cover the C-terminal of C4, V5, C5, and gp41 N terminus (nucleotides 7626 to 7986, HXB2 numbering). Both the gag and env cDNA fragments were generated by using the Expand kit under the same conditions described for the V3 region except that 2.0 mM MgCl2 was used for the env fragments. The following PCR program was used for amplification in a Stratagene 40 Robocycler: 1 cycle of 94°C for 2 min and 45 s, 53°C for 45 s, and 68°C for 1 min; 40 cycles of 94°C for 45s, 53°C for 45 s, and 68°C for 1 to 4 min (1-min increase every 10 cycles); and 1 cycle of 94°C for 45 s, 53°C for 45 s, and 68°C for 7 min.
HTA.
V1/V2-HTAs were carried out by using a single-strand 35S-labeled probe derived from either the HIV-1 JR-FL or Ba-L cloned genome as described by Kitrinos et al. (26). An aliquot of 8 μl of unpurified RT-PCR product was combined with 1 μl of probe, 1 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), and 0.1 μM concentrations of V1 and V2 primers in a total volume of 10 μl. The probe and duplex PCR product were denatured at 95°C for 2 min, followed by a 5-min incubation at room temperature to allow formation of heteroduplexes. Heteroduplexes were resolved by electrophoresis through a 6% nondenaturing polyacrylamide gel (acrylamide-bisacrylamide [37.5:1]) in 1× Tris-borate-EDTA buffer. Dried gels were exposed to X-ray film and a phosphorimaging screen (Molecular Dynamics).
V3-HTAs were carried out as described by Nelson et al. (40, 41). Briefly, annealing reactions were set up as in the V1/V2-HTA but with either a clade B V3 probe (JN27) and the V3L4 primer or the clade C V3 probe (JN33) and the V3R5 primer. Screening by HTA with a clade B R5-consensus sequence probe (JN27) detects variants with sequence changes away from the R5-like consensus (41). Sequence analysis can then be used to identify variants with changes indicative of CXCR4 usage. Additional screening with a heterologous clade C V3-HTA probe (JN33) enhances the detection of point mutations that distinguish V3 genotypic variants. The heteroduplexes were resolved by electrophoresis in a 12% polyacrylamide gel under the same conditions as the V1/V2-HTA. Dried gels were exposed to X-ray film and a phosphorimaging screen. All amplifications and HTA analyses were repeated to document the presence of the same variants at the same relative concentration as a control to validate the quality of sampling. All samples that displayed a single genotypic variant had RNA levels comparable to the samples that gave acceptable sampling of multiple variants.
Sequence analysis.
V1/V2 RT-PCR products were gel purified by using a Qiagen gel purification kit. V3 RT-PCR products were purified by using a QIA-quick PCR purification kit (Qiagen). Purified products were either directly sequenced or cloned by using the pT7Blue-3 Perfectly Blunt cloning kit (Novagen). Individual clones were amplified by colony PCR with the appropriate primers and screened by HTA with the appropriate probe. Two to ten clones representing each variant were selected for sequencing. Sequences were determined by using ABI dye terminator sequencing (Perkin-Elmer Corp.) and analyzed with MacVector 7.0 (Genetics Computer Group, Madison, Wis.).
Uncorrected V1/V2 nucleotide pairwise distances were calculated with DISTANCES (Genetics Computer Group (GCG) Wisconsin package, version 10.1-GCG). The sequence relatedness of V1/V2 clones was further analyzed by neighbor-joining. Length differences were removed from sequence alignments used in phylogenetic analysis.
Bulk sequencing of the G1, G2, E1, and E2 primer set products was obtained by direct sequencing of purified RT-PCR products by using ABI dye terminator sequencing (Perkin-Elmer Corp.). The two gag fragments from each subject were aligned by using GCGSEQALIGN and combined into one sequence spanning nucleotides 2546 to 3515 (HXB2 numbering). Analysis of the two env sequences was performed by concatenating the sequence fragments representing each isolate. The phylogenetic analysis of the gag region was performed by a neighbor-joining tree.
Sets of sequences representing subtype B isolates from the United States (123 and 133 sequences spanning the above regions of gag and env sequences, respectively) were obtained from the Los Alamos HIV Sequence Database. A consensus sequence was generated for each fragment from multiple sequence alignments created with GCG Pileup. Amino acid positions were identified by aligning this consensus sequence with the corresponding HIV-1 HXB2 protein sequence. Amino acid sequence identity scores between all pairs of unaligned sequences were determined by using GCG Gap.
Nucleotide sequence accession number.
The sequences reported here have been submitted to GenBank under accession numbers AY705242 through AY705348.
RESULTS
Identification of subjects.
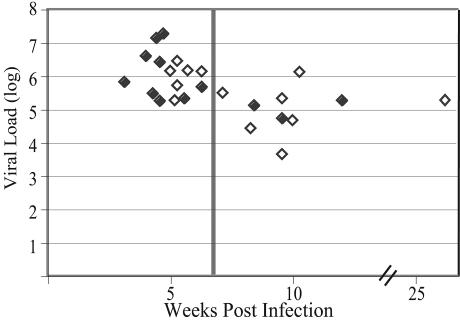
The primary infection cohort of 26 subjects includes 3 (11%) females and 23 (89%) males. Likely modes of transmission established by patient history include heterosexual (n = 7), homosexual (n = 18), and oral-genital (n = 1). The mode of transmission was not known for one subject (Z30). Subject characteristics are given in Table 1. HIV-1-specific antibodies, measured by ELISA, were absent at baseline in subjects Z14, Z24, and Z30. The remaining subjects had evolving Western blots except for subjects Z04 and Z09. In a controlled study of primary infection in macaques, all animals resolve their viremia by 7 weeks postinfection (52). Subjects estimated as being at <7 weeks postinfection had significantly higher viral loads than those at >7 weeks postinfection (P = <0.001 [Wilcoxon rank sum test]) (Fig. 1), supporting both the primary infection diagnosis and timing estimates.
FIG. 1.
Relationship between viral load and estimated time since infection. Log10 viral load (copies/milliliter) for the sample analyzed is plotted versus the estimated time after infection, assuming infection was 2 weeks prior to the onset of symptoms of acute infection. A vertical line is included at week 7 to indicate groups of subjects staged to before or after this time. Filled symbols are subjects whose virus showed a single V1/V2 variant (as assessed by HTA); open symbols are subjects whose virus showed multiple V1/V2 variants.
A cohort of 17 chronic stage subjects enrolled at the University of North Carolina-Chapel Hill to assess the effects of retroviral treatment on the development of HIV-1 associated cognitive disorders was used for comparison. All samples assayed were obtained at times when the subjects were not receiving treatment. This cohort consisted of 14 males and 3 females.
Virus complexity during primary infection.
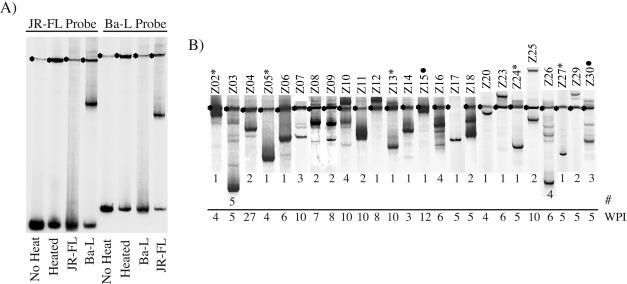
The number of coexisting viral variants within subjects was assessed by analyzing the variable regions 1 and 2 (V1/V2) of the HIV-1 env gene by HTA. A single V1/V2 viral variant was detected in approximately one-half of primary infection subjects (12 of 25), with two to five variants being detected in the remainder (mean = 2.0, median = 2) (Fig. 2). The complexity of the V1/V2-HTA pattern obtained from primary infection subjects was compared to that seen in the 17 chronic-stage subjects. Only one subject from the chronic cohort displayed a single V1/V2 variant, with the remainder having 2 to 10 variants (mean = 5.1, median = 4) (Fig. 3). Thus, although the complexity of the virus population was reduced in the primary infection cohort relative to the chronically infected cohort, ca. 50% of primary infection patients had multiple variants present.
FIG. 2.
V1/V2-HTA patterns of subjects during primary HIV-1 infection. (A) Control lanes with the probes for V1/V2 diversity analysis (JR-FL and Ba-L). Probes were run alone with or without the denaturing step or with their own or alternate probe's PCR product. The asterisks indicate the single-stranded probe, and the lower bands are the probe homoduplex. (B) Blood plasma samples were used as the source of viral RNA which was then used as a template to amplify the V1/V2 region of env using PCR. Heteroduplexes were formed with either the JRFL or BAL V1/V2 probe, which were then resolved in a polyacrylamide gel. The identifier of each subject is shown above the lane. The band with the asterisk represents the position of the single-stranded HTA probe. The weeks postinfection (WPI) and the numbers of V1/V2 variants detected (#) are indicated at the bottom of each lane.
FIG. 3.
Comparison of number of V1/V2 variants detected by HTA with time after infection. Each datum point represents one subject from the primary infection cohort. The estimated time postinfection is plotted versus the number of V1/V2 variants detected for the subject (Fig. 2). Subjects in the primary infection cohort are compared to a separate group of subjects during the chronic stage of their HIV-1 infection. Although not resolved in the figure, there are nine individuals identified with single variants prior to week 7.
There was no statistically significant difference between the distribution of the number V1/V2 variants per person in primary infection samples obtained between 3 to 7 weeks and 8 to 14 weeks postinfection (P = 0.115) (Fig. 3). There was no relationship between viral diversity and the detection of early antibodies (not shown). Diversity was evident in one of the three ELISA-negative subjects (Z14, Z24, and Z30), with Z30 displaying three variants. These data indicate that the V1/V2-HTA patterns provide one measure of the complexity of the virus early after transmission and that multiple genotypic variants are frequently present.
Multiple V1/V2 variants were not equally distributed across the different modes of transmission (Fig. 2). Homosexual transmission resulted in infection with a single variant in only 6 of 18 (33%) instances, while heterosexual transmission to a male resulted in only single variants being transmitted (5 of 5) (P = 0.014 [Fisher exact test]). In the two females infected by heterosexual contact, one (Z15) had a single variant and the other (Z30) displayed multiple variants.
Intrapatient genetic distances and detection of dual infections.
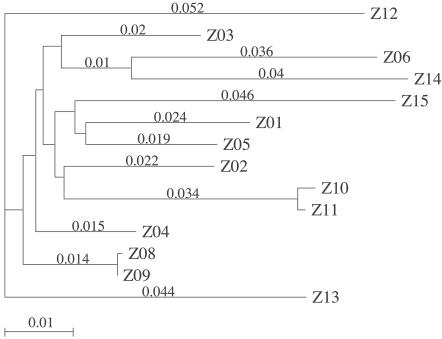
The presence of multiple V1/V2 variants during primary infection could be the result of multiple independent transmission events occurring in close succession. In these instances, the genetic distance between viral variants would be greater than that expected within a single subject. The individual V1/V2 viral variants were cloned, and the genetic distance was calculated from a subset of subjects representing the transmission of either single (Z06 and Z24) or multiple (Z03, Z10, Z11, Z16, Z18, Z25, Z26, and Z30) variants. Subjects were selected from both before and after the 7-week division. Either 6 or 10 clones from each of the two independent RT-PCRs were cloned from Z06 and Z24, respectively. All clones from Z06 (12 of 12) were identical to the nucleotide level. In subject Z24, 18 of 20 clones were identical to the nucleotide level, with the remaining 2 each having a single nucleotide change. Thus, the variability within an HTA band in these two primary infection subjects that had single HTA variants is essentially zero and therefore represents homogeneous populations. Similarly, when multiple variants were transmitted, sequence analysis of clones representing intraband comparisons showed that 90% of the clones were either identical to or differed by a single nucleotide from the inferred consensus sequence for that band, again indicating that each HTA variant largely represented a clonal infection.
The genetic distances between HTA variants within a subject (interband) were calculated in the subset of subjects examined with multiple variants. Identified variants were cloned and sequenced. The majority (six of eight) of subjects had interband genetic distances for these V1/V2 segments that were consistent with the transmission of multiple variants from a single donor (Fig. 4), with interband distances ranging from 0.23 to 7.3% (mean of 2.4%). However, the interband distances in two subjects (Z25 and Z26) were indicative of dual infection (Fig. 4). Subject Z25 had an interband genetic distance of 21%, suggesting dual infection from two independent transmission events, each resulting in the transfer of a single variant. The interband genetic distances in subject Z26 ranged from 2 to 19%, suggesting one transmission event of a single variant and another of multiple variants.
FIG. 4.
Phylogenetic analysis of V1/V2 clones. RT-PCR products from separate reactions independently cloned after gel purification. Variant clones were identified by HTA screening. Where possible, at least two independent clones of each variant were sequenced. For two subjects, Z06 and Z24, the single variant was independently cloned and sequenced 6 to 10 times from each RT-PCR product. The neighbor-joining tree was made with aligned sequences with gaps ignored. All sequences are from the blood. Identifiers indicate subject (e.g., Z10), clone (e.g., g3), and HTA band (e.g., 0.1, 0.2, etc.), respectively. For subjects Z06 and Z24, which each had a single variant, the HTA band number is indicated as “0.” For these two subjects, the number of clones with identical sequences is shown in parentheses.
Viral tropism during primary infection.
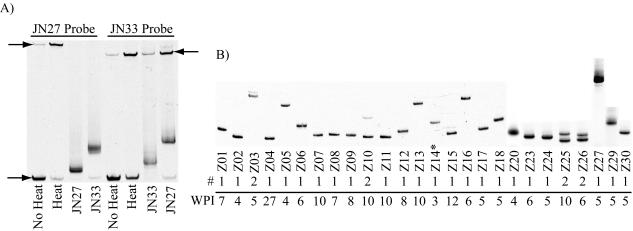
Viral cell tropism and V3 diversity at primary infection can be inferred from V3-HTA and sequence analysis. The V3-HTA data for the 26 primary infection subjects are shown in Fig. 5. The HTA probe (clade B or C) revealing the greatest number of variants is shown. Eighty-five percent of the subjects (22 of 26) had one detectable V3 variant, with no one having more than two variants. All subjects with two V3 variants also exhibited multiple V1/V2 variants. In contrast, a mean of 2.5 V3 variants was detected in the chronic cohort (median = 2, range = 1 to 4) (data not shown).
FIG. 5.
V3-HTA patterns of subjects during primary HIV-1 infection. (A) Control lanes with the clade B (JN27) and clade C (JN33) V3 probes alone or against their PCR products. The single-stranded probe is indicated by the top arrows, and the lower arrow indicates the probe homoduplex. (B) Viral RNA from blood plasma was used as a template to amplify the V3 region of env using PCR. Heteroduplexes were formed between the PCR product and either the pJN27 (clade B) or pJN33 (clade C) V3 HTA probes. The resulting heteroduplexes were resolved in a polyacrylamide gel. Subject identifiers are shown at the bottom of the each lane. WPI, weeks postinfection; #, number of V3 variants detected.
Sequence analysis of either the bulk PCR product or clones of the PCR products showed that all but one subject harbored R5-like virus (not shown). Subject Z14 had the characteristic X4-like 11R substitution within V3. CXCR4 usage was confirmed by growth of a virus isolate from Z14 peripheral blood cells in MT2 cells (not shown).
Characterization of linked transmissions.
Contact tracing revealed that primary infection subject Z10 was the source of virus for subject Z11. Only 5 days separate their calculated day of infection. The direction of transmission is supported by delayed seroconversion in subject Z11 relative to Z10. Although this linked transmission is not apparent as comigrating bands in the V1/V2 HTA analysis, sequence analysis of Z10 and Z11 V1/V2 clones revealed significant sequence similarity outside the length-variable regions (not shown). Sequence differences within the V1/V2 length-variable regions of Z10 and Z11 are consistent with their different V1/V2-HTA patterns. In the V3 region, only the lower of the two Z10 V3 variants was detectable in Z11 (Fig. 5). The V3 sequence of this variant was identical in Z10 and Z11, as were the bulk protease and RT sequences (not shown). Phylogenetic analysis confirmed this link, with Z10 and Z11 sequences from env and gag being more closely related to each other than to other clade B primary infection sequences (Fig. 4 and 6). The sequence relatedness supports the contact tracing, whereas the HTA analysis indicates the potential for contraction of the viral population at transmission.
FIG. 6.
Phylogenetic analysis of plasma gag sequences. The sequence from the bulk PCR product covering two overlapping regions of gag p17 and p24 were combined for this analysis. Subject numbers are indicated. Branch numbers represent number of nucleotide changes per 100 bases.
Unexpectedly, subjects Z08 and Z09 had very similar V1/V2-HTA patterns. Further interviews indicated a recent shared sexual partner. The translated V1/V2 sequences of Z08 and Z09 are identical, even through the length-variable regions. The nucleotide sequence varies by a single synonymous change. Subjects Z08 and Z09 also share an identical V3 variant by both HTA and sequence analysis of the PCR product. Finally, bulk sequencing of PCR product spanning the majority of the gag gene showed close branching (Fig. 6). The presence of such closely related V1/V2 (Fig. 4), V3 (Fig. 5), and gag variants (Fig. 6) suggests a common source of infecting virus and, in these cases, the transmission of the same variant.
Analysis of compartmentalization during primary infection.
Paired blood plasma, SP, and/or CSF samples were examined in both the V1/V2 and the V3 env regions by HTA in a subset of subjects to determine whether compartmentalization between these regions exists during primary infection. Virus was detected in these compartments in all cases examined. This analysis included six semen samples and five CSF samples from 31 to 72 days postinfection. Thus, it appears the infecting virus can populate these compartments quickly after infection.
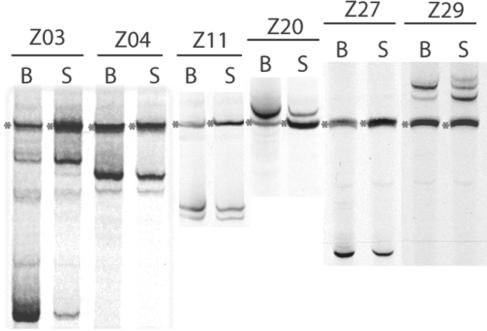
The V1/V2-HTA for the blood/SP comparison for six subjects is shown in Fig. 7. Evidence for compartmentalization is present only in subject Z03, with shared variants present at differing relative abundance. The difference seen in Z03 was confirmed by quantitative phosphorimager analysis. Phosphorimager data were collected in duplicate by analyzing the HTA banding pattern from two separate RT-PCRs. In the blood, the fastest-migrating variant constitutes 75% (averages of 76 and 74%) of the total viral population, and the slower-migrating variant represents 10% (averages of 10 and 9%). In the seminal compartment, the slower-migrating variant represents 72% (averages of 70 and 74%) of the total population and the faster migrating variant 14% (averages of 13 and 14%). The two other cases with multiple V1/V2 variants in the blood had similar variant representation in the semen (Z04 and Z11). No differences were detected in the V3 region in any of the samples (data not shown). Thus, to a large extent, variants in the blood are found early in the semen, with some differences in the relative proportion of V1/V2 variants.
FIG. 7.
Comparison of V1/V2 env variants in blood and semen of subjects during primary HIV-1 infection. Viral RNA extracted from paired blood plasma (B) or SP (S) was used as a template to amplify the V1/V2 region of env using PCR. Subject identifiers are above the lane. Heteroduplexes were formed and analyzed as described in the legend of Fig. 2.
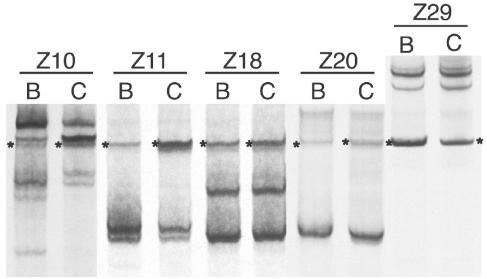
The V1/V2 variants found in the blood plasma versus the CSF for five subjects are shown in Fig. 8. Only subject Z10 showed any evidence of compartmentalization, with a few minor blood V1/V2 variants not detected in the CSF. The multiple V1/V2 variants present in the three other subjects were present in similar proportions in both the blood and CSF. Again, no differences were detected in the V3 region (data not shown). The lack of compartmentalization between the periphery and the central nervous system suggests that during primary infection, these two compartments equilibrate relatively quickly.
FIG. 8.
Comparison of V1/V2 env variants in blood and CSF of subjects during primary HIV-1 infection. Viral RNA extracted from paired blood plasma (B) or CSF (C) was used as a template to amplify the V1/V2 region of env using PCR. Subject identifiers are above the lane. Heteroduplexes were formed and analyzed as described in the legend of Fig. 2.
Selection of a vaccine candidate among viruses isolated during primary infection.
Significant effort is being made to develop HIV-1 vaccine candidates. Although cross-subtype reactivity has been demonstrated in cytotoxic T lymphocytes (CTLs) from individuals infected with different subtypes (2, 15, 17, 35, 62), the cross-subtype reactivity is typically not as strong as intra-subtype reactivity and frequently shows subtype preferences. This observation is supported by the finding that certain CTL epitopes are better conserved within one subtype versus across subtypes (51). To facilitate the development of a subtype B vaccine, viral sequences from this primary infection cohort were identified with high amino acid similarity and identity to the subtype B consensus sequence. This strategy is designed to identify functional viral genes that minimize the amino acid genetic distance to extant sequences. Sequences from a primary infection cohort by definition represent currently circulating variants and would have the advantage that the resulting vaccine candidate will represent a currently circulating transmissible variant. We have previously used this strategy to identify subtype C vaccine candidate genes (61).
Sequences from the gag (p17 and p24) and env (V4 through gp41) genes were obtained for subjects Z01 to Z18. Two additional primary infection subjects from another cohort were also included (qho515 and qho692). Sequences from these primary infections were compared to each other and to a collection of 123 and 133 gag and env sequences, respectively, obtained from the Los Alamos HIV Sequence Database. Pairwise alignments of these regions were made, and the distributions of amino acid identity scores resulting from all pairwise comparisons, including those for the consensus sequence, were determined for both the translated Gag and Env (Fig. 9) sequence fragments.
FIG. 9.
Genetic distances of sequences from viruses present in the primary infection cohort to a large set of HIV-1 sequences. Regions of the gag (A) and env (B) genes were amplified by RT-PCR, and the purified PCR products were sequenced by using one of the PCR primers. The amino acid sequence was inferred from the nucleotide sequences. Pairwise comparisons were made for percent amino acid identity between each sequence and the clade B consensus sequence (○) and to a large set of HIV-1 sequences. For the large comparison, the mean value (+), the inner quartile range (box), and the entire range (line) are shown.
Isolate Z04 had the highest percent identity score with the subtype B consensus sequence in both Gag and Env. Due to these high percent identity scores, the full-length gag and env genes from the Z04 isolate have been cloned for use in a subtype B HIV-1 vaccine. These genes are derived from a replication-competent clinical isolate that will facilitate the testing of immune responses.
DISCUSSION
The study of transmitted virus population complexity during early primary HIV-1 infection has been limited by the difficulty of identifying subjects before the seeding population has significantly evolved. Multiple factors, including mode of transmission, source of inoculum, complexity of the virus population in the donor, and recipient factors, likely affect what and how many variants are transmitted. The conflicting results of existing primary infection studies are likely accounted for by the inability to control for all of these variables in the transmission events of the study population. We have used the V1/V2-HTA as a tool to evaluate the complexity of the virus population in a well-defined primary infection cohort.
Diversification of the transmitted virus in response to HIV-1-specific host immune responses can complicate the interpretation of population diversity in primary infection studies. SIV studies can provide temporal profiles regarding host immunological responses and thus allow one to estimate the time required for viral diversification. SIV Env-specific neutralizing antibodies in macaques are present 7 weeks postinfection. Diversification of the V1/V2 region in macaques is not detected until after 12 weeks after mucosal infection (52). Strong HIV-1-neutralizing antibodies have been detected 6 to 8 weeks postinfection in humans (50). These similarities suggest the timeframe of diversification of the V1/V2 region in macaques and humans may be similar. Our recent study with longitudinal samples from humans suggests V1/V2 diversification starts in the 8- to 11-week timeframe (unpublished observations). All but two samples in our study were obtained within this timeframe. Thus, we believe this cross-sectional analysis of human subjects in primary infection allows us to evaluate characteristics of the virus present at the earliest time points after transmission and likely represent the infecting virus. It is clear that a delay in identifying subjects in primary infection will result in a bias toward the detection of multiple variants as a result of env diversification due to the humoral response. In this regard we note that the subjects staged as most proximal to the transmission event had single variants (Fig. 1 and 3). However, this group also included most of the heterosexual transmissions to males, which appears to favor the transmission of single variants (33) (see below).
The preponderance of studies examining HIV-1 transmission show a mostly homogeneous population. Primary infection studies have focused on env variable domains to measure viral diversity. These variable domains are not equally diverse. In subtype B, the V1/V2 region tends to be the most variable while the V3 region is the most conserved of the variable domains. When we examined the V1/V2 region during primary infection, just >50% of subjects displayed multiple variants. However, when the V3 region was examined, only 17% of subjects displayed multiple variants. All subjects with multiple V3 variants also had multiple V1/V2 variants. This suggests the V1/V2 region can reveal more diversity in transmitted variants than the V3 region. Therefore, we believe V1/V2 HTA is a more robust tool for describing a complex viral population than V3 HTA, which we have also used previously in this context (46). The detection of multiple variants in 50% of subjects is a minimum estimate since transmission of two variants with the same V1/V2-HTA pattern would be recorded as a single variant. This underestimate may be corrected by also examining diversity in the V4/V5 region. As with all primary infection studies, the timing of infection is critical to an accurate assessment of primary infection viral diversity. Underestimating the time since transmission can result in an overestimate of the complexity of the transmitted variants since the immune response drives the viral population to greater complexity.
The phylogenetic analysis of V1/V2 clones supported the transmission of multiple variants from a single donor. The genetic distance between variants was too extensive to have occurred during the time of infection. Dual infection was identified in two subjects. Subject Z25 was infected by two single variant transmission events. Subject Z26 had genetic distances consistent with a transmission event of a single variant transmission and transmission event of multiple variants. Half (two of four) of the subjects with multiple variants by V3-HTA were dually infected. This suggests that diversity in the V3 region during primary infection may serve as a useful screen in the search for dual infections that ultimately must be identified through sequence analysis.
Zhang et al. (66) detected a higher level of diversity in gag versus env during primary infection by examining sequences from DNA clones generated from these genes. By HTA, we detected a much higher level of heterogeneity in the env gene than implied by the Zhang et al. study (66). This difference may be explained by the different techniques used. The HTA samples all of the genomes and templates that are present in the PCR amplification, allowing major and minor V1/V2 populations to be evident. Detection of minor populations may be missed when a limited number of clones are sequenced since each genome or template is sampled as one sequence. Also, diversification due to CTL selection likely precedes diversification by antibody selection (which presumably drives V1/V2 diversification), and the greater gag diversity reported may reflect the effect of the early host CTL response. In addition, Learn et al. (30) have suggested that multiple variants are typically transmitted but then undergo selection, resulting in a homogeneous population. We do not observe a decrease in V1/V2 env diversity during the early period of infection either in this cross-sectional analysis (Fig. 3) or in longitudinal studies in macaques (52).
Several studies have shown that R5 variants are most often transmitted, although X4 variants can be transmitted (59). Our observation of one infection with an X4 variant is consistent with this infrequent occurrence. The low transmission frequency of X4-like virus in this cohort was similar to the 3% frequency found in a larger UCSD cohort (99 subjects) (K. Ritola, R. Swanstrom, D. Richman, and S. Little, unpublished data). The low viral diversity in the V3 region and high prevalence of R5-like variants support the transmission of low complexity R5-like V3 variants but do not resolve the nature of the apparent bias for transmission of R5 over X4 variants.
Multiple models could account for our observations of primary infection diversity. Transient periods of hyperinfectiousness in the donor or hypersusceptibility in the recipient, the sex of the recipient, or the transmission of virus through infected cells could all explain the presence of multiple V1/V2 variants. If the infection inoculum represents cell-free virus, then, using the Poisson distribution, the probability of infection would have to approach 1 to give the pattern of 1 versus 2 versus 3 variants seen in this cohort. This probability of infection is orders of magnitude greater than current estimates (5, 18, 22, 58).
Long et al. (33) have proposed a link between the sex of the newly infected person and the number of variants transmitted. They examined the diversity in the V1/V2 and V3 regions in a primary infection cohort of heterosexually infected men and women in Kenya. Transmission of multiple variants was evident in 63% of the females, whereas no males were found to harbor multiple variants. They concluded that the sex of the recipient is the primary determinant of the number of variants transmitted. However, in our study we did not observe exclusive transmission of single variants in homosexual men.
We found that 12 of 18 (67%) men harbored multiple variants after homosexual transmission events. In contrast, none of five men infected through heterosexual contact harbored multiple variants, while one of two women had multiple V1/V2 variants, a finding consistent with the study by Long et al. (33). These data apparently contradict the model that the gender of the recipient is the principal determinant of initial viral diversity in HIV-1 infection. An alternative hypothesis may explain both our own observations and those of Long et al. (33). First, it may be the characteristics of the exposed mucosal surface that determine the initial diversity of the infecting virus population. Both rectal and vaginal mucosal surfaces contain no keratinized epithelium and hence provide an infecting inoculum with a different environment than does the penile skin. We propose that the involvement of the recipient's mucosal surface greatly increases the likelihood that infected cells participate in transmission. In this model, women would typically transmit single variants to men, while men would typically transmit multiple variants to women or men.
Transmission solely by cell-free virus in the context of low transmission probabilities should result in the presence of single variants much more frequently than multiple variants. This is inconsistent with our observation that homosexual transmission more frequently results in the transmission of multiple variants. Transmission via infected cells in semen could account for both the transmission of multiple variants and the observation that transmission is a low-frequency event. Zhu et al. (68) have previously shown that seminal cells can be the source of infecting virus. Jung et al. (24) have shown that HIV-1-infected spleen cells each harbor between 1 to 8 proviruses, with an average 3.2 proviruses per cell. These numbers are consistent with our data for the number of variants transmitted from a seminal inoculum. There was a range of one to five variants, with an average of two variants per subject. In this model, the rare transmission event from semen can involve a single infected cell that contains multiple viral variants initiating the new infection.
The presence of virus in the central nervous system and the seminal compartment has been well documented during chronic and end-stage infection. Our data indicate that these compartments are quickly populated by the infecting strain, at least as measured by V1/V2-HTA and V3-HTA. During acute primary infection, predominantly the same viral variants were detected in the periphery and the CSF and/or SP, suggesting that these variants rapidly and efficiently enter these compartments. Evidence for some early compartmentalization was detected in two subjects: in the SP of Z03 and in the CSF of Z10. These differences could be the result of different rates of penetration or expansion between the compartments. The presence of identical R5-like variants in the periphery and CSF supports the macrophage-tropic requirement for central nervous system infection described by Gorry et al. (21). Viral variants apparently become compartmentalized later during the course of infection.
There is some interest in considering primary infection isolates as vaccine candidates, especially given the possibility that they may have distinctive properties (13). We have previously used a sequence survey approach of subtype C HIV-1, primary infection subjects to identify vaccine sequences near the consensus sequence (61). Given that CTL epitopes tend to be more conserved within a subtype (51), it may be useful to develop subtype-specific vaccines. To this end, we have identified isolate Z04 as having a sequence profile close to that of the subtype B consensus (Fig. 9). Although this does not represent a consensus sequence, it does allow the use of viral genes derived from a replication competent isolate that will not represent an outlier among the currently circulating subtype B viral sequences.
In conclusion, we have shown that the V1/V2-HTA represents a useful tool to assess the viral diversity present during primary infection and that multiple variants are frequently present. We hypothesize that the sex of the transmitter and the nature of the exposed mucosal surface rather than the sex of the recipient influences the number of variants transmitted, with multiply infected cells in semen playing an important role in transmission. Compartmentalization between the periphery and the seminal or central nervous system compartment is not evident during primary infection and therefore must develop as the infection progresses. The potential role of a multiply infected cell in transmission events may have important implications for the design of a vaccine and/or a microbicide.
Acknowledgments
We thank Myron Cohen for helpful discussions.
This study was supported by NIH grants R01-AI44667, K23-AI01781, GCRC-RR00046, K24AI01608, and R01-DK49381 and the UNC Center for AIDS Research (P30-AI50410). K.R. was supported in part by NIH Training Grant T32-AI007419.
REFERENCES
- 1.Amedee, A. M., N. Lacour, J. L. Gierman, L. N. Martin, J. E. Clements, R. Bohm, Jr., R. M. Harrison, and M. Murphey-Corb. 1995. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J. Virol. 69:7982-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. Luo, N. N′Gandu, H. Sheppard, B. H. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 4.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, H., P. K. Sen, R. W. Helms, P. L. Vernazza, S. A. Fiscus, J. J. Eron, B. K. Patterson, R. W. Coombs, J. N. Krieger, and M. S. Cohen. 2001. Viral burden in genital secretions determines male-to-female sexual transmission of HIV-1: a probabilistic empiric model. AIDS 15:621-627. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Daar, E. S., S. Little, J. Pitt, J. Santangelo, P. Ho, N. Harawa, P. Kerndt, J. V. Glorgi, J. Bai, P. Gaut, D. D. Richman, S. Mandel, S. Nichols, et al. 2001. Diagnosis of primary HIV-1 infection. Ann. Intern. Med. 134:25-29. [DOI] [PubMed] [Google Scholar]
- 8.Daar, E. S., T. Moudgil, R. D. Meyer, and D. D. Ho. 1991. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. N. Engl. J. Med. 324:961-964. [DOI] [PubMed] [Google Scholar]
- 9.Delwart, E. L., and C. J. Gordon. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12:348-354. [DOI] [PubMed] [Google Scholar]
- 10.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart, E. L., H. W. Sheppard, B. D. Walker, J. Goudsmit, and J. I. Mullins. 1994. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 68:6672-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 13.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 14.Dickover, R. E., E. M. Garratty, S. Plaeger, and Y. J. Bryson. 2001. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durali, D., J. Morvan, F. Letourneur, D. Schmitt, N. Guegan, M. Dalod, S. Saragosti, D. Sicard, J. P. Levy, and E. Gomard. 1998. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J. Virol. 72:3547-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enose, Y., M. Okada, T. Sata, W. Ma, T. Igarashi, K. Ibuki, E. Ido, and M. Hayami. 1997. Restriction of viral population by intravaginal infection of simian immunodeficiency viruses in macaque monkeys. Arch. Virol. 142:37-51. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari, G., W. Humphrey, M. J. McElrath, J. L. Excler, A. M. Duliege, M. L. Clements, L. C. Corey, D. P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiebig, E. W., D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871-1879. [DOI] [PubMed] [Google Scholar]
- 20.Furuta, Y., T. Bergstrom, G. Norkrans, and P. Horal. 1994. HIV type 1 V3 sequence diversity in contact-traced Swedish couples at the time of sexual transmission. AIDS Res. Hum. Retrovir. 10:1187-1189. [DOI] [PubMed] [Google Scholar]
- 21.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, R. H., M. J. Wawer, R. Brookmeyer, N. K. Sewankambo, D. Serwadda, F. Wabwire-Mangen, T. Lutalo, X. Li, T. vanCott, and T. C. Quinn. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149-1153. [DOI] [PubMed] [Google Scholar]
- 23.Haynes, B. F., G. Pantaleo, and A. S. Fauci. 1996. Toward an understanding of the correlates of protective immunity to HIV infection. Science 271:324-328. [DOI] [PubMed] [Google Scholar]
- 24.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 25.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63-76. [DOI] [PubMed] [Google Scholar]
- 26.Kitrinos, K. M., N. G. Hoffman, J. A. Nelson, and R. Swanstrom. 2003. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 77:6811-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kliks, S., C. H. Contag, H. Corliss, G. Learn, A. Rodrigo, D. Wara, J. I. Mullins, and J. A. Levy. 2000. Genetic analysis of viral variants selected in transmission of human immunodeficiency viruses to newborns. AIDS Res. Hum. Retrovir. 16:1223-1233. [DOI] [PubMed] [Google Scholar]
- 28.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamers, S. L., J. W. Sleasman, J. X. She, K. A. Barrie, S. M. Pomeroy, D. J. Barrett, and M. M. Goodenow. 1993. Independent variation and positive selection in env V1 and V2 domains within maternal-infant strains of human immunodeficiency virus type 1 in vivo. J. Virol. 67:3951-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindback, S., A. C. Karlsson, J. Mittler, A. Blaxhult, M. Carlsson, G. Briheim, A. Sonnerborg, H. Gaines, et al. 2000. Viral dynamics in primary HIV-1 infection. AIDS 14:2283-2291. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 34.Ludlam, C. A., J. Tucker, C. M. Steel, R. S. Tedder, R. Cheingsong-Popov, R. A. Weiss, D. B. McClelland, I. Philp, and R. J. Prescott. 1985. Human T-lymphotropic virus type III (HTLV-III) infection in seronegative haemophiliacs after transfusion of factor VIII. Lancet ii:233-236. [DOI] [PubMed] [Google Scholar]
- 35.Lynch, J. A., M. deSouza, M. D. Robb, L. Markowitz, S. Nitayaphan, C. V. Sapan, D. L. Mann, D. L. Birx, and J. H. Cox. 1998. Cross-clade cytotoxic T-cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J. Infect. Dis. 178:1040-1046. [DOI] [PubMed] [Google Scholar]
- 36.Machado, D. M., E. L. Delwart, R. S. Diaz, C. F. de Oliveira, K. Alves, B. D. Rawal, M. Sullivan, M. Gwinn, K. A. Clark, and M. P. Busch. 2002. Use of the sensitive/less-sensitive (detuned) EIA strategy for targeting genetic analysis of HIV-1 to recently infected blood donors. AIDS 16:113-119. [DOI] [PubMed] [Google Scholar]
- 37.McNearney, T., Z. Hornickova, B. Kloster, A. Birdwell, G. A. Storch, S. H. Polmar, M. Arens, and L. Ratner. 1993. Evolution of sequence divergence among human immunodeficiency virus type 1 isolates derived from a blood donor and a recipient. Pediatr. Res. 33:36-42. [DOI] [PubMed] [Google Scholar]
- 38.Moore, J. P., Y. Cao, D. D. Ho, and R. A. Koup. 1994. Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J. Virol. 68:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder-Kampinga, G. A., C. Kuiken, J. Dekker, H. J. Scherpbier, K. Boer, and J. Goudsmit. 1993. Genomic human immunodeficiency virus type 1 RNA variation in mother and child following intra-uterine virus transmission. J. Gen. Virol. 74(Pt. 9):1747-1756. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, J. A. E., F. Baribaud, T. Edwards, and R. Swanstrom. 2000. Patterns of changes in human immunodeficiency virus type 1 V3 sequence populations late in infection. J. Virol. 74:8494-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, J. A. E., S. A. Fiscus, and R. Swanstrom. 1997. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J. Virol. 71:8750-8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak, P., A. C. Karlsson, L. Naver, A. B. Bohlin, A. Piasek, and A. Sonnerborg. 2002. The selection and evolution of viral quasispecies in HIV-1 infected children. HIV Med. 3:1-11. [DOI] [PubMed] [Google Scholar]
- 43.Ou, C. Y., C. A. Ciesielski, G. Myers, C. I. Bandea, C. C. Luo, B. T. Korber, J. I. Mullins, G. Schochetman, R. L. Berkelman, A. N. Economou, J. J. Witte, L. J. Furman, G. A. Satten, K. A. MacInnes, J. W. Curran, H. W. Jaffe, L. I. Group, and E. I. Group. 1992. Molecular epidemiology of HIV transmission in a dental practice. Science 256:1165-1171. [DOI] [PubMed] [Google Scholar]
- 44.Pang, S., Y. Shlesinger, E. S. Daar, T. Moudgil, D. D. Ho, and I. S. Chen. 1992. Rapid generation of sequence variation during primary HIV-1 infection. AIDS 6:453-460. [DOI] [PubMed] [Google Scholar]
- 45.Pilcher, C. D., J. J. Eron, Jr., P. L. Vemazza, M. Battegay, T. Harr, S. Yerly, S. Vom, and L. Perrin. 2001. Sexual transmission during the incubation period of primary HIV infection. JAMA 286:1713-1714. [DOI] [PubMed] [Google Scholar]
- 46.Ping, L. H., M. S. Cohen, I. Hoffman, P. Vernazza, F. Seillier-Moiseiwitsch, H. Chakraborty, P. Kazembe, D. Zimba, M. Maida, S. A. Fiscus, J. J. Eron, R. Swanstrom, and J. A. Nelson. 2000. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J. Virol. 74:8946-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renjifo, B., M. Chung, P. Gilbert, D. Mwakagile, G. Msamanga, W. Fawzi, and M. Essex. 2003. In-utero transmission of quasispecies among human immunodeficiency virus type 1 genotypes. Virology 307:278-282. [DOI] [PubMed] [Google Scholar]
- 49.Resch, W., N. Parkin, E. L. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T-cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybarczyk, B. J., D. Montefiori, P. R. Johnson, A. West, R. E. Johnston, and R. Swanstrom. 2004. Correlation between env V1/V2 region diversification and neutralizing antibodies during primary infection by simian immunodeficiency virus sm in rhesus macaques. J. Virol. 78:3561-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scarlatti, G., T. Leitner, E. Halapi, J. Wahlberg, P. Marchisio, M. A. Clerici-Schoeller, H. Wigzell, E. M. Fenyo, J. Albert, M. Uhlen, and P. Rossi. 1993. Comparison of variable region 3 sequences of human immunodeficiency virus type 1 from infected children with the RNA and DNA sequences of the virus populations of their mothers. Proc. Natl. Acad. Sci. USA 90:1721-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schacker, T., A. C. Collier, J. Hughes, T. Shea, and L. Corey. 1996. Clinical and epidemiologic features of primary HIV infection. Ann. Intern. Med. 125:257-264. [DOI] [PubMed] [Google Scholar]
- 55.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodora, D. L., F. Lee, P. J. Dailey, and P. A. Marx. 1998. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retrovir. 14:171-181. [DOI] [PubMed] [Google Scholar]
- 57.Sutthent, R., S. Foongladda, S. Chearskul, N. Wanprapa, S. Likanonskul, U. Kositanont, S. Riengrojpitak, S. Sahaphong, and C. Wasi. 1998. V3 sequence diversity of HIV-1 subtype E in infected mothers and their infants. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:323-331. [DOI] [PubMed] [Google Scholar]
- 58.Trask, S. A., C. A. Derdeyn, U. Fideli, Y. Chen, S. Meleth, F. Kasolo, R. Musonda, E. Hunter, F. Gao, S. Allen, and B. H. Hahn. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J. Virol. 76:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verhofstede, C., E. Demecheleer, N. De Cabooter, P. Gaillard, F. Mwanyumba, P. Claeys, V. Chohan, K. Mandaliya, M. Temmerman, and J. Plum. 2003. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J. Virol. 77:3050-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson, C., L. Morris, M. F. Maughan, L. H. Ping, S. A. Dryga, R. Thomas, E. A. Reap, T. Cilliers, J. van Harmelen, A. Pascual, G. Ramjee, G. Gray, R. Johnston, S. A. Karim, and R. Swanstrom. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retrovir. 19:133-144. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, S. E., S. L. Pedersen, J. C. Kunich, V. L. Wilkins, D. L. Mann, G. P. Mazzara, J. Tartaglia, C. L. Celum, and H. W. Sheppard. 1998. Cross-clade envelope glycoprotein 160-specific CD8+ cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res. Hum. Retrovir. 14:925-937. [DOI] [PubMed] [Google Scholar]
- 63.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103-110. [DOI] [PubMed] [Google Scholar]
- 64.Wolfs, T. F., G. Zwart, M. Bakker, M. Valk, C. L. Kuiken, and J. Goudsmit. 1991. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology 185:195-205. [DOI] [PubMed] [Google Scholar]
- 65.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]