Abstract
XRCC1 is a molecular scaffold protein that assembles multi-protein complexes involved in DNA single-strand break repair1,2. Here, we show that biallelic mutations in human XRCC1 are associated with ocular motor apraxia, axonal neuropathy, and progressive cerebellar ataxia. XRCC1-mutant patient cells exhibit not only reduced rates of single-strand break repair but also elevated levels of protein ADP-ribosylation; a phenotype recapitulated in a related syndrome caused by mutations in the XRCC1 partner protein PNKP3-5 and implicating hyperactivation of poly (ADP-ribose) polymerase/s as a cause of cerebellar ataxia. Indeed, remarkably, genetic deletion of Parp1 rescued normal cerebellar ADP-ribose levels and reduced the loss of cerebellar neurons and ataxia in Xrcc1-defective mice, identifying a molecular mechanism by which endogenous single-strand breaks trigger neuropathology. Collectively, these data establish the importance of XRCC1 protein complexes for normal neurological function and identify PARP1 as a therapeutic target in DNA strand break repair-defective disease.
A forty-seven year old woman of East Indian descent and non-consanguineous parents was diagnosed at age forty-one with cerebellar atrophy, gait and limb ataxia, ocular motor apraxia, and peripheral neuropathy (Fig. 1a, b). Prenatal and early developmental history was completely normal and difficulties with balance and gait were first noticed at twenty-eight years, but this was not fully investigated until age forty. Magnetic resonance imaging (MRI) and cerebellar examination revealed progressive cerebellar atrophy (Fig. 1c) and multiple ataxic abnormalities including dysmetria, dysdiadochokinesis, and dysarthria, and nerve conduction studies revealed chronic length-dependent sensory-motor predominantly axonal peripheral neuropathy (Fig. 1b, Supplementary Information). After ruling out more than ten known spinocerebellar at axias by genetic and metabolic screening (Supplementary Information) exome sequencing of the proband identified compound heterozygous mutations in XRCC1 (NM_006297). The mutations were confirmed by Sanger sequencing as c.1293G>C (p.K431N) and c.1393C>T (p.Q465*) and were present in trans, with the unaffected sibling of the proband heterozygous for c.1293G>C (Fig. 1d). c.1393C>T has not previously been described in the population, whereas c.1293G>C was previously detected in heterozygous state in four individuals of South Asian descent (ExAC Consortium, Cambridge, MA). After ruling out other rare gene variants on the basis of their presence in the unaffected sibling, presence in homozygous state in unaffected in-house controls and/or ExAC, and lack of functional and/or disease relevance no other candidate causative mutations remained.
Figure 1. XRCC1 mutations are associated with cerebellar ataxia, ocular motor apraxia and axonal neuropathy.
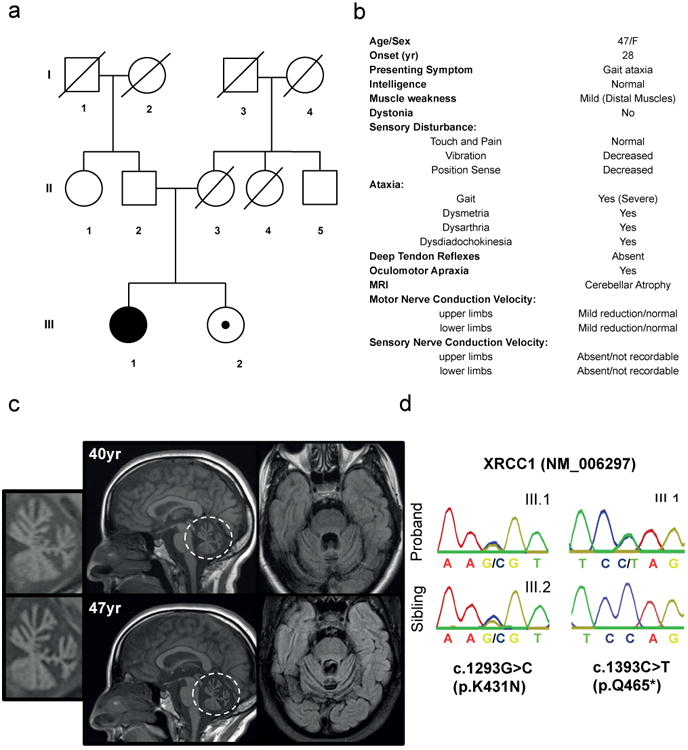
a, Pedigree of the proband (III.1; black circle) and unaffected sibling (III.2; circle/black dot). b, Proband clinical features (see Supplementary Information for full description). c, Proband MRI at 40yr and 47yr. Sagittal T1 weighted images (middle) demonstrating vermian atrophy and Axial FLAIR images (right) demonstrating atrophy of the cortex of the vermis and cerebellar hemispheres. The cerebellum is circled and insets (left) are magnifications highlighting the cerebellar atrophy. d, Sanger sequencing confirming the mutations c.1293G>C (p.K431N) and c.1393C>T (p.Q465*) in the proband (III.1) and c.1293G>C (p.K431N) in the unaffected sibling (III.2).
The c.1393C>T mutation is located within exon 12 and creates a premature stop codon at amino acid 465, most likely triggering nonsense mediated mRNA decay. The c.1293G>C mutation is located at the end of exon 11 and is also part of the donor splice site for intron 11, most likely affecting splicing and inducing premature stop codons/nonsense-mediated decay and/or encoding XRCC1 with the missense mutation, K431N. Consistent with this, reduced total levels of XRCC1 mRNA were observed in patient cells, as well as aberrant splicing of XRCC1 transcripts if nonsense-mediated decay was inhibited with cycloheximide (Extended Data Fig. 1).
To establish the pathogenic impact of the biallelic XRCC1 mutations we examined patient fibroblasts and lymphoblastoid cells (LCLs) for levels of XRCC1 protein by indirect immunofluorescence and Western blotting. The level of XRCC1 in the patient primary fibroblasts was greatly reduced when compared to wild type primary human fibroblasts (1BR) by indirect immunofluorescence and was not measurably higher than in human RPE-1 cells in which XRCC1 was deleted by CRISPR-Cas9 (Fig. 2a). However, Western blotting suggested that patient fibroblasts and LCLs both retained a small amount (∼5%) of residual XRCC1 (Fig. 2b, Extended Data Fig. 2a). Indeed, this was confirmed using XRCC1 siRNA, which reduced the anti-XRCC1 signal on Western blots of patient fibroblasts even further (Fig. 2b, bottom). Levels of DNA ligase IIIα (Lig3α) were also greatly reduced (by >80%) in patient cells, consistent with the established impact of XRCC1 on the cellular stability of this partner protein (Fig. 2b, top, Extended Data Fig. 2b)6,7. Since germ-line deletion of Xrcc1 in mouse is embryonic lethal8 we suggest that the small amount of XRCC1 remaining in the patient was important for embryonic viability. Consistent with this idea, embryonic viability in mice is supported by a little as ∼10% of normal Xrcc1 levels9.
Figure 2. Patient XRCC1 mutations reduce XRCC1 levels and recruitment into chromatin.
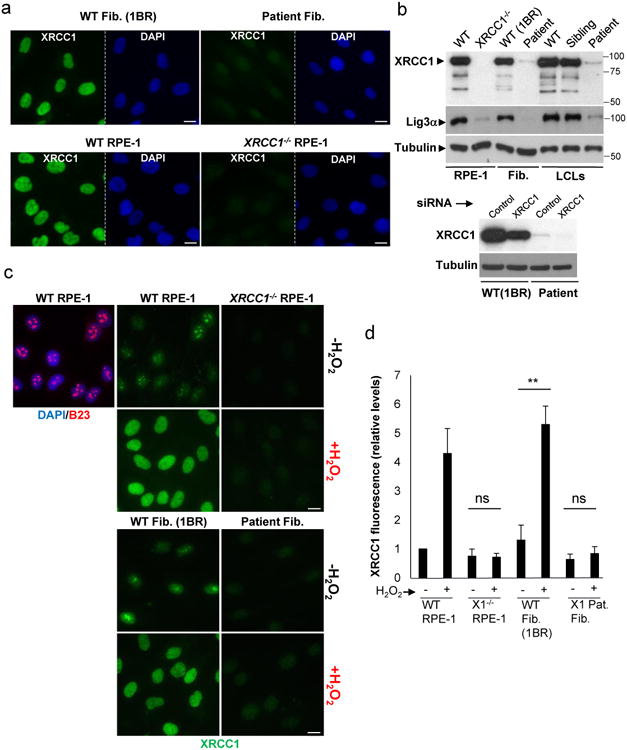
a, XRCC1 levels measured by immunofluorescence in wild type (“WT”) 1BR fibroblasts, WT RPE-1 cells, XRCC1 patient fibroblasts, and XRCC1-/- RPE-1 cells. b, Top, XRCC1 & Lig3α levels measured in the above cells by Western blotting and additionally in WT, XRCC1-patient, and sibling LCLs. The source data are included in Supplementary Figure 1. Bottom, WT or patient fibroblasts were transfected with non-targeting or XRCC1 siRNA and immunoblotted as above. c, XRCC1 chromatin binding measured by immunofluorescence in the indicated cells before and 10 min after treatment with 1 mM H2O2. d, Quantitation of XRCC1 in chromatin (excluding nucleoli) from >1000 cells per sample using ScanR software. Data are the mean (+/-1SD) of three independent experiments. Statistical analysis (two-tailed t-test) is indicated (**p<0.01; “ns”, not significant). Representative ScanR images are in Extended Data Fig. 3a. Scale bars; 10 μm.
To determine whether the residual XRCC1 in patient cells can engage in single-strand break repair (SSBR) we quantified the extent to which it bound oxidized chromatin. XRCC1 was primarily detected in nucleoli in undamaged wild-type RPE-1 cells and normal primary 1BR fibroblasts, following the extraction of soluble proteins with detergent, but was rapidly recruited into global nuclear chromatin following treatment with H2O2 (Fig. 2c, d); a physiological source of oxidative single-strand breaks (SSBs)10. In contrast, little or no XRCC1 recruitment into chromatin was detected in XRCC1-patient fibroblasts by high-resolution or high-content imaging (Fig. 2c, d, Extended Data Fig. 3a). Similar results were observed following treatment with camptothecin (CPT), a topoisomerase poison that induces SSBs triggered by abortive topoisomerase I activity (Extended Data Fig. 3b).
Importantly, the defect in XRCC1 recruitment in patient fibroblasts was accompanied by a delay in the kinetics of DNA single-strand break repair (SSBR) following H2O2 treatment (Fig. 3a). This phenotype was recapitulated in XRCC1-/- RPE-1 cells and is consistent with the established molecular role of XRCC12. In contrast, we failed to detect a major difference in double-strand break repair in XRCC1-patient fibroblasts, as measured by γH2AX immunostaining following ionising radiation (Fig. 3b, Extended Data Fig. 4). In agreement with the defect in SSBR, XRCC1-patient LCLs exhibited a four-fold increase in sister chromatid exchange; a hyper-recombination phenotype resulting from elevated homologous recombination triggered by unrepaired SSBs in S/G2 phase of the cell cycle (Fig. 3c)11.
Figure 3. Patient XRCC1 mutations reduce single-strand break repair.
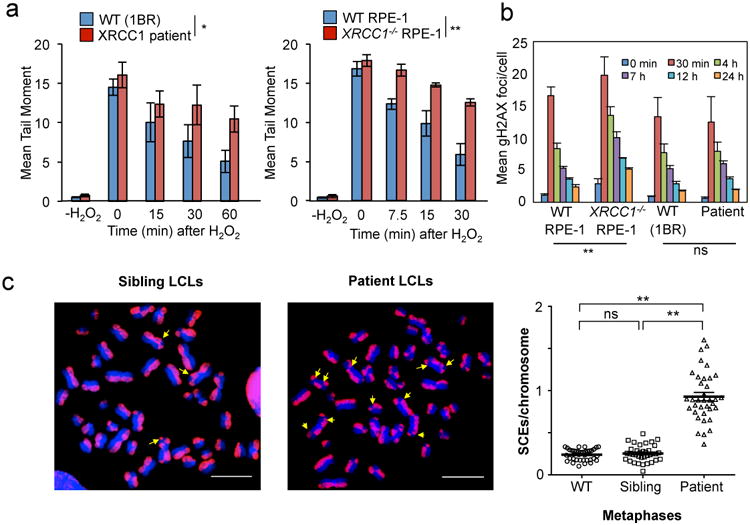
a, DNA strand breaks quantified in the indicated fibroblasts (left) or RPE-1 cells (right) by alkaline comet assays before and at the indicated times after H2O2 treatment. Data are the mean (+/-SEM) comet tail moments (an arbitrary unit-measure of DNA strands) of three independent experiments. Statistical analyses (two-way ANOVA) are indicated (*p<0.05; **p<0.01). b, DSBs quantified as γH2AX foci in the indicated cell lines before and at the indicated times after ionising radiation (2 Gy). Data are the mean γH2AX foci per cell (+/-1SD) from three independent experiments (∼1000 cells/sample/experiment). Statistical analysis as above. Representative images are in Extended Data Fig. 4. c, SCE frequencies per chromosome (mean +/-1SEM) quantified in control and XRCC1 patient LCLs (36 metaphases per genotype). Representative metaphases (arrows, SCEs) are shown, left. Statistical analysis; two-tailed t-test (**p<0.01; ns, not significant). Scale bars; 10 μm.
XRCC1 is a scaffold protein that assembles SSBR multi-protein complexes and, importantly, components of these complexes are mutated in the cerebellar ataxias spinocerebellar ataxia with axonal neuropathy-1 (SCAN1; mutated in TDP1)12,13, ataxia oculomotor apraxia-1 (AOA1; mutated in Aprataxin)14, and ataxia oculomotor apraxia-4 (AOA4; mutated in PNKP)3,4. Indeed, it is striking that the pathology of the XRCC1 patient combines features of each of these diseases, consistent with the role played by XRCC1 in coordinating their activity. The discovery that XRCC1 is itself mutated in cerebellar ataxia is thus significant because it demonstrates the importance of these complexes in preventing neurodegeneration in humans.
To investigate the mechanism(s) by which unrepaired SSBs trigger neuropathology we considered the possibility that persistent unrepaired SSBs might result in prolonged activity of the SSB sensor protein, PARP1. This hypothesis was prompted by the observation that excessive synthesis of poly (ADP-ribose) and/or excessive depletion of NAD+ by PARP1 is neurotoxic and associated with ischemia reperfusion injury15,16. Consistent with this idea, whilst ADP-ribose was rapidly detected in both wild type and patient fibroblasts following H2O2 treatment it persisted at a higher level in the latter cells during subsequent incubation in drug-free medium (Fig. 4a, left). This was also evident in XRCC1-/- human RPE-1 cells, confirming that this phenotype was induced by loss of XRCC1. Elevated ADP-ribose levels were also detected in XRCC1 patient fibroblasts and XRCC1-/- RPE-1 cells following treatment with camptothecin (CPT) (Fig. 4a, right). Indeed, the difference in ADP-ribose levels between wild type and XRCC1-mutant cells was even greater following CPT than following H2O2 treatment. The type of SSB induced by CPT has been linked previously with SSBR-defective neurodegenerative disease and is a possible source of pathogenic SSBs in SSBR-defective individuals13,17. Consistent with this idea, CPT-induced ADP-ribose levels were also elevated in fibroblasts from a patient with ataxia oculomotor apraxia-4 (AOA4); the cerebellar ataxia resulting from mutation of the XRCC1 protein partner, PNKP (Fig. 4b, Extended Data Fig. 5). Importantly, the elevated ADP-ribose observed in CPT-treated XRCC1-/- RPE-1 cells, XRCC1- patient cells, and PNKP-patient cells was entirely dependent on PARP1 activity (Fig. 4c, Extended Data Figs 6, 7). Moreover, this phenotype was rescued by the introduction of wild-type recombinant XRCC1 into XRCC1 patient fibroblasts by electroporation (Fig. 4d, Extended Data Fig. 8).
Figure 4. XRCC1 mutation elevates ADP-ribosylation in cells and cerebellum.
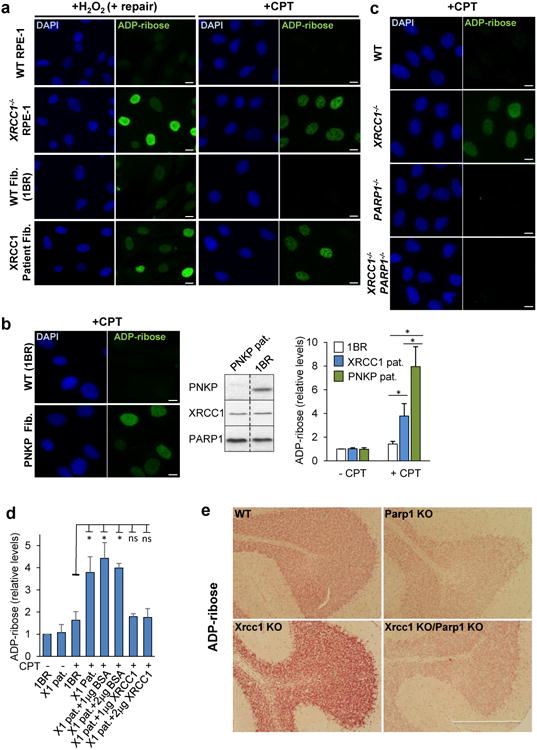
a, ADP-ribosylation in wild type (WT) 1BR fibroblasts, WT RPE-1 cells, XRCC1-patient fibroblasts, and XRCC1-/- RPE-1 cells following H2O2 or CPT treatment. b, Left, ADP-ribosylation levels in 1BR or PNKP patient fibroblasts following CPT treatment. Middle, PNKP, XRCC1, and PARP1 levels in 1BR and PNKP patient fibroblasts. The source data are included in Supplementary Figure 1. Right, ADP-ribose levels quantified by ScanR imaging in 1BR, XRCC1-mutant patient, and PNKP patient fibroblasts before and after CPT treatment. Data are mean (+/-1SD) ADP-ribose levels (relative to untreated 1BR cells) from three independent experiments (>1000 cells/sample). Statistically significant differences (two-tailed t-test) are indicated (*p<0.05). c, ADP-ribosylation levels in the indicated RPE-1 cells following CPT treatment. d, ADP-ribosylation levels quantified by ScanR imaging before and after CPT treatment in 1BR and XRCC1 patient fibroblasts (“X1 pat.”) with or without transfection with 1 μg or 2 μg of BSA or recombinant human XRCC1 protein. Statistical analyses as above. Representative ScanR images are in Extended Data Figure 8. e, ADP-ribosylation levels measured by immunohistochemistry in cerebellar sections from WT mice or mice deleted (“KO”) of Xrcc1 and/or Parp1. Scale bars in IF images are 10 μm and in histology images are 1 mm.
Next, to examine directly whether hyperactive PARP1 triggers cerebellar ataxia in the absence of efficient SSBR we employed a mouse model in which Xrcc1 was conditionally deleted in brain (Xrcc1Nes-Cre)7. Xrcc1Nes-Cre mice exhibit pronounced cerebellar histopathology including increased apoptosis of cerebellar granule neurons, greatly reduced numbers of cerebellar interneurons and decreased electrophysiological spike activity in Purkinje cells (Ref. 4 & Extended Data Fig. 9). Moreover, consistent with the pathology of the XRCC1-mutant patient and other SSBR-defective patients, Xrcc1Nes-Cre mice exhibit cerebellar ataxia7. Strikingly, we detected elevated levels of ADP-ribose in the cerebellum of Xrcc1Nes-Cre mice suggesting that the loss of Xrcc1 can trigger Parp1 hyperactivation in brain even at endogenous levels of SSBs (Fig. 4e). Indeed, the deletion of Parp1 ablated both the elevated level of ADP-ribose and the characteristic loss of cerebellar interneurons in Xrcc1Nes-Cre mice, thereby increasing neuronal density in the molecular layer ∼4-fold to wild type levels (Fig. 5a, b). This did not reflect an impact of Parp1 deletion on the rate of SSBR because the latter was similarly slow in XRCC1-/- and XRCC1-/-/PARP1-/- RPE-1 cells (Extended Data Fig. 10). Rather, these data demonstrate that in the absence of Xrcc1-dependent SSB repair Parp1 is hyperactivated, resulting in the loss and/or dysfunction of cerebellar neurones. Finally, to examine whether Parp1 deletion also rescued the cerebellar ataxia observed in Xrcc1Nes-Cre mice we compared Xrcc1Nes-Cre and Xrcc1Nes-Cre/Parp1-/- mice for their performance on an accelerating rotarod. Indeed, remarkably, whereas Xrcc1Nes-Cre mice were profoundly ataxic and unable to remain on the rotarod for more than a few seconds the additional deletion of Parp1 improved the rotarod performance of Xrcc1Nes-Cre mice >30-fold, increasing their mean retention time to ∼30 sec (Fig. 5c).
Figure 5. Parp1 deletion restores normal interneuron density and reduces cerebellar ataxia in XRCC1Nes-Cre mice.
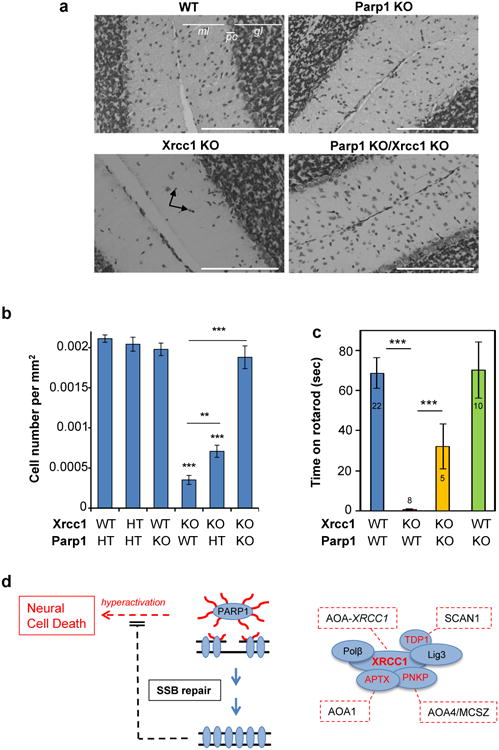
a, Representative cerebellar sections from wild type mice or mice deleted (“KO”) of Xrcc1 and/or Parp1 stained with Nissl to detect neurones. Scale bars are 200 μm. gl; granule layer; ml, molecular layer; pc, Purkinje cells. Black arrows indicate interneurons. b, Interneuron density in the molecular layer of cerebella of the indicated genotype. Data are the mean (+/-SEM) Nissl-positive cells per μm2 of 5-10 mice per genotype. T-test comparisons are indicated (**p<0.01, ***p<0.001). c, Motor coordination in mice of the indicated genotype measured on a rotarod (mouse numbers analysed indicated in/above the bars). Statistical tests as above. d, Model for PARP1 hyperactivation and neural death triggered by unrepaired SSBs. PARP1 activation at SSBs triggers auto- and trans- protein ADP-ribosylation (red wavy lines). XRCC1 binds poly(ADP-ribose) and assembles SSBR protein complexes. Mutated XRCC1 or partner proteins results in delayed SSBR and PARP1 ‘hyperactivation’, resulting in cyotoxic levels of poly(ADP-ribose) and/or NAD+ depletion. SSBR proteins and their associated cerebellar ataxias are highlighted/boxed in red. SCAN1; spinocerebellar ataxia with axonal neuropathy-1 (TDP1-mutated). AOA1; ataxia oculomotor apraxia-1 (APTX-mutated). AOA4; ataxia oculomotor apraxia-4 (PNKP-mutated). AOA-XRCC1; ataxia oculomotor apraxia-XRCC1 mutated18.
Collectively, these data identify elevated ADP-ribose levels as a biomarker of PARP1 hyperactivity and as a cause of cerebellar ataxia induced by unrepaired SSBs (Fig. 5d). This scenario might also extend to other more common neurodegenerative diseases because elevated levels of oxidative stress and DNA strand breakage are also implicated in diseases such as Alzheimer's disease, Huntington's disease, and Parkinson's disease17-19. Intriguingly, patients with mutations in the XRCC1 partner protein PNKP exhibit not only ataxia but also microcephaly and seizures, and we have more recently identified a second patient with rare putative pathogenic mutations (<0.01% allele frequency and not present in ExAC in homozygous state) in XRCC1 that exhibits this same combination of phenotypes12,13. Consistent with this, Xrcc1Nes-Cre mice also present with seizures, raising the prospect that PARP1 hyperactivation may induce not only ataxia, but also more severe pathologies.
Finally, these data identify PARP1 as a possible drug-target for the treatment of cerebellar ataxias associated with unrepaired SSBs. Inhibition of PARP1 with currently available chemical inhibitors may not be useful in this context, however, because these inhibitors “trap” PARP1 on DNA20 and do not mimic PARP1 genetic deletion (Extended Data Fig. 10). However, the development of selective inhibitors of PARP1 that prevent DNA binding by this enzyme may have significant therapeutic potential.
Data analysis and statistics
The number of experimental repeats and statistical tests (conducted in Excel or GraphPad) are indicated in the relevant figure legends. The means of population averages from multiple independent experiments (+/-SD or SEM) are indicated. Where appropriate, power calculations were conducted using online statistical resources. Samples were not blinded but the collection and analysis of microscopic data was automated and free of user bias. No animals/samples were omitted from data points/data analyses.
Methods
Whole-exome sequencing
Whole-exome library preparation, exon capture and sequencing were performed at the Genome Québec Innovation Center (Montréal, QC, Canada) as previously described19. Genomic DNA was captured using the SureSelect Human 50Mb All Exon kit v5 (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed on an Illumina HiSeq2000 (Illumina, San Diego, CA, USA) with paired-end 100-bp reads. A mean coverage of 137× was obtained and 97% of the bases were covered at more than 10×. Read alignment, variant calling, and annotation were done with a pipeline based on BWA, SAM tools, Annovar, and custom annotation scripts. All sequences were aligned to Human genome Hg19. We excluded variants with minor allele frequency greater than 5% in either the 1000 genomes project (http://browser.1000genomes.org/index.html) or the 6500 NHLBI EVS (http://evs.gs.washington.edu/EVS), and seen in more than 30 samples from our in-house database (containing approximately 2000 samples). The WES data was further filtered to keep protein-damaging variants (nonsense, missense, frameshift, indel, and splice variants).
Antibodies and chemicals
The antibodies employed in this study were anti-XRCC1 rabbit polyclonal (Millipore;ABC738), anti-Lig3α (TL25) rabbit polyclonal20, anti-PNKP (SK3195) rabbit polyclonal21, rabbit Fc-fused Anti-pan-ADP-ribose binding reagent (Millipore; MABE1016), anti-poly (ADP-ribose) rabbit polyclonal (Trevigen; 4336), anti-α-tubulin rat polyclonal (Abcam; ab6160), anti-BrdUrat monoclonal, crossreacting with CldU, (BioRad; OBT0030G), anti-nucleophosmin (B23) mouse monoclonal (Invitrogen; 325200), anti-PARP1 mouse monoclonal (Serotec; MCA1522G) and anti-γH2AX mouse monoclonal (Millipore; 05-636). The secondary antibodies employed for Western blotting were HRP-conjugated goat anti-rabbit (Bio-Rad; 170-6515), goat anti-mouse (Bio-Rad; 170-6516) and rabbit anti-rat (Abcam; ab6734) and for indirect immunofluorescence were goat anti-mouse or anti-rabbit Alexa 488 (Invitrogen; A11001 and A31628), goat anti-rabbit Alexa 568 (Invitrogen; A11036), donkey anti-mouse Alexa 647 (Invitrogen; A39571) and goat anti-rat Alexa 568 (Invitrogen; A11077). Camptothecin (CPT) was purchased from Sigma and Hydrogen Peroxide (H2O2) was obtained from Fischer Scientific. Veliparib (ABT-888) was purchased from Selleckchem and KU0058948 hydrochloride from Axon.
Cell lines
All cell lines were tested for absence of mycoplasma. Wild type human hTERT RPE-1 cells (ATCC; CRL4000)(denoted “RPE-1” for simplicity) and their XRCC1-/- derivative (XRCC1-/- RPE-1 cells) were cultured in Dulbecco's Modified Eagle's Medium (DMEM/F12; Sigma) supplemented with 10% fetal calf serum and 0.01 mg/ml hygromycin B in a humidified atmosphere of 5% CO2 at 37°C. Wild type control cells were 1BR3 (denoted 1BR in the text for simplicity) primary human fibroblasts and the lymphoblastoid cell line (LCL) 11-27 isolated from an normal unaffected control. XRCC1 patient primary fibroblasts (identifier number; 5596502b) were generated from a patient skin biopsy and the LCL cell lines HEP15-00082 and HEP15-00083 were obtained from fresh blood from, respectively, the unaffected sibling and affected patient by EBV transformation. Appropriate patient consent was provided for preparation of primary fibroblasts. Primary human fibroblasts from a PNKP-mutated patient with cerebellar ataxia have been described previously5. Primary human fibroblasts were grown in Minimum Essential Media (MEM; Gibco) containing 15% fetal calf serum, 2 mM glutamine, and the antibiotics penicillin (100 units/ml) and streptomycin (100 μg/ml) at low oxygen (5%) at 37°C. LCLs were cultured in RPMI medium (Gibco) containing 10% FBS, 2 mM glutamine and penicillin/streptomycin, in a humidified atmosphere of 5% CO2 at 37°C.
Generation of gene-edited RPE-1 cells
Guide sequences were identified using either E-CRISP (http://www.e-crisp.org/E-CRISP/) or CRISPR direct (http://crispr.dbcls.jp). For XRCC1 gene editing we chose the 23-mer CRISPR complementary guide RNA sequences 5′-CCGCCUCCGCCAUGUCGUGUCCU-3′ & 5′-AGGGACACGACAUGGCGGAGGCGG-3′ (PAM underlined) spanning XRCC1 ORF nucleotides 12-34, and employed the 58-mer synthetic oligonucleotides (XCr2F; 5′-TTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGACACGACATGGCGGAGG & XCr2R; 5′-GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACCCTCCGCCATGTCGTGTC) (Eurofins) encoding 18 bp Tru-guide22 versions of the guide (underlined) minus the PAM. For PARP1 gene editing we chose the 20-mer ‘Tru-guide’ sequences 5′-GCACCCUGACGUUGAGGUGG-3′ and 5′-CCACCUCAACGUCAGGGUGC-3′ (PAM underlined) spanning nucleotides 195-214 of the human PARP1 ORF, and employed the 57-mer synthetic oligonucleotides PARP1-2F: 5′-TTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGCACCCTGACGTTGAGG-3′ & PARP1-2R:5′-GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACCCTCAACGTCAGGGTGC-3′ encoding the 17 bp Tru-guide versions of the guide (underlined) minus the PAM.
The relevant oligonucleotide guide pairs were annealed and extended into a 98-mer oligonucleotide duplexes using Phusion polymerase (NEB) and then subcloned into the guide RNA vector (Addgene; #41824)23 using Gibson Assembly(NEB). hTERT RPE-1 cells were co-transfected with the relevant guide construct/s separately (XRCC1-/-, PARP1-/-) or together (XRCC1-/-/PARP1-/-) and with the Cas9 expression construct Addgene #4181523 using a NEON Transfection System (Invitrogen). 24 h later, the transfected cells were selected in medium containing 0.5 mg/ml G418 for 5 days and subcloned into 96-well plates. Once at sufficient cell density the subclones were analyzed for expression of the relevant protein/s by indirect immunofluorescence. Absence of the relevant protein/s in selected clones was then confirmed by Western blotting. For XRCC1-/-, PARP1-/-, and XRCC1-/-/PARP1-/- RPE-1 cells we selected clones #3, #G7, and #D1 for further work, respectively.
Complementation of XRCC1 patient cells with recombinant XRCC1 protein
Recombinant human XRCC1 harbouring a C-terminal decahisitidine tag (denoted XRCC1-His) was expressed in E. coli from pET16b-XH and purified by metal-chelate affinity chromatography and gel filtration6. 1 or 2 μg of control bovine serum albumin (BSA) or purified XRCC1-His was electroporated into 1×105 XRCC1 patient fibroblasts using a NEON Transfection System (Invitrogen) according to the manufacturer's protocol. 18 hours later the cells were treated with CPT for 45 min, fixed, and immunostained for levels of XRCC1 and ADP-ribose as indicated.
RNA extraction, cDNA synthesis and qPCR analysis
LCLs were treated with either 100 μg/ml Cycloheximide (Sigma) or vehicle alone for 4 h and total RNA extracted with RNAeasy Kit (Qiagen) essentially as described by the manufacturer but with an additional 15 min DNAse I (Promega) digest of the samples on the column. 1 μg total RNA was annealed to oligodT(15) primer and reverse transcribed using M-MuLV RT (NEB) for 2 h at 42°C. After RNAse A digest, the cDNA was purified using PCR purification kit (Qiagen) and 1/40 of the eluate used per reaction. Three replicate qPCR reactions using ABsolute qPCR SYBR Low ROX (Thermo) were performed per experiment in a MX3005P (Agilent) thermocycler and analysed using MxPro software (Agilent). The fold change was calculated from ΔCt values relative to actin and ΔΔCt values relative to WT untreated for three independent experiments. Primers were:
XRCC1 exon10 forward: CAACACCCCCAAGTACAGC
XRCC1 exon 10 reverse: AGTCCAGCACCCACTCCTTAC
XRCC1 exon11 forward: TCCAGCAGTGAGGAGGATG
XRCC1 intron11 reverse: AGGCAAGAGTGGGAAGTTTG
XRCC1 exon 12 reverse: AGTGGGCTTGGTTTTGGTC
Actin forward: CTCGTCATACTCCTGCTTGC
Actin reverse: GAAGTGTGACGTGGACATCC
XRCC1 cDNA cloning
cDNA prepared as above from patient cells treated with cycloheximide was used for Phusion polymerase (NEB) amplification of full length XRCC1 transcripts using primers ATGCCGGAGATCCGCCTCCG and GGCTTGCGGCACCACCCCAT. PCR products were purified using Gel extraction kit (Qiagen), cloned using TOPO cloning kit (Thermo Fischer Scientific) and plasmids originating from single colonies purified using Miniprep kit (Qiagen) and sequenced by Sanger sequencing (Beckman Coulter).
siRNA
Wild type and patient primary fibroblasts were reverse-transfected with Lipofectamine RNA iMAX (Life Technologies) as indicated by the manufacturer, using non-targeting siRNA (ON-TARGETplus, Dharmacon) and siXRCC1 SMARTpool (Dharmacon).
Western blotting
PBS-washed cells were lysed in Laemmli buffer, heated for 10 min at 95°C, and sonicated for 30 sec using Bioruptor® Pico (Diagenode). Protein concentrations were determined using the BCA assay (Pierce). Samples were subjected to SDS PAGE, proteins transferred onto nitrocellulose membrane and detected by immunoblotting using the relevant primary and horseradish peroxidase-conjugated secondary antibodies. Peroxidase activity was detected by ECL reagent (GE Healthcare) and Amersham Hyperfilm ECL (GE Healthcare). Non-saturated film exposures were digitalized using an EPSON perfection 2400 photo scanner and quantified using ImageJ software.
Immunofluorescence and microscopy
Cells cultured on glass coverslips were fixed in 4% paraformaldehyde for 10 min at room temperature, permeabilized in methanol/acetone solution (1:1), blocked in 10% foetal calf serum and incubated with primary antibodies for 60 min at room temperature. Following rising in PBS, coverslips were incubated with secondary antibodies at room temperature for 60 min. Finally, after washing in PBS, nuclei were counterstained with DAPI (Sigma) and coverslips mounted using anti–fading mounting reagent (Vectashield, Vector Laboratories). To measure chromatin retention of proteins, cells were pre-extracted in cold 0.2% Triton X-100 for 2 min on ice prior to fixation as above. High-resolution microscopy of fixed samples was carried out on a Zeiss AxioObserver.Z1 microscope, equipped with oil immersion objectives (Plan-Apochromat 63×/1.4 and 100×/1.4), Hamamatsu ORCA-Flash4.0 LT camera and ZEN 2 core imaging software. Automated wide-field microscopy was performed on an Olympus ScanR system (motorized IX83 microscope) with ScanR Image Acquisition and Analysis Software, 20×/0.45 (LUCPLFLN 20× PH) and 40×/0.6 (LUCPLFLN 40× PH) dry objectives and Hamamatsu ORCA-R2 digital CCD camera C10600. Where indicated, cells were counterstained with DAPI.
ADP-ribosylation
Levels of ADP-ribosylation were measured in the indicated cell lines by indirect immunofluorescence using Anti-pan-ADP-ribose binding reagent (Millipore). Cells were treated where indicated with 150 μM H2O2 for 10 min and then incubated in drug-free medium for 60 min to allow DNA repair or were incubated with 30 μM CPT for 45 min. ADP-ribose levels were quantified by Olympus ScanR imaging and plotted relative to the ADP-ribose level in untreated cells. For cerebellar sections, mice (P17-P23) were anaesthetized using 0.2 mg/g Euthanal (Vetoquinol UK Ltd) and perfused transcardially with PBS followed by 4% paraformaldehyde. Brains were postfixed in 4% paraformaldehyde for 48 h and stored in 25% sucrose/PBS until moulding and freezing. 7 μm sagittal sections were prepared using a cryostat (Leica CM1850) and immunohistochemistry was conducted essentially as described7. Briefly, slides were washed in PBS and heated until boiling in antigen retrieval buffer (Nacalai Tesque, Histo VT One). Endogenous peroxidase was blocked by incubating slides in 0.6% H2O2 in methanol. After incubating in blocking solution (5% goat serum, 1% BSA, 0.4% Triton-X-100 in PBS) rabbit anti-poly (ADP-ribose) primary antibody (Trevigen) was applied overnight. Anti-rabbit Biotin-SP-conjugated AffiniPure Goat secondary antibody (JacksonImmuno Research, 1:500) was incubated on the slides for 1 hr followed by ABC reagent (Vectstain Elite ABC kit, Vector Laboratories) according to manufacturer's instructions. The chromogen was developed using VIP reagent (Vector VIP peroxide substrate kit, Vector laboratories, Inc.; SK-4600). Images were obtained using a Nikon Eclipse E400 mounted with Electronic Digital Eyepiece Camera CMOS (C-mount UK).
Sister chromatid exchanges
Lymphoblastoid cell lines were incubated in medium containing 8 μM chlorodeoxyuridine, 32 μM thymidine, 10 μM fluorodeoxyuridine and 200 μM cytidine (such that 20% of incorporated thymidine is replaced with chlorodeoxyuridine)11,24 for 20 h, washed twice in media and then allowed to grow for another 20 h in medium containing 10 μM thymidine. Cells were treated with 100 ng/ml colcemid for 1 h, swollen in 75 mM KCl for 5 min at 37°C and fixed in Carnoy's fixative before preparation of metaphase spreads. Slides were allowed to air dry, rehydrated in PBS, incubated in 2 M HCl for 30 min, washed twice in 100 mM borate buffer pH 8.5 for 10 min and blocked in 10% foetal calf serum in PBS for 30 min. Further immunofluorescence and imaging was performed as above.
SSBR & DSBR assays
Alkaline comet assays were performed essentially as described25 and inducing DNA breaks with 50 μM H2O2 (RPE-1 cells) or 25 μM H2O2 (primary fibroblasts) for 10 min on ice. Data are plotted as the average comet tail moment (an arbitrary unit-measure of DNA strand breaks) of 100 cells per sample and are the mean (+/-SEM) of three independent experiments. For DSB repair assays, cells were irradiated with 2 Gy using Gammacell 1000 machine, and γH2AX quantified at the indicated times, afterwards. Data are the average number of γH2AX foci per cell from ∼1000 cells/sample, scored by OIympus ScanR software, and are the mean (+/-1SD) of three independent experiments.
Mouse maintenance and analysis
Animals were maintained and used under the auspices of UK Home Office project licence number 70/8300. The generation of Parp1-/- and Xrcc1Nes-Cre mice have been reported previously7,26. Intercrosses between Parp1-/- and Xrcc1+/loxp mice were maintained in a mixed C57/Bl6 × S129 strain and housed on a 12 h light/dark cycle with lights on at 7 am. Temperature and humidity were maintained at 21°C (+/-2°C) and 50% (+/-10%), respectively. All experiments were carried out under the UK Animal (Experimental Procedures) Act, 1986. Genomic DNA was extracted from biopsied tail using the REDExtract-N-Amp™ Tissue PCR Kit in accordance to the manufacturer's instructions (Sigma). For Parp1, the following primers were used: PARP1 F1 (5′-GTT GTG AAC GAC CTT CTG GG-3′), PARP1 R1 (5′-CCT TCC AGA AGC AGG AGA AG-3′) and PARP R2 (5′-GCT TCA GTG ACA ACG TCG AG-3′). PCR products were generated by an initial denaturation step at 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 54°C for 40 s and 72°C for 180 s. Xrcc1 was amplified using the forward PC1 (5′-TAT GCT TGC TGT ACA GGG ATT GGG-3′) and reverse PC2 (5′-TGG ACC ATG AAA AAG CTG TGT GC-3′) primers. A 400 bp cre PCR product was generated using the forward Cre-3 (5′-CTG CCA CGA CCA AGT GAC AGC-3′) and reverse Cre-4 (5′-ACC TGC GGT GCT AAC CAG CG-3′) primers. Both Xrcc1 and Cre PCR products were amplified using the following PCR conditions: initial denaturation for 5 min at 94°C followed by 35 cycles of 94°C for 30 s, 59°C for 45 s and 72°C for 45 s.
Nissl staining of paraffin-embedded sections
Brains were removed at the indicated times and placed in 10% neutral buffered formalin (3.7% formaldehyde, 3.5 g/l NaH2PO4, 6.5 g/l Na2HPO4) for 24 h and then transferred to PBS. Paraffin embedding, sagittal sectioning and Nissl staining were carried out by Propath UK Ltd (Hereford, UK) and UCL IQPath (Institute of Neurology, London, UK). Briefly, deparaffinised sectioned were stained in 0.1% cresyl violet solution (0.1% cresyl violet, 0.3% glacial acetic acid) for 3-10 minutes, rinsed in water, then 95% ethanol for 30 s to 5 min, 100% ethanol for 2 × 5 min and xylene for 2 × 5 min before mounting. Nissl-positive interneurons were counted within three randomly-selected 16875 μm2 regions of each molecular layer, and the number of cells per μm2 was calculated.
Rotarod analysis
To evaluate motor coordination/cerebellar ataxia, an accelerating Panlab Rotarod (Harvard Apparatus Ltd., UK) was used. The apparatus was composed of a rod with a diameter of 30 mm divided into five opaque methacrylate arnite barriers, separating the rod into five 50 mm sections. Mice were trained for three successive attempts on the day prior to assessment. For assessment, mice were positioned on the rod facing away from the experimenter whilst the rod was stationary. Once activated, the rod accelerated at 0 to 40 rpm at an acceleration rate of 5 min. Mice were rested for 15 min between each of the three trials and then positioned back on the rod for the next assessment. The time spent on the rod was recorded for each mouse and the mean time was calculated from the three independent trials.
Purkinje cell recordings
Vermis parasagittal slices (200 μm) were taken from P13-P17 mice using a vibroslicer (VT1200S, Leica Microsystems, Germany) in ice-cold artificial cerebrospinal fluid (ACSF) containing (in mM): 125 NaCl, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 26 NaHCO3, 1 MgCl2, 2 CaCl2 (bubbled with 95% O2 and 5% CO2, pH 7.3). Slices were maintained in ACSF at 34-35°C for 45 min and then at 23-25°C for electrophysiology. Voltage-clamp recordings of Purkinje cells were performed in cell-attached mode using a Multiclamp 700A amplifier (Molecular Devices) with pipettes (5-7 MΩ) containing 150 mM NaCl. Signals were filtered at 10 kHz and digitized at 50 kHz and analysis of spike activity performed offline using Clampfit (Molecular Devices).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Extended Data
Extended Data Figure 1. Aberrant splicing of XRCC1 pre-mRNA in XRCC1 patient cells.
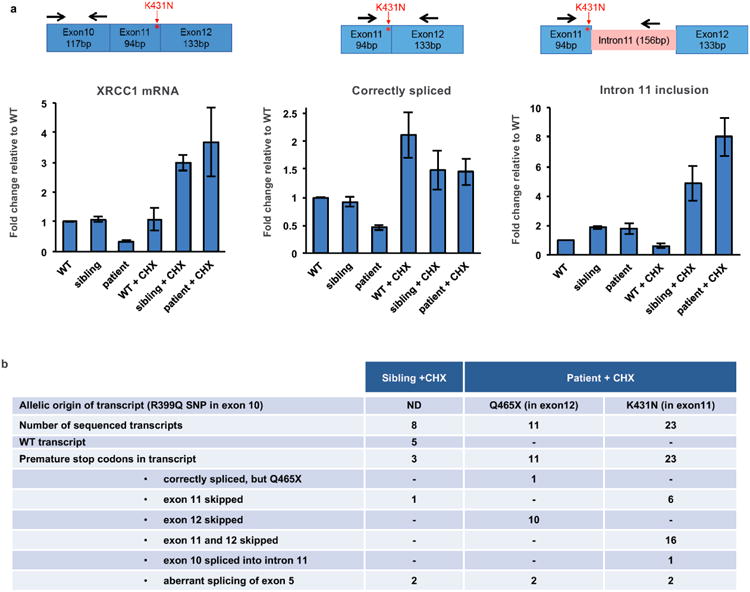
a, qPCR analysis of cDNA prepared from polyA-tailed RNA from wild type (WT), unaffected sibling, and patient LCLs. Cells were mock-treated or treated with cycloheximide (CHX) to inhibit nonsense-mediated decay. Cartoons show the position of primers employed to quantify total XRCC1 mRNA (left), mRNA with correctly spliced exon11/12 junction (middle) or intron 11 retention (right). Fold change was calculated from ΔΔCt values relative to actin and untreated WT from three independent experiments (mean +/-SEM). b, Summary of splicing defects observed in sequenced XRCC1 transcripts from sibling and patient cells treated with CHX. Whole XRCC1 cDNA was amplified from oligodT-primed reverse-transcribed RNA and individual transcripts cloned and sequenced. The allelic origin of individual patient transcripts was assigned using a R399Q SNP in the Q465X allele.
Extended Data Figure 2. XRCC1 and Lig3a protein levels in XRCC1-/- RPE-1 cells and XRCC1 patient cells.
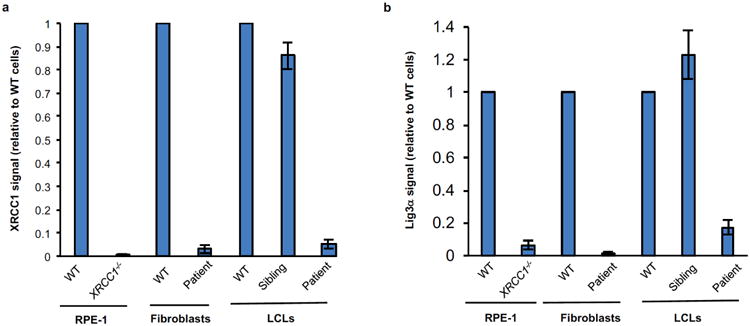
Quantification of XRCC1 (a) and Lig3α (b) protein levels in the indicated cell lines by Western blotting. Data are the mean signal intensity from three independent experiments (+/-SEM) normalized to tubulin and to the respective wild type (WT) sample for each cell type. The anti-XRCC1 signal detected in XRCC1-/- RPE-1 cells reflects non-specific background.
Extended Data Figure 3. Reduced XRCC1 recruitment into damaged chromatin in XRCC1 patient cells.
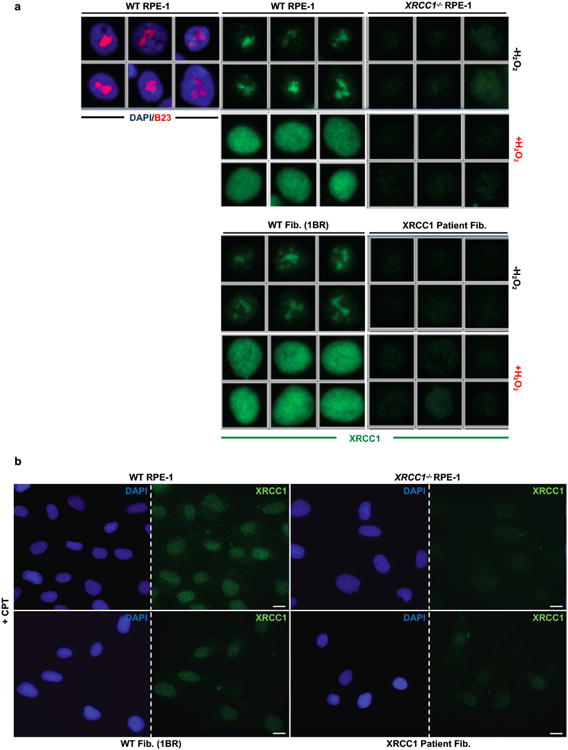
a, XRCC1 recruitment into chromatin was compared in the indicated cell lines by ScanR high content imaging before and 10 min after treatment with 1 mM H2O2. Cells were pre-extracted with detergent prior to fixation and immunostaining. Representative images of the ScanR (Olympus) data used for the quantification in Fig. 2d are shown. b, XRCC1 recruitment into chromatin using high resolution Zeiss microscope was compared in the indicated cell lines after treatment 45 min with 30 μM CPT. Cells were pre-extracted with detergent prior to fixation and immunostaining as above. Scale bars in images are 10 μm.
Extended Data Figure 4.
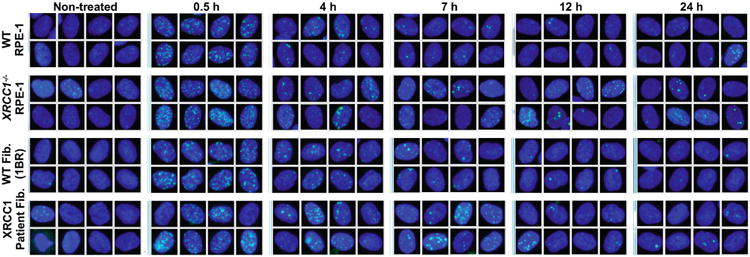
Representative images from ScanR high-content imaging of gH2AX foci (green) in the nuclei (blue) of the indicated cells at the indicated times after ionising radiation (2 Gy). Quantification is shown in Fig. 3b.
Extended Data Figure 5. Elevated ADP-ribosylation in XRCC1-/- RPE-1 cells, XRCC1 patient fibroblasts, and PNKP patient fibroblasts.
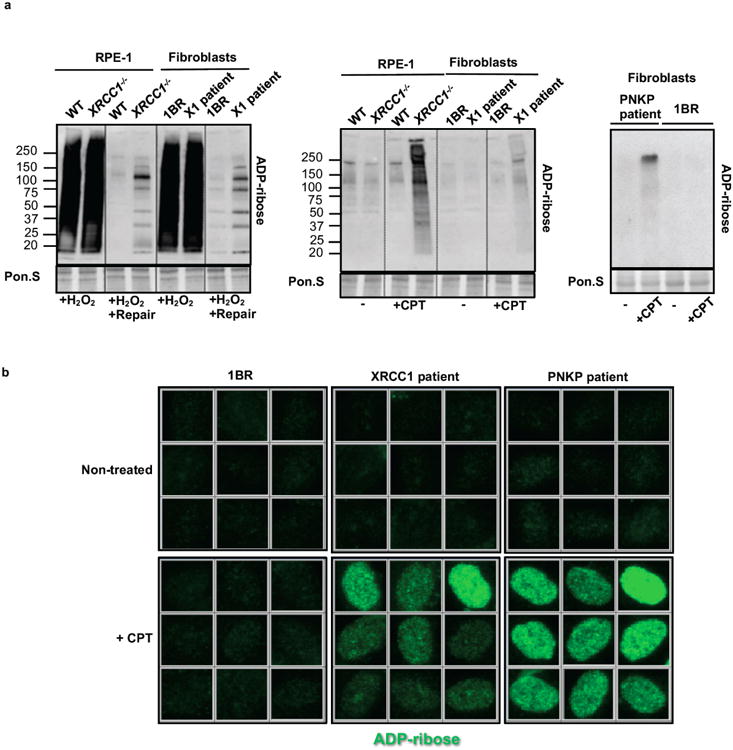
a, ADP-ribosylated proteins were detected in cell extracts from wild type RPE-1 cells (“WT”), XRCC1-/- RPE-1 cells, wild type 1BR fibroblasts, XRCC1 patient fibroblasts (“X1 patient”), and PNKP patient fibroblasts treated as in Fig. 4a by Western blotting and Anti-pan-ADP-ribose binding reagent. b, Levels of ADP-ribosylation in 1BR wild type, XRCC1 patient, and PNKP patient fibroblasts measured before and after CPT treatment by indirect immunofluorescence as indicated in Fig. 4b. Representative ScanR images are shown.
Extended Data Figure 6. Elevated ADP-ribose levels in CPT-treated XRCC1-/- RPE-1 cells are PARP1 dependent.
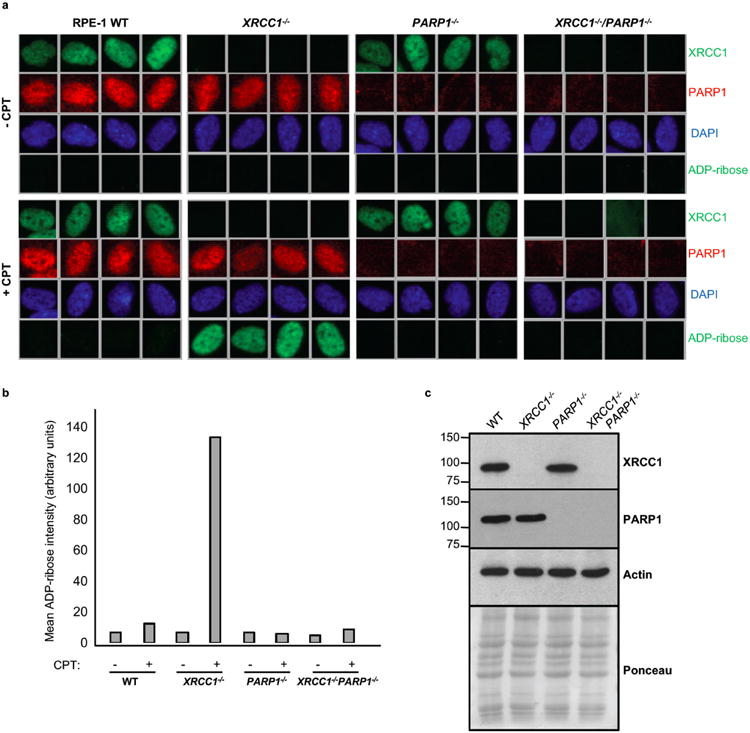
a, Representative ScanR images of wild type (“WT”), XRCC1-/-, PARP1-/-, and XRCC1-/-/PARP1-/- RPE-1 cells before and after 30 μM CPT treatment stained by indirect immunofluorescence for XRCC1, PARP1, DAPI or ADP-ribose. b, Quantification of ADP-ribose intensity in the nucleus from >3500 cells per sample from a (data from single experiment). c, Western blot showing XRCC1 and PARP1 protein levels in the indicated RPE-1 cell lines.
Extended Data Figure 7. Elevated ADP-ribose levels in CPT-treated XRCC1 patient and PNKP patient cells are prevented by PARP inhibitors.
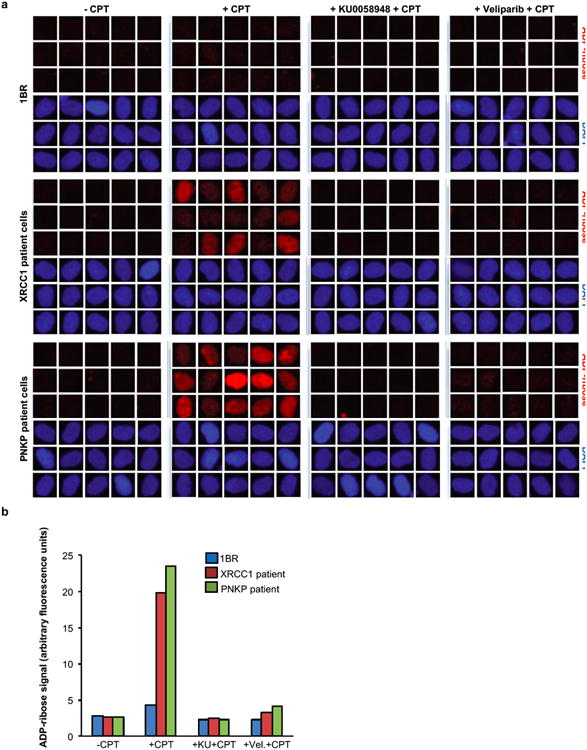
Levels of ADP-ribose in wild type 1BR fibroblasts, XRCC1 patient fibroblasts and PNKP patient fibroblasts were measured before and after 45 min 30 μM CPT treatment, and after 45 min 30 μM CPT treatment in the presence of 10 μM of the indicated PARP inhibitor (also including 1 hour pre-treatment). Cells were pre-extracted with detergent prior to fixation and immunostaining with Anti-pan-ADP-ribose binding reagent. Representative ScanR images are shown in a and quantification from >1500 cells per sample, from a single experiment, are shown in b.
Extended Data Figure 8.
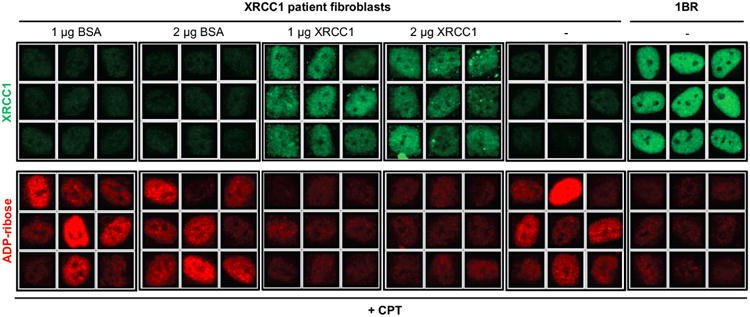
Levels of ADP-ribosylation were quantified before and after CPT treatment by high content imaging in wild type 1BR fibroblasts, XRCC1 patient fibroblasts, and XRCC1 patient fibroblasts transfected by electroporation in the presence of 1 mg or 2 mg of control BSA or purified recombinant human XRCC1-His protein. Representative ScanR images are presented. Quantification of ADP-ribosylation is shown in Fig. 4d.
Extended Data Figure 9. Reduced cerebellar Purkinje cell firing frequency in Xrcc1Nes-Cre (KO) mice.
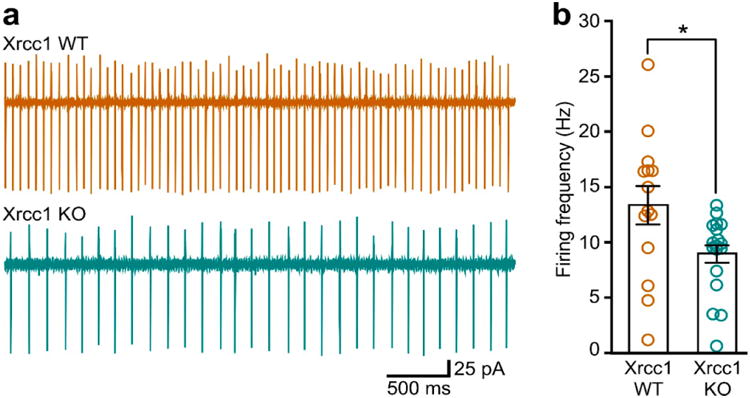
a, Representative traces from cell-attached recordings. b, Summary of data. Bars and error bars indicate mean (+/-SEM) firing frequencies for Xrcc1 WT (13.35 ± 1.73, n = 14 cells) versus Xrcc1Nes-Cre mice (8.96 ± 0.82, n = 18 cells)(*p<0.05, t-test). Circles are mean firing frequencies for each cell.
Extended Data Figure 10.
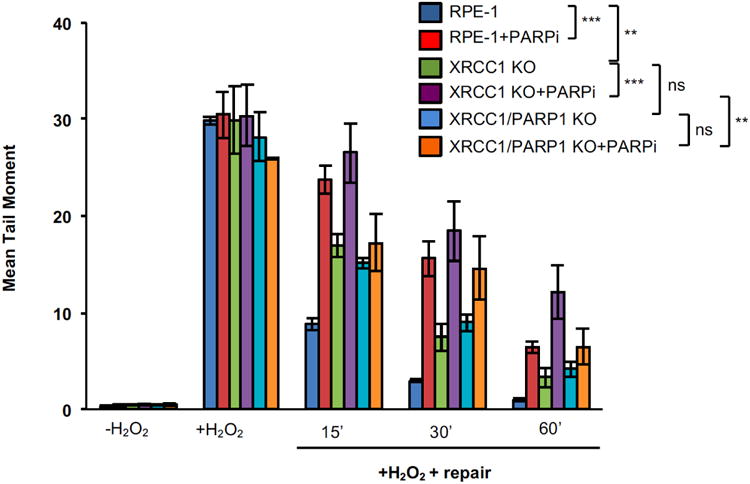
Chromosomal single-strand break repair rates in the indicated RPE-1 cell lines following treatment with 50 μM H2O2 in the presence/absence of 10 μM of the PARP inhibitor (PARPi) Veliparib, as measured by alkaline comet assay. Data are the mean tail moment of 100 cells per sample per experiment and are the average (-/+SEM) of three independent experiments. Two-way ANOVA analysis between relevant genotypes is presented (ns= not significant, **= p<0.01, ***= p<0.001).
Supplementary Material
Acknowledgments
We would like to thank the patient and her family for their contribution to this study. This work was selected for study by the Care4Rare Canada (Enhanced Care for Rare Genetic Diseases in Canada) Consortium Gene Discovery Steering Committee: Kym Boycott (lead; University of Ottawa), Alex MacKenzie (co-lead; University of Ottawa), Jacek Majewski (McGill University), Michael Brudno (University of Toronto), Dennis Bulman (University of Ottawa), and David Dyment (University of Ottawa). We especially thank David Dyment (University of Ottawa) for his advice and discussion. We thank Saskia vanderVelde-Visser and Janneke Schuurs-Hoeijmakers at the Radboud Nijmegen University Medical Centre for EBV transformation of the patient and sibling LCLs. We also thank Sherif El-Khamisy and Andrew Ridley for preliminary analyses and assistance with the mouse work.
Funding. This work was funded by a MRC Programme Grant (MR/J006750/1) to KWC, a “Science without Borders” postdoctoral fellowship (CAPES Foundation, Ministry of Education, Brazil, BEX9769-13-7) to NH, and funding to GY from Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, Children's Hospital of Eastern Ontario Foundation and the Hospital for Sick Children. PMcK acknowledges the NIH (NS-37956, CA-21765), the CCSG (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital for support.
Footnotes
Author contributions. H.H generated gene-edited RPE-1 cell lines and designed and conducted the immunofluorescence, high-content imaging (Olympus ScanR), protein complementation, and DSB repair experiments. N.H analysed XRCC1 patient cells by Western blotting and RT-qPCR and designed and conducted sister chromatid exchange and comet assays. S.L.R, E.K., and L.J, designed and conducted mouse behaviour and histopathology experiments. S.R and K.S designed and conducted electrophysiology experiments. P.H. prepared recombinant XRCC1. Z.Z generated CRISPR guide constructs. W.G conducted preliminary CPT/ADP-ribose experiments. G.M.S.M provided PNKP patient fibroblasts. P.J.M and Z-Q.W provided mouse models. G.Y. identified and oversaw genetic analysis of the XRCC1 patient and J.W. and M.T. conducted exome analysis. K.W.C conceived and managed the project and wrote the manuscript with H.H and N.H. All authors edited the manuscript.
The authors declare that there is no conflict of interest.
References
- 1.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 2.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 3.Shen J, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bras J, et al. Mutations in PNKP cause recessive ataxia with oculomotor apraxia type 4. Am J Hum Genet. 2015;96:474–479. doi: 10.1016/j.ajhg.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulton C, et al. Progressive cerebellar atrophy and polyneuropathy: expanding the spectrum of PNKP mutations. Neurogenetics. 2012 doi: 10.1007/s10048-012-0351-8. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tebbs RS, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 9.Tebbs RS, Thompson LH, Cleaver JE. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair (Amst) 2003;2:1405–1417. doi: 10.1016/j.dnarep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Bradley MO, Kohn KW. X-ray induced DNA double strand break production and repair in mammalian cells as measured by neutral filter elution. Nucleic Acids Res. 1979;7:793–804. doi: 10.1093/nar/7.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillehay LE, Thompson LH, Minkler JL, Carrano AV. The relationship between sister-chromatid exchange and perturbations in DNA replication in mutant EM9 and normal CHO cells. Mutat Res. 1983;109:283–296. doi: 10.1016/0027-5107(83)90053-2. [DOI] [PubMed] [Google Scholar]
- 12.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 13.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 14.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 15.Eliasson MJ, et al. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 16.Chiarugi A. Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- 17.Katyal S, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci. 2014 doi: 10.1038/nn.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marras C, et al. Nomenclature of genetic movement disorders: Recommendations of the international Parkinson and movement disorder society task force. Mov Disord. 2016;31:436–457. doi: 10.1002/mds.26527. [DOI] [PubMed] [Google Scholar]
- 19.Tetreault M, et al. Whole-exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum Genet. 2015;134:981–991. doi: 10.1007/s00439-015-1577-y. [DOI] [PubMed] [Google Scholar]
- 20.Cappelli E, et al. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 21.Breslin C, Caldecott KW. DNA 3′-phosphatase activity is critical for rapid global rates of single-strand break repair following oxidative stress. Mol Cell Biol. 2009;29:4653–4662. doi: 10.1128/MCB.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinkel D, Thompson LH, Gray JW, Vanderlaan M. Measurement of sister chromatid exchanges at very low bromodeoxyuridine substitution levels using a monoclonal antibody in Chinese hamster ovary cells. Cancer Res. 1985;45:5795–5798. [PubMed] [Google Scholar]
- 25.Breslin C, et al. Measurement of chromosomal DNA single-strand breaks and replication fork progression rates. Meth Enzymol. 2006;409:410–425. doi: 10.1016/S0076-6879(05)09024-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang ZQ, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


