Abstract
Prior work by others has shown that insertion of ψ (i.e., leader) sequences from the Moloney murine leukemia virus (MLV) genome into the 3′ untranslated region of a nonviral mRNA leads to the specific encapsidation of this RNA in MLV particles. We now report that these RNAs are, like genomic RNAs, encapsidated as dimers. These dimers have the same thermostability as MLV genomic RNA dimers; like them, these dimers are more stable if isolated from mature virions than from immature virions. We characterized encapsidated mRNAs containing deletions or truncations of MLV ψ or with ψ sequences from MLV-related acute transforming viruses. The results indicate that the dimeric linkage in genomic RNA can be completely attributed to the ψ region of the genome. While this conclusion agrees with earlier electron microscopic studies on mature MLV dimers, it is the first evidence as to the site of the linkage in immature dimers for any retrovirus. Since the Ψ+ mRNA is not encapsidated as well as genomic RNA, it is only present in a minority of virions. The fact that it is nevertheless dimeric argues strongly that two of these molecules are packaged into particles together. We also found that the kissing loop is unnecessary for this coencapsidation or for the stability of mature dimers but makes a major contribution to the stability of immature dimers. Our results are consistent with the hypothesis that the packaging signal involves a dimeric structure in which the RNAs are joined by intermolecular interactions between GACG loops.
Retroviral proteins package their genomic RNAs with very high selectivity. This selectivity results from the recognition by the viral Gag protein of cis-acting packaging signals (generically called ψ) in the viral RNA. While this recognition event is far from understood in molecular terms, all available evidence indicates that the major elements in ψ signals are located near the 5′ end of the genomic RNA (10).
In all normal retrovirus particles, the genomic RNA is in dimeric form (10). It consists of two identical, positive-strand monomers, joined together by a thermolabile linkage. The linkage presumably consists of a limited number of base pairs. After the assembled virion is released from the cell, the viral protease (PR) is activated and cleaves the viral proteins into a series of smaller proteins (62). This set of cleavages, termed maturation of the particle, convert the particle into infectious form. The cleaved proteins in turn cause a change in the conformation of the dimeric RNA in the particle, frequently resulting in a more stable dimeric linkage than that present in immature virions (24, 26, 61) The change in conformation of the RNA has been called maturation of the dimers. It seems likely that dimer maturation is due to the action of the nucleocapsid (NC) protein on the RNA: this protein is known to possess nucleic acid chaperone activity (the ability to catalyze the conformational rearrangement of nucleic acids into the structure with the maximum number of base pairs), and is tightly associated with the genomic RNA in the mature virion (15, 20, 45, 55, 60).
Neither the location nor the structure of the linkage between the two monomers is known with precision. Many years ago, electron micrographs of partially denatured RNA extracted from mature retrovirus particles showed that the most stable linkage is near, but not at, the 5′ end of the genome (7, 8, 31, 37, 47). This region has therefore been termed the dimer linkage site (DLS). In contrast, however, there is no information in the literature on the site of the linkage in immature dimers.
More recently, it was found that transcripts of the 5′ region of retroviral genomes could dimerize spontaneously in vitro under conditions of high ionic strength (11, 14, 51, 57). A conserved sequence feature of this region is the presence of a stem-loop with a palindromic four- or six-base sequence in the loop. This motif has been termed the kissing loop (10). One obvious possible mechanism for dimer formation would be base-pairing between the loops of two monomers, and convincing evidence has been marshaled that this occurs during dimerization of transcripts in vitro. Furthermore, examination of the sequence of a kissing loop shows that it could assume two alternative dimeric conformations, one more stable than the other: in the less stable conformation (the kissing complex), only the palindromic loops would pair with each other. In the more stable structure, the bases comprising the stems in the kissing loop structure would come apart and form an extended dimer, in which bases in both the stems and the loops are paired intermolecularly. Thus, sequences present in retroviral DLS regions suggest a model not only for the structure of the dimer, but even for the existence of two forms of dimer, one with a more stable linkage than the other. Indeed, experimental systems have been devised demonstrating that the transcripts can form two alternative species of dimer, and the viral NC protein has been shown to be capable of converting the more labile species into the more stable species (20, 45, 56).
Transcripts offer appealing, tractable model systems for experimental analysis and have proven to be a fruitful source of new concepts about viral RNAs. However, the validity of these concepts can ultimately be assessed only by studies of authentic RNAs obtained from virus particles. In the present work, we used a genetic approach for the identification of RNA sequences involved in the immature and mature dimeric linkages in retroviral RNAs. With an experimental strategy pioneered by Adam and Miller (1), we placed viral sequences from the ψ/DLS regions of Moloney murine leukemia virus (MLV) and related viruses into the 3′ untranslated region (UTR) of a nonviral mRNA. We then expressed these mRNAs, along with MLV genomes, in mammalian cells. Under these conditions, the ψ signal in the mRNA is recognized by the MLV proteins, and the mRNA is packaged into virions.
The principal focus of our experiments has been the analysis of the state of these mRNAs within virions. We find that the mRNAs are packaged as dimers; in general, the dimers of these mRNAs exhibit the same thermostabilities as the dimers of the parental genomic RNAs. Therefore, the dimeric linkage in viral RNAs can indeed be attributed to the sequences that we inserted. This is the first direct experimental evidence that the linkage in immature dimers, like that in mature dimers, is in the ψ/DLS region of retroviral genomes.
In addition, the insertion of DLS sequences into a nonviral, reporter mRNA enabled us to begin to explore the relationship between dimerization and packaging and to dissect the sequences involved in immature and mature dimeric linkages. We quantitated RNA copy numbers by real-time reverse transcription-PCR and found that mRNA with an insert of viral sequence is packaged far less efficiently than the full-length, parental genomic RNA. The dimeric character of these packaged mRNA molecules provides strong support for the hypothesis that they are incorporated into virions in pairs, perhaps as a preformed immature dimer. However, we also analyzed mRNAs in which the kissing loop structure has been destroyed. These RNAs do not form the normal immature dimeric linkage but are nevertheless incorporated in pairs. One hypothesis that is consistent with these results is that assembly is normally initiated by the binding of Gag protein molecules to a very labile “preimmature” dimer. The possible structure of this hypothetical dimer is discussed.
MATERIALS AND METHODS
Plasmid constructs.
The helper virus constructs used in these experiments include the wild-type MLV plasmid pRR88 (28); the D32L mutant (at the active site of MLV PR) of pRR88 (26); ψ− MLV, which is derived from pPAM3 (41) between the 5′ end and the XhoI site at nucleotide 1560, with the remainder from pRR88 (46); and ψ− PR−, which was constructed by replacing the region of the ψ− construct that was derived from pRR88 with the corresponding region from the D32L PR− mutant (46). The ψ− constructs contain a deletion between nucleotides 212 and 563. In all of these constructs, the plasmid backbone is that of pGCcos3neo, a derivative of pSV2neo (59).
Our studies used a number of plasmids encoding hygromycin phosphotransferase (hph) mRNA with variations in the viral sequences inserted in the 3′ UTR. The plasmids used here are listed in Fig. 1. Inserts all contained the correct viral sequence. In all of these plasmids, hph mRNA is driven by the cytomegalovirus immediate-early promoter. Two of these constructs have also been described before, pCMVhygMoψ and pCMVhyg (54). pCMVhygMoψ is designated hph (−151)-1560 and pCMVhyg is termed hph no ψ in the present work. hphHaSV contains nucleotides −151 to 942 of Harvey sarcoma virus (HaSV) in its 3′UTR and was constructed from a plasmid originally obtained from D. Lowy (National Cancer Institute) (65). hph (−151)-725 is a truncation of the (−151)-1560 insert, made by digestion with BstEII followed by blunt-end filling and religation.
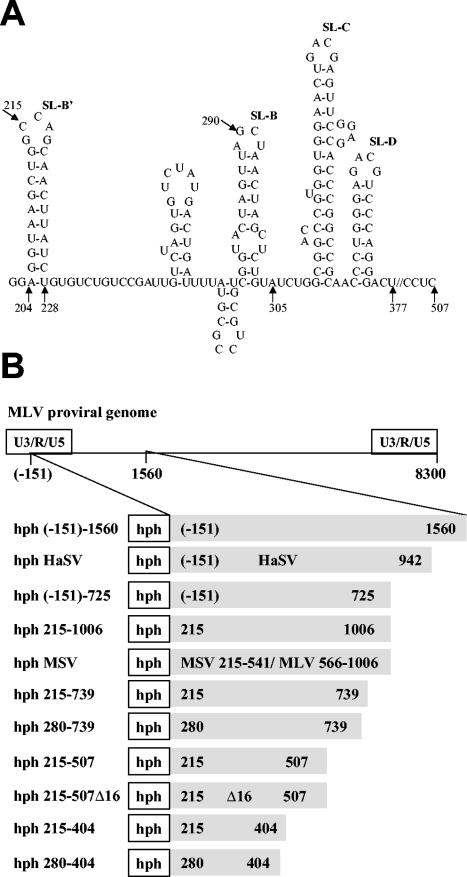
FIG. 1.
MLV ψ region. (A) Sequence of nucleotides 202 to 377, showing predicted stem-loops (SLs). (B) Chimeric RNAs analyzed in the present study. The gray portion of each line denotes the viral insert placed in the 3′ UTR of the hph mRNA.
Other hph plasmids were constructed by PCR synthesis of a viral insert by using primers with an XbaI site at the 5′ end and an XhoI site at the 3′ end and replacement of the viral insert in hph (−151)-1560 with the new product. The following hph plasmids were made by PCR with the 5′-oligonucleotide primer (5′GCTCTAGACCAGCAACTTATCTG): hph 215-1006, hph 215-739, hph 215-507, hph 215-507Δ16, hph 215-404, and hph MSV. Plasmids hph 280-739 and hph 280-404 were made with the 5′ primer 5′GCTCTAGAGTACTAGTTAGCTAAC. Plasmids hph 215-1006 and hph MSV were made with the 3′ primer 5′CGATCTCGAGGATCGAGGCG. Plasmids hph 215-739 and hph 280-739 were made with the 3′ primer 5′CGATCTCGAGCAGAAGGTAAC. Plasmids hph 215-507 and hph 215-507Δ16 were made with the 3′ primer 5′CGATCTCGAGGCGGGAACTG. Plasmids hph 215-404 and 280-404 were made with the 3′ primer 5′CGATCTCGAGGTCGGGCCAC.
The template for these PCRs was hph (−151)-1560 except that the hph plasmid containing the murine sarcoma virus (MSV)-MLV chimeric insert (designated hph MSV) was constructed by PCR with the same primers as for hph 215-1006 but with the pψ+ (1) plasmid as a template for the synthesis of the viral insert. The pψ+ plasmid contains nucleotides 215 to 541 from MSV 124 and nucleotides 566 to 1038 from MLV, but the 3′ end of the insert that we placed in hph MSV is at nucleotide 1006. In one plasmid, hph 215-507Δ16, nucleotides 290 to 305 were fortuitously deleted during the PCR. We also analyzed RNA of pLXSH (42) (a kind gift of Vineet Kewal Ramani).
Cell culture.
293T cells were transfected with hph plasmids with the calcium phosphate method (29), with 5 μg of DNA on 60-mm plates seeded 1 day earlier with 3 × 105 cells. Pools of stably transfected cells were selected with 200 μg of hygromycin B (Calbiochem) per ml.
To analyze the state of hph mRNA within virions, we transfected hygromycin-resistant cells with one of the helper constructs described above, using the calcium phosphate method with 10 μg of DNA on 100-mm plates seeded 1 day earlier with 106 cells. Controls included mock transfections, in which the cells were transfected with pGCcos3neo DNA, which does not encode viral proteins. The cells were washed after overnight exposure to the DNA, and culture fluids were collected 24, 48, and 72 h after the wash.
Virus preparation and RNA isolation.
Culture fluids were filtered through 0.45-μm filters (Nalge), and virions were then collected by centrifugation through a 20% sucrose cushion as described (46). Pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% sodium dodecyl sulfate, 100 mM NaCl, and 100 μg of proteinase K per ml). Lysis was at 37°C for 30 min, followed by phenol-chloroform extraction and ethanol precipitation with 0.3 M sodium acetate (pH 5.2) with 0.02% linear acrylamide as the carrier.
In experiments measuring packaging efficiency, cellular RNA was isolated as follows: 10-cm plates were gently washed with 4 ml of phosphate-buffered saline. Cells were lysed directly in the plate with 7 ml of Trizol reagent (Invitrogen). The Trizol mix was transferred to a tube and incubated 5 min at 25°C; 0.2 ml of chloroform per ml of Trizol was added, shaken for 15 s, incubated for 2 min at 25°C, and then centrifuged for 15 min at 4°C. The aqueous phase was then transferred to a fresh tube, and the RNA was precipitated by isopropanol. Amounts of cellular RNA were determined by measuring absorbance at 260 nm.
Northern blotting.
Nondenaturing Northern blot analysis was performed as previously described (26, 33). RNA riboprobes were synthesized by transcription with either T7 or T3 RNA polymerase (Promega), in accord with the manufacturer's instructions, with [α-32P]CTP (Amersham). An MLV-specific riboprobe template complementary to the MLV pol gene was made by BglII digestion of plasmid pMXH (22). An MLV ψ riboprobe template was made by XbaI digestion of plasmid hph 215-739. An hph-specific riboprobe template was made by HindIII digestion of hph(no ψ). An HaSV-specific riboprobe template, complementary to the v-rasH gene, was made by XbaI digestion of a Bluescript (Stratagene)-based plasmid containing the insert of pBS9 (19).
Dissociation of dimeric RNAs.
RNA pellets were resuspended in R buffer (26). RNA was incubated at various temperatures for 8 min. One-tenth volume of 10X RNA gel loading dye (26) was then added to each RNA sample, and the sample was loaded directly onto the agarose gel. Percent dimerization was determined by phosphorimaging or densitometry analysis of these native Northern blots. (These two methods of quantitation were compared on several samples and found to give virtually identical results.) Dimeric RNA was defined as the radioactivity in the lane from the top of the lane until just above the dissociated monomeric RNA band. The dissociated monomeric RNA was defined as the clear monomeric band as well as any significant radioactivity below this position. The percent dissociation was determined for each lane as the amount of monomeric RNA/(monomeric plus dimeric) RNAs. The value of this fraction at the highest temperature used was then set to 100% despite the persistence of some radioactivity in the dimeric region of the blot. The Tm is the temperature at which the percent dissociation was 50%. In all graphs of dissociation (except bar graphs), all of the results were obtained in the same experiment. In contrast, bar graphs show the means of the Tms obtained in multiple experiments, with error bars indicating the standard deviations.
Real-time reverse transcription-PCR.
Copies of MLV genomic RNA and hph mRNA were enumerated by real-time reverse transcription-PCR with the ABI 7700 instrument (Applied Biosystems). After digestion with RNase-free DNase (Promega), the RNA was treated with 2.7 M guanidinium isothiocyanate and precipitated with ethanol in the presence of linear polyacrylamide. The precipitate was redissolved in water containing 1 mM dithiothreitol (Promega) and 0.7 U of RNase Out (Invitrogen)/μl. RNAs used in the construction of the standard curves were, for MLV RNA, a 5-kb transcript of pMXH DNA (22), and for hph RNA, a 0.5-kb transcript that was generated from a T7 promoter-containing PCR product with an hph-specific primer set (25). The RNA concentrations in these standards were measured by their absorbance at 260 nm.
Tenfold serial dilutions of the RNAs were first copied into DNA in 30 μl containing 5 mM MgCl2, 0.05 mM of each deoxynucleoside triphosphate, 1 mM dithiothreitol, 0.15 μg of random primers (Promega), 1× PCR buffer II (Invitrogen), 20 U of RNase Out (Invitrogen), and 20 U of Superscript II reverse transcriptase (Invitrogen). These reactions were performed for 15 min at 25°C, 40 min at 42°C, and 10 min at 85°C; 30 μl of Taqman PCR 2× Universal Master Mix (Applied Biosystems), containing 600 nM each of the forward and reverse primers and 100 nM Taqman probe, was then added to each tube, and the tubes were heated for 10 min at 95°C and then exposed to 45 cycles of 15 s at 95°C and 1 min at 60°C. Control reactions containing buffer without reverse transcriptase were run for each sample; these controls gave values significantly lower than 8 × 102 copies (the lowest value used in constructing the standard curve) for either hph or MLV RNAs.
The MLV-specific primers were 5′AGGAAAGCGGTATCGCTGGAC (forward primer, nucleotides 2399 to 2419) and 5′GAGTGGGTGACCTTACCGGT (reverse primer, complementary to nucleotides 2464 to 2445) and the probe, modified at its 5′ end with 6-carboxy fluorescein (FAM) and at its 3′ end with 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA), was 5′ACGGATCGCAAAGTACATCTAGC (nucleotides 2421 to 2443). hph RNA was quantitated as described (25). Measurements were also made with a primer-probe set detecting MLV Ψ sequences: forward primer, 5′TCCAATAAACCCTCTTGCAG, nucleotides 44 to 63; reverse primer, 5′AGGAGACCCTCCCAAGGAAC, complementary to nucleotides 107 to 88; probe, modified as above, 5′TTGCATCCGACTTGTGGTCTCGC (nucleotides 64 to 86). This reagent set detects hph RNAs containing Ψ sequences as well as MLV RNA and gave results nearly identical, within a factor of 2, to the larger of the two results, hph specific and MLV specific, when used with samples containing both hph RNA and MLV.
Immunoblotting.
Quantities of virions in virus preparations were compared by immunoblotting analysis with rabbit anticapsid (CA) antiserum. Relative amounts of virus in different preparations were determined by comparison with a standard curve, generated from serial dilutions of a single virus preparation in the same immunoblotting experiment. RNA copy numbers were corrected by these small (<2-fold) differences in the levels of virus in different preparations produced in parallel.
RNA folding.
Sequences were entered into Zuker's mFold program (67) with no constraints. The sequences for the dimeric structures were entered as duplicate sequences joined by approximately 75 N (or A) residues.
RESULTS
Packaging of hph mRNAs in virions.
Previous work has analyzed the effects on packaging of insertion of sequences from the 5′ region of a retroviral genome into the 3′UTR of a nonviral mRNA. It was found (1, 2, 54) that mRNAs with these insertions are encapsidated into virions formed by the homologous retroviral Gag protein. The primary purpose of the experiments presented here was to determine the state of these RNAs within the virions. In all of these experiments (except that in Fig. 2D), 293T cells were first stably transfected with a plasmid expressing hph mRNA that contains an insert derived from MLV or a related virus (shown in Fig. 1). After the stably transfected cells were isolated by selection with hygromycin, they were transiently transfected with an MLV-derived proviral clone encoding wild-type MLV Gag, and virus was collected and analyzed.
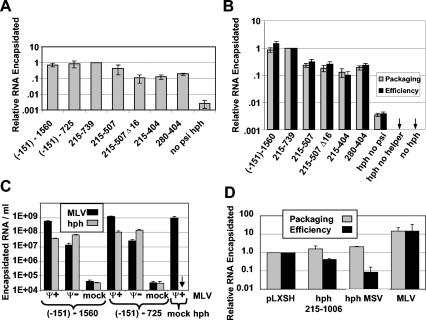
FIG. 2.
Packaging levels and efficiencies of chimeric and viral RNAs. (A) Packaging of hph mRNA by ψ− MLV as a function of the viral insert. Cells that had been stably transfected with hph constructs were transiently transfected with ψ− MLV. The amount of hph RNA in virus particles was then measured and normalized to that in hph 215-739. The results shown are averages from several experiments. (B) Packaging levels and efficiencies of hph RNAs. The amounts of hph RNA in virions and in cell lysates were measured after transfection as in A; efficiency represents the ratio of the amount in virions to the amount in the lysate. Results were normalized to that for hph 215-739. Data are averages of multiple measurements from a single experiment but are representative of several experiments. (C) Packaging of hph and MLV RNAs. Cells that had been stably transfected with hph plasmids (indicated in the hph line below the figure) were transiently transfected with ψ+ (wild type), ψ−, or no (mock) MLV as indicated in the MLV line below the figure. The amounts of hph and MLV RNA in viral pellets from these cells were measured. The results shown are averages of several experiments. (D) Packaging levels and efficiencies of pLXSH genomic RNA, hph RNAs, and MLV genomic RNA. Cells were transiently cotransfected with a mixture of ψ− MLV DNA and either pLXSH, hph 215-1006, hph MSV, or wild-type MLV DNA. Two days later, supernatants were collected and cells were lysed. In samples containing pLXSH, hph 215-1006, or hph MSV, the amounts of hph RNA in virions and cells were measured; in the sample with MLV, MLV RNA was measured. The results are normalized to pLXSH. In all cases (panels A to D), the values have been corrected for minor differences in the CA protein content of different samples.
We began these studies by determining the level of packaging of the RNAs in MLV particles, using real-time reverse transcription-PCR. hph mRNAs containing substantial stretches of MLV-derived sequence were packaged by ψ− MLV. Virions were collected and quantitated by immunoblotting, so that equal amounts of virus could be compared with respect to hph mRNA content. As shown in Fig. 2A, the levels in the viral pellets of hph mRNAs containing nucleotides −151 to 1560 or nucleotides 215 to 739 were very similar to each other (for ease of comparison, the values are all normalized to that of hph 215-739). They were nearly 1,000 times higher than that of hph mRNA lacking a viral insert in its 3′ UTR (no ψ). In turn, the level of this ψ− mRNA in the pellet was ≈30-fold higher than that in a “mock” pellet from culture fluid of cells that were not expressing MLV Gag (data not shown). These two controls provide strong evidence that the hph RNAs in the pellets are associated with virus particles rather than with cell debris and that they are packaged in the particles in response to their ψ sequences. In addition, they show that extensions either 5′ or 3′ of the core 215 to 739 ψ sequence have no significant effect on the packaging of hph mRNA. Packaging of hph mRNAs with other viral inserts was also measured here but will be discussed below.
It seemed possible that the differences in encapsidation of the hph mRNAs were really due to differences in the steady-state level of these RNAs within the cells. To test this possibility, we performed an additional experiment in which we measured the levels of these RNAs in lysates of the cells from which the virus particles were produced. Figure 2B shows the normalized amounts of hph mRNA in the virions and also the efficiency of encapsidation (the level in the virions divided by the level in the cell lysate). We found that their levels in the different lysates were virtually identical; this is evident from the fact that, for each construct, the normalized value for efficiency is very similar to the normalized packaging level.
We also compared the levels of hph RNA in virions with the levels of MLV genomic RNA. Cells expressing hph mRNA with the −151 to 1560 insert were transfected with ψ− or ψ+ MLV proviral clones. Virions produced by these cells were collected, and their RNA was analyzed by real-time reverse transcription-PCR. As shown in Fig. 2C, the RNA preparations from the particles produced from either ψ+ or ψ− MLV contained ≈4 × 107 to 6 × 107 hph (−151)-1560 RNA molecules per ml of culture supernatant (the copy numbers have been corrected for small differences in the amount of virus in the different preparations, as described in Materials and Methods). In contrast, the MLV genomic RNA in the virions produced by ψ+ MLV was present at ≈6 × 108 copies of MLV genomic RNA per ml of culture supernatant. This value is similar to that present in an MLV preparation containing no hph RNA (mock hph) and ≈50-fold higher than that in the virus preparation made with ψ− MLV. As shown in the figure, the packaging of hph mRNA with the (−151)-725 insert was very similar to that observed with the (−151)-1560 insert. Thus, while ψ sequences cause the encapsidation of hph mRNA molecules, the amounts of these RNAs packaged are significantly lower than that of MLV genomic RNA. These results are in contrast with those obtained with neo mRNAs by Adam and Miller (1), who reported that their mRNAs were packaged to the same extent as viral RNA; we do not know the reason for this difference.
It is interesting (Fig. 2C) that the MLV and hph RNAs do not appear to be in significant competition with each other for inclusion in virions, since nearly as much hph RNA is packaged by ψ+ MLV RNA as by ψ− MLV RNA. Conversely, the level of ψ+ MLV RNA is not significantly affected by the presence of hph RNA; hph packaging also had no detectable effect on the level of ψ− MLV RNA (data not shown).
We also analyzed the encapsidation of RNAs containing chimeric ψ sequences, derived partly from MLV and partly from the MSV 124 isolate of MSV, as previously described by Adam and Miller (1). These RNAs included hph MSV, which is identical to the ψ+ RNA described by these authors except that the viral insert is 32 nucleotides shorter at its 3′ end and neo has been replaced by hygro, and a retroviral vector, pLXSH, which contains hph and the same ψ sequence as hph MSV (42). Unlike hph MSV, however, pLXSH RNA is an authentic retroviral genome, containing all of the cis-acting sequences required for the retroviral replication cycle and containing its ψ sequences in their normal locations near the 5′ end of the RNA rather than in ithe 3′ untranslated region.
As shown in Fig. 2D, the packaging of pLXSH RNA was approximately the same as that of hph MSV. In addition, we measured the packaging of hph 215-1006 RNA, which is identical to hph MSV RNA except for the divergence between MSV and MLV (discussed below). This RNA was also packaged to the same extent as hph MSV. The levels of all of these RNAs in the virus particles were ≈10-fold lower than that of MLV genomic RNA. Thus, the deficiency in encapsidation of hph mRNAs that contain packaging signals, relative to MLV genomic RNA, does not appear to be due to the unusual configuration of these RNAs, in which ψ is in the 3′ untranslated region.
We also measured the steady-state levels of these RNAs in virus-producing cells. We found that these hph-containing RNAs were actually more abundant in the cells than MLV genomic RNA, as indicated in Fig. 2D by the fact that the efficiency of encapsidation of the latter was at least 10-fold higher than that of the hph-containing RNAs. It should be noted that this experiment was performed by transiently cotransfecting 293T cells with ψ− MLV plasmid and the hph expression plasmid (or wild-type MLV plasmid). Under these conditions, we expect that both plasmids will be taken up by the same cells, so that efficiencies of encapsidation of hph RNA and MLV RNA can be compared directly. These results indicate that the relative deficiency in packaging of the hph-containing RNAs is not due to their scarcity in the cell.
hph mRNAs are packaged as dimers.
It was of considerable interest to determine whether ψ-containing mRNA molecules in virions are, like authentic viral RNAs, held in a dimeric linkage. We therefore extracted RNA from particles formed by transfection of either ψ− or ψ− PR− MLV into cells containing hph mRNA with the large [(−151)-1560] MLV insert and analyzed it by electrophoresis under nondenaturing conditions. Lanes 15 and 22 of Fig. 3A show the results obtained by Northern analysis with an hph probe. It can be seen that the hph-reactive RNA species migrated as a very diffuse band (labeled DD for diffuse dimer) under these conditions. We also heated these RNA preparations to different temperatures before electrophoresis; the results are shown in Fig. 3, lanes 16 to 21 and 23 to 28. We found that the hph mRNA packaged by ψ− PR− MLV (Fig. 3, lanes 16 to 19) migrated slightly more rapidly, as a broad but discrete band (labeled D for dimer), after exposure to 50°C. As the RNA was incubated at progressively higher temperatures, an increasing fraction of it was converted to a discrete, rapidly migrating, band. The mobility of this band corresponds to a molecular size of ≈3 kb (determined by comparison to a standard curve [not shown] with RNA markers of known lengths on the same gel). Finally, when the preincubation temperature was raised to 60°C (Fig. 3, lane 20), nearly all of the hph RNA was shifted into this band. Exposure to 65°C produced no further change in this pattern. The data suggest that hph mRNA is packaged as a dimer; in turn, this dimer may be part of a larger complex (the very diffuse material seen in the unheated sample [Fig. 3, lane 15]); alternatively, the dimers may be heterogenous in mobility because they are in a variety of different conformations. Incubation at 50°C disrupted the larger complex (or reduced the conformational heterogeneity of the dimers), revealing the dimer band; incubation at higher temperatures causes the dissociation of the dimer to the 3-kb hph mRNA monomer.
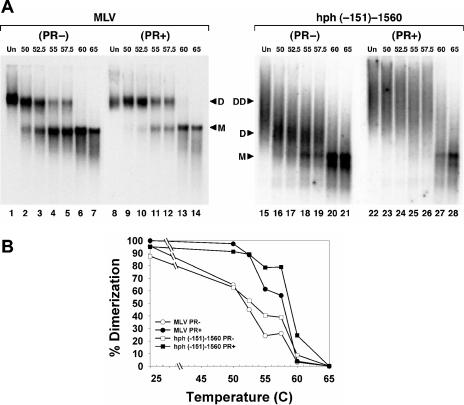
FIG. 3.
Dissociation of dimeric RNAs of MLV and of hph (−151)-1560 mRNA following extraction from virions. (A) Nondenaturing Northern analysis, with MLV-specific (lanes 1 to 14) or hph-specific (lanes 15 to 28) probes. Lanes 1 to 7, PR− MLV; 8 to 14, wild-type MLV; 15 to 21, hph mRNA packaged by PR− ψ− MLV; 22 to 28, hph mRNA packaged by ψ− MLV. RNA preparations were heated to the indicated temperatures before electrophoresis. DD, diffuse dimers; D, dimers; M, monomers. (B) Graph of dissociation data from panel A.
The effects of heat on the hph mRNA encapsidated in mature particles by ψ− PR+ MLV are shown in lanes 23 to 28 of Fig. 3A. In this case, heating to temperatures as high as 57.5°C (Fig. 3A, lanes 23 to 26) had no significant effect on the broad, heterodisperse hph RNA pattern. However, when the RNA was exposed to 60°C (Fig. 3A, lane 27), it was abruptly shifted to the 3-kb band. Incubation at 65°C (Fig. 3A, lane 28) produced the same pattern as at 60°C.
For comparison with these results, we also monitored the dissociation of the dimers of MLV genomic RNA extracted from ψ+ PR− and ψ+ PR+ particles. As can be seen in Fig. 3A, lanes 1 to 7, MLV RNA from PR−particles migrated as a discrete band after exposure to 37°C (Fig. 3A, lane 1); incubation at temperatures between 50 and 57.5°C (Fig. 3A, lanes 2 to 5) caused the shift of progressively increasing amounts of this RNA into a new band migrating considerably faster than that seen in the 37°C sample. This shift represents the conversion of immature dimeric RNA into monomeric RNA; the conversion was complete by 60°C (Fig. 3A, lane 6), and the monomer band was unaffected by a further increase in temperature to 65°C (Fig. 3A, lane 7).
MLV RNA extracted from PR+ particles (Fig. 3A, lanes 8 to 14) showed a somewhat different dissociation pattern. In this case, the dimers were not significantly dissociated until the temperature was raised to 55°C (Fig. 3A, lane 11). However, they were completely converted to monomers by incubation at 60°C (Fig. 3A, lane 13); again, increasing the temperature to 65°C had no additional effect on these monomers (Fig. 3A, lane 14). This difference in the thermostability of MLV RNAs from PR− and PR+ virions is in good agreement with our earlier findings (26).
The appearance of monomeric RNAs as a result of stepwise increases in temperature is shown graphically in Fig. 3B. It is striking to compare the effects of progressive increases in temperature on encapsidated hph RNAs with those on MLV RNAs. When either hph or MLV RNAs were extracted from immature virions, they were partially converted to free monomers by temperatures as low as 50°C, and further increases in temperature raised the proportion of monomers; the conversion to monomers was complete at 60°C. In contrast, RNAs from mature virions were largely unaffected by heat until the temperature was raised to 55°C (genomic RNA) or 57.5°C (hph RNA); they were then converted relatively abruptly to monomers. The congruence of the responses of hph and MLV RNAs to heating is strong evidence that the hph RNAs in virions are joined in a dimeric linkage similar or identical to the normal linkage of MLV genomic RNAs. In some, but not all, experiments, a discrete dimeric species of hph RNA was seen after exposure of the RNA to intermediate temperatures (for example, see Fig. 3A, lanes 16 to 19, and Fig. 5A below).
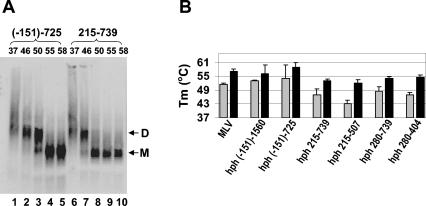
FIG. 5.
Dissociation of dimeric RNAs of hph mRNAs containing truncated MLV-derived inserts. (A) Dissociation of dimeric RNAs of hph (−151)-725 (lanes 1 to 5) and hph 215-739 (lanes 6 to 10) mRNAs packaged by ψ− PR− MLV, monitored by nondenaturing Northern analysis. (B) Tms of hph mRNAs containing truncated MLV inserts, packaged by ψ− MLV or ψ− PR− MLV. The figure also shows the Tms of PR− and wild-type MLV genomic RNAs. Gray bars, RNAs from PR−virions; black bars, RNAs from PR+ virions. Values shown in panel B are averages from several experiments.
Dimeric linkage in encapsidated hph RNA is a function of the viral insert.
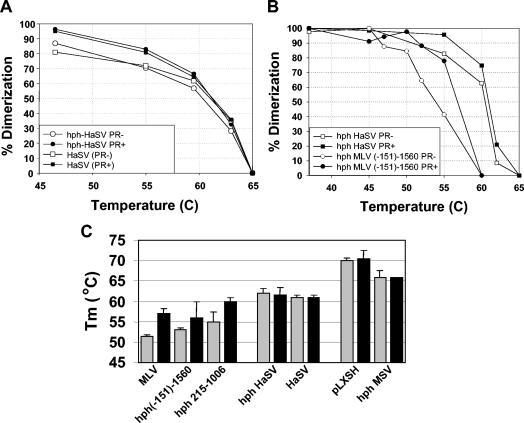
The dimeric RNAs of different gammaretroviruses differ from each other in thermostability. Thus, the dimers of HaSV genomic RNA are quite stable, with little if any difference between HaSV dimers from immature virions and those from mature virions (21). We therefore inserted nucleotides (−151)-942 of HaSV into the 3′ UTR of hph mRNA. We found that, as in Fig. 3, the encapsidated hph mRNA migrated as a diffuse band upon electrophoresis under nondenaturing conditions, but was converted to a monomer upon heating (data not shown). Quantitation of the appearance of the monomeric band, along with quantitation of dissociation of authentic HaSV genomic dimers, led to the results shown in Fig. 4A. It can be seen that the complexes containing hph mRNA with the HaSV insert exhibited the same thermostability as dimers of HaSV genomic RNA. The complexes isolated from immature and mature virions had similar or identical thermostabilities; both were more thermostable than those with MLV sequences in their 3′ UTR (Fig. 4B).
FIG. 4.
Dissociation of dimeric RNAs of hph RNAs compared with genomic RNAs of HaSV, MLV, and pLXSH. (A) Graph of dissociation of HaSV RNA and of hph HaSV mRNA packaged by ψ− MLV or ψ− PR− MLV. (B) Graph of dissociation of hph HaSV mRNA and of hph (−151)-1560 mRNA packaged by ψ− MLV or ψ− PR− MLV. (C) Tms of MLV genomic RNA and hph mRNAs containing MLV-derived inserts; HaSV genomic RNA and hph HaSV RNA; and pLXSH genomic RNA and hph MSV RNA (which contains the same MLV-MSV chimeric ψ region as pLXSH). Gray bars, RNAs from PR− virions; black bars, RNAs from PR+ virions. Values shown in panel C are averages from several experiments.
We also tested the state of hph MSV RNA containing a chimeric ψ insert derived partly from MLV and partly from MSV (Fig. 2) following isolation from immature and mature virions (1). We found that this mRNA was in complexes that were extremely thermostable (Fig. 4C); as with HaSV, there was little if any difference in stability between the complexes from immature and mature virions. The figure shows that these properties were specifically conferred on the mRNA by the MSV insert, since the corresponding nucleotides from MLV (nucleotides 215 to 1006) yielded far more fragile dimers, in which virus maturation induced significant stabilization. Similar properties, i.e., very high thermostability, with no detectable stabilization associated with virus maturation, were also observed in the dimeric RNAs of the pLXSH vector (Fig. 4C), which contains the same chimeric ψ region as hph MSV.
Packaging and dimerization of hph mRNAs containing truncated MLV inserts.
We also analyzed the effects of a limited number of 5′ and 3′ truncations of the MLV ψ sequence on the packaging of hph mRNAs. We found (Fig. 2A) that truncation of the insert from nucleotides 215-739 to nucleotides 215-404 reduced the level of hph mRNA in virions about 10-fold. In addition, we analyzed the dimeric linkages in hph mRNAs containing truncated inserts. One example of these results is shown in Fig. 5A. It can be seen that dimers of hph (−151)-725 mRNA are significantly more thermostable than those of hph 215-739 mRNA following isolation from immature virions. These data imply that sequences between nucleotides −151 and 215 contribute to the stability of immature dimers.
In all cases in which hph mRNAs were extracted from PR+ particles, they were found to be packaged as dimers; as shown in Fig. 5B, these dimers all had rather similar thermostabilities, although dimers whose inserts began at nucleotide −151 appeared to be slightly more stable than those beginning with nucleotide 215 or 280. In contrast, there were distinct differences in the stabilities of the dimers isolated from PR− particles. It was found that immature dimers formed by nucleotides 215-739 were somewhat labile. Immature dimers containing either nucleotides (−151)-725 or nucleotides 215-1006 (Fig. 4C) were more stable than those containing only nucleotides 215-739; thus, viral sequences on either the 5′ or the 3′ side of the nucleotide 215-739 “core” can serve to stabilize immature dimeric linkages. The 125 bases between nucleotides 280 and 404 are evidently sufficient for the formation of a dimeric linkage.
Stem-loop B is crucial for the stability of immature dimers but not mature dimers.
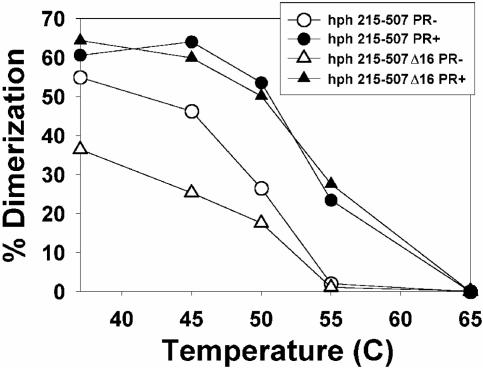
As noted above, one of the elements thought to contribute to retroviral dimerization is the kissing loop (nucleotides 278 to 303, also referred to as stem-loop B of MLV RNA). We generated an hph mRNA containing MLV nucleotides 215 to 507 but with a deletion of nucleotides 290 to 305. As shown in Fig. 2A, the deletion reduced the encapsidation of the RNA ≈5-fold. When the RNA was encapsidated by PR+ MLV, it was present in dimers with the same thermostability as that of control hph mRNAs containing stem-loop B (Fig. 6, upper curves, and Table 1). However, when it was packaged by PR− MLV, less than half of the hph RNA lacking stem-loop B was dimeric, even without heating above 37°C (Fig. 6, lower curves, and Table 1). (The percent initial dimer values for this RNA shown in Table 1 have relatively large standard deviations, so that one might doubt the significance of the effect of the deletion. However, in each individual experiment, there was a significant difference between the percent initial dimers of the deleted and undeleted RNAs.) Thus, nucleotides 290 to 305 (i.e., the deleted bases) are evidently crucial for the stability of the immature dimeric linkage but not the mature linkage.
FIG. 6.
Dissociation of dimers of hph 215-507 with or without a deletion of nucleotides 290 to 305, following packaging by ψ− MLV or ψ− PR− MLV.
TABLE 1.
Effect of stem-loop B on thermostability of encapsidated hph 215-507 RNAa
| RNA |
Tm (°C)
|
% initial dimers
|
||
|---|---|---|---|---|
| PR− | PR+ | PR− | PR+ | |
| 215-507 | 43 ± 1.5 | 52 ± 1.5 | 62 ± 7.5 | 79 ± 18 |
| 215-507Δ16 | <37 | 51.5 ± 2.2 | 35 ± 8 | 68 ± 7 |
The Tms of hph 215-507 dimers with or without the deletion of nucleotides 290 to 305 were determined following extraction from either ψ− or PR− ψ− virions. The table shows means and standard deviations for Tm and for percent dimers in the sample incubated at 37°C. A Tm could not be obtained for the PR− sample containing the deletion because it contained <50% dimers following incubation at 37°C.
DISCUSSION
The results presented here can be briefly summarized as follows. First, hph mRNA molecules containing large inserts from the 5′ end of MLV-related genomes in their 3′ UTR are packaged by MLV Gag, as previously reported by others for neo mRNAs. However, our measurements show that the level of these mRNAs in the virions is only ≈7 to 10% of that of genomic RNA (Fig. 2C). Second, the hph mRNAs are apparently packaged as dimers; the thermostabilities of these dimers indicate that the monomers are joined by the same linkage as that in dimers of the genomic RNA from which the insert was derived. Thus, mRNA molecules containing HaSV- or MSV-derived inserts have a considerably more stable dimeric linkage than those containing MLV inserts. While the dimers formed from MLV-derived sequences are significantly stabilized upon virion maturation, those from HaSV or MSV sequences are not. Third, elimination of stem-loop B (the kissing loop) from the hph mRNA insert dramatically weakens the linkage between the mRNAs in immature virions but has little effect on its stability in mature virions. Fourth, sequences between nucleotides 280 and 404 of MLV RNA are sufficient for the formation of mature dimers with nearly the same stability as those of genomic RNA. In turn, these data have several important implications concerning the packaging and dimerization of retroviral genomic RNAs.
Packaging of nonviral mRNA molecules containing ψ/DLS sequences.
We found that MLV sequences between nucleotides 215 and 739 are sufficient to increase the encapsidation of hph mRNA nearly 1,000-fold over background packaging (Fig. 2A) but still ≈10- to 15-fold less than MLV genomic RNA (Fig. 2C). One explanation for this deficit in packaging of these mRNAs could be that the great majority of hph mRNA molecules are sequestered in polyribosomes, unavailable for incorporation into nascent virions. Another possibility is that a ψ sequence in the 3′ UTR of an mRNA is less effective for encapsidation than the same sequence placed near the 5′ end of an RNA molecule, as in genomic RNA. However, the fact that pLXSH genomic RNA is also packaged ≈10-fold less than MLV genomic RNA (Fig. 2D) suggests that neither of these hypotheses is correct. It is also conceivable that hph sequences (unlike neo sequences) (1) inhibit encapsidation, perhaps via long-range interactions with the ψ moiety of the chimeric mRNA. Finally, MLV genomic RNA may contain additional sequences, located outside the 5′ end region, that contribute to efficient encapsidation. We also noted that removal of nucleotides 404 to 739 from hph 215-739 reduced encapsidation ≈10-fold (Fig. 2A); thus, nucleotides 215 to 404 increase the packaging of hph mRNA ≈50-fold above the background. This is quite consonant with the fact that deletion of nucleotides 212 to 563 from genomic RNA reduces its packaging 50- to 100-fold (Fig. 2C) (54).
mRNA molecules containing ψ/DLS sequences are encapsidated in pairs.
The fact that the encapsidated hph mRNAs are in dimers even when the majority of particles do not contain them (Fig. 2C) is strong evidence that they are incorporated into virions in pairs rather than independently of each other. One simple explanation for this result is that packaging of ψ-containing RNAs depends upon their dimerization; this would be in accord with previous suggestions (10) that some dimeric structure is an element of the packaging signal. Sakuragi and colleagues (58) recently described elegant experiments with human immunodeficiency virus type 1 whose results lend support to this proposal.
Sequences near the 5′ end of the MLV genome are sufficient for the normal immature and mature dimer linkages.
This basic conclusion is quite consistent with earlier electron microscopic localization of the mature linkage (7, 8, 31, 37, 47). To our knowledge, however, this is the first information to be presented concerning the locus of the immature dimer linkage. This linkage is of particular interest because genomic RNAs are, presumably, normally packaged as immature dimers; it is the immature linkage which may be part of the packaging signal.
The MLV ψ region (extending approximately from nucleotides 215 to 563) contains three conserved stem-loops: stem-loop B (the putative kissing loop, comprising nucleotides 278 to 303) and stem-loops C and D (nucleotides 310 to 352 and 355 to 374, respectively) (Fig. 1A). The loops of stem-loops C and D both consist of the sequence GACG; a pair of stem-loops with this loop sequence are conserved throughout the gammaretroviruses (36), and these stem-loops are important elements in the encapsidation signal (6, 23, 43, 44, 66). A more proximal stem-loop, termed stem-loop B′, is present at nucleotides 204 to 228; like stem-loop B, this stem-loop could engage in loop-loop base-pairing interactions (48). Studies with transcripts have supported a role for stem-loop B in dimer formation (27); significantly, stem-loops C and D also contribute to the dimerization of MLV transcripts (16) and can independently form a remarkably stable dimer involving pairing between the CG portions of the GACG tetraloops (18, 34). In addition, stem-loop B′ has been reported to contribute to the dimerization of MLV transcripts (40, 48) and, in combination with stem-loop B, to the stability of mature MLV dimers (40).
One advantage of the experimental system used in the present work is that the viral sequences do not perform any known function in the hph mRNAs and can thus be varied at will. We exploited this property by making a series of truncations of the viral inserts. The truncations removed sequences from either side of the core ψ region (nucleotides 215 to 739 of MLV) and also eliminated sequences within this region. We found (Fig. 5B) that extension of this region in the 5′ direction from nucleotide 215 to −151 slightly increased the thermostability of the linkage between hph mRNAs extracted from mature virions. Therefore, sequences within this 5′ region (which includes stem-loop B′) evidently make a small but significant contribution to the stability of mature dimers or can independently form a linkage that is slightly more stable than that formed in the core ψ region.
We also found (Fig. 5B) that an extension on the 5′ side (to nucleotide −151) of the core sequence is required for the normal stability of the mRNA dimers from immature particles. Again, this result is consistent with the possibility that stem-loop B′ contributes to the stability of immature MLV dimers. Similarly, extension of the insert from nucleotides 739 to 1006 increases the stability of immature dimers (Fig. 4C); thus, sequences 3′ of the core can also enhance the stability of these dimers.
It should be noted that the correspondence between the dissociation temperature of the hph dimers and genomic RNA dimers suggests that the viral inserts in the hph RNAs are the site of the strongest linkage in genomic dimers. However, there may well be other, weaker linkages elsewhere in genomic RNAs, as suggested by Ortiz-Conde and Hughes (49). Indeed, the results of Tchenio and Heidmann (63) lend strong support to this idea.
Kissing loop is essential for the immature but not the mature dimer linkage.
We also found that a deletion eliminating stem-loop B dramatically reduced the stability of the mRNA dimers extracted from immature virions. This result suggests that base-pairing between the palindromic stem-loop B loops in the two monomers is important in the immature dimeric linkage. However, it is striking that the mRNAs containing this deletion must still be packaged in pairs, since they form dimeric linkages with normal thermostability when the particle undergoes maturation (Fig. 6, Table 1). Therefore, the linkage responsible for the pairwise coencapsidation of ψ/DLS-containing RNAs is independent of stem-loop B; the linkage that gives mature dimers their stability also does not require stem-loop B.
One hypothesis that is consistent with all these results is that Gag proteins normally select dimers for encapsidation. These dimers could be formed by intermolecular base-pairing between one or both of the GACG loops (18, 34). While pairing between the stem-loop B loops and/or stem-loop B′ loops might lend additional stability to these hypothetical preimmature dimers, it is not necessary for their formation. We also found that stem-loop B is not absolutely necessary for encapsidation but does enhance the level of RNA packaged (Fig. 2A). It is striking to note that that MLV NC protein binds with high affinity to an RNA comprising stem-loops B, C, and D but not to RNAs containing only two of them (18).
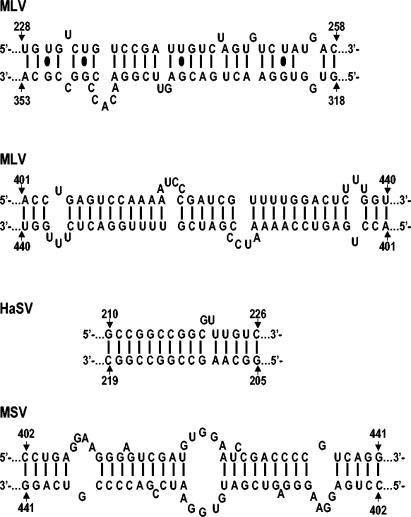
Presumably, the stabilization in RNA dimers accompanying maturation represents the formation of a structure containing more intermolecular base pairs than the immature dimer. The available data do not allow us to identify these base pairs, but the sequences within the core MLV ψ region can be folded in silico into remarkably extensive base-paired structures. Two of these potential linkages are shown in Fig. 7 below.
FIG. 7.
Regions of possible extended base-pairing in MLV, HaSV, and MSV.
Dimers formed from HaSV or MSV sequences differ from MLV dimeric RNAs.
It would also be of interest to understand the RNA dimers formed by HaSV- and MSV-derived inserts. In both of these cases, there is little if any difference in the thermostabilities of dimers from immature particles and those from mature particles. This suggests that in MSV and HaSV, there is no kinetic trap arresting dimerization at an intermediate stage until free NC is available in the mature virion, as is seen with MLV, human immunodeficience virus type 1, and avian retroviruses.
Figure 8 shows sequence alignments between the putative ψ/DLS regions of MSV (Fig. 8A) and HaSV (Fig. 8B) and that of MLV. As pointed out previously (32, 53), there is very little sequence divergence between MLV and MSV except that nucleotides 408 to 458 of MLV are replaced in the MSV genome by nucleotides 408 to 439. (This MSV-specific sequence is closely related to the murine VL30 sequence BVL-1 [GenBank accesssion no. X17124] [32].) In contrast, there is almost no sequence similarity between the HaSV and MLV sequences (beyond the 5′-terminal ≈220 nucleotides, which are derived from MLV, as is the 3′ end of the genome) (12, 19), except for HaSV nucleotides 289 to 320 and 329 to 351, which closely resemble MLV nucleotides 309 to 340 and 349 to 371, respectively: these are stem-loops C and D, each crowned by a GACG tetraloop. It is remarkable that the HaSV sequence, with so little identity to that of MLV, is encapsidated efficiently by MLV Gag proteins; this observation again highlights the importance of stem-loops C and D in encapsidation, although HaSV and MLV RNAs may well have additional resemblances in three-dimensional structure that are not evident in sequence alignments.
FIG. 8.
Alignments between MLV sequences and (A) MSV sequences and (B) HaSV sequences. In both panels A and B, the top line is the MLV sequence. The GACG loops in stem-loops C and D are indicated by horizontal lines above the sequence.
Inspection of the MSV-specific sequence does not yield any obvious answers to the questions of why these RNAs form very stable dimers or why, unlike MLV, they form stable dimers even in immature particles. However, it is interesting that a 40-base stretch containing the MSV-specific replacement (MSV nucleotides 402 to 441) contains extensive self-complementarity (Fig. 7). This could lead to intermolecular base-pairing; the structure has a single large bulge and several smaller bulges and contains 18 G:C and 10 A:U base pairs in a total length of 40 bases. The corresponding region in MLV (MLV nucleotides 401 to 440) is also part of a very long imperfect palindrome and could form an intermolecularly base-paired structure containing 16 G:C and 16 A:U base pairs (see Fig. 7).
We previously noted (21) that HaSV RNA can potentially form a stem-loop consisting of nucleotides 205 to 226 and that the loop sequence in this stem-loop, GGCC (nucleotides 213 to 216), is a palindrome. (This hypothetical stem-loop would have a bulge at nucleotides 220 to 221. Rasmussen et al. [52] concluded, on the basis of structure probing of HaSV transcripts, that the stem actually comprises nucleotides 204 to 232, with a bulge between nucleotides 220 and 226.) This structure could well be a kissing loop and serve as an initiation site for dimerization; in fact, replacement of G with A at nucleotide 217 drastically impairs dimerization of HaSV-derived transcripts (64). In addition, the bulge might destabilize the stem, so that the kissing complex is spontaneously converted to an extended dimer structure, without the need for virus maturation or free NC protein. Indeed, Kolb et al. have shown that bulged residues can destabilize stems, so that loop-loop helices can proceed to more stable structures (35). The double-stranded region in this extended dimer, in which nucleotides 205 to 219 of one strand are paired with nucleotides 210 to 226 of the other, would contain 12 G:C and two A:U base pairs, with only a single two-base bulge in one of the strands (shown in Fig. 7). It is interesting that this HaSV stem-loop is in the same position as stem-loop B′ of MLV (MLV nucleotides 204 to 228).
We also observed that hph mRNA is significantly more abundant in cell extracts than MLV genomic RNA, despite the fact that it is packaged to a significantly lower extent than the latter (Fig. 2D). The relative inefficiency of ψ+ hph mRNA packaging might suggest that some MLV genomic RNA molecules are somehow protected from incorporation into polyribosomes, so that they are available for encapsidation into nascent virions. This would be in agreement with an earlier proposal by Levin and Rosenak (39).
It is interesting to compare our results with those of prior studies. We found that the mRNAs and MLV genomic RNA do not significantly compete with each other for encapsidation, since the level of hph mRNA was virtually unaffected by rescue with wild-type rather than ψ- genomic RNA, and conversely, packaging of hph mRNA did not reduce the level of viral RNA (Fig. 2C). Similar results were reported by Adam and Miller (1) and, in analogous experiments in an avian system, by Linial and colleagues (2, 3).
We also found that deletion of stem-loop B from hph mRNA reduces its encapsidation (Fig. 2A); however, when the deleted RNAs are isolated from mature virions, they are in dimers with the same stability as the intact control RNAs (Fig. 6, Table 1). Several studies have shown that the deletion or alteration of the kissing loop from human immunodeficiency virus type 1 genomic RNA has analogous effects, reducing packaging to some extent without affecting the stability of the mature encapsidated dimers (9, 13, 38, 50). Another laboratory reported an effect of a kissing loop deletion on the stability of mature dimers (30). To our knowledge, however, no data have been presented on the properties of immature human immunodeficiency virus type 1 dimers containing these mutations. In addition, studies with avian retroviruses show that the analogous structure (the L3 stem-loop) is not absolutely required for encapsidation or replication, although the palindromic loop does confer a moderate selective advantage (4, 5, 17).
In summary, we have found that insertion of the ψ-DLS region from MLV-related viruses into a nonviral mRNA is sufficient for encapsidation of this mRNA in dimeric form. This result is a genetic demonstration that the most stable dimeric linkage in RNAs of MLV-related viruses is in this region of the RNA. This conclusion is consistent with prior work on mature dimers but is the first evidence that the most stable linkage in immature dimers is also in this region. It should be possible to extend this experimental approach to the analysis of dimeric linkages to the single-nucleotide level.
Acknowledgments
We thank Wei-Shau Hu, David Thomas, and William Fu for generous help with real-time PCR and for the use of the real-time PCR instrument. We also thank Dusty Miller, Douglas Lowy, and Vineet KewalRamani for reagents.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronoff, R., and M. Linial. 1991. Specificity of retroviral RNA packaging. J. Virol. 65:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. D., B. O. Kealoha, and M. L. Linial. 1999. An Mpsi-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 73:8926-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, J. D., and M. L. Linial. 2000. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 74:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks, J. D., A. Yeo, K. Green, F. Cepeda, and M. L. Linial. 1998. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 72:6190-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beasley, B. E., and W. S. Hu. 2002. cis-Acting elements important for retroviral RNA packaging specificity. J. Virol. 76:4950-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, W., Y. H. Chien, S. Chattopadhyay, P. K. Vogt, M. B. Gardner, and N. Davidson. 1978. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 25:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, W., and N. Davidson. 1976. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7:595-607. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout, B., and J. L. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz, R., J. Fisher, and S. P. Goff. 1996.RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 11.Bieth, E., C. Gabus, and J. L. Darlix. 1990. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 18:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, U. H., M. Lai, T. Y. Shih, I. M. Verma, E. M. Scolnick, P. Roy-Burman, and N. Davidson. 1979. Heteroduplex analysis of the sequence relationships between the genomes of Kirsten and Harvey sarcoma viruses, their respective parental murine leukemia viruses, and the rat endogenous 30S RNA. J. Virol. 31:752-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darlix, J. L., C. Gabus, M. T. Nugeyre, F. Clavel, and F. Barre-Sinoussi. 1990. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 216:689-699. [DOI] [PubMed] [Google Scholar]
- 15.Darlix, J. L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 16.De Tapia, M., V. Metzler, M. Mougel, B. Ehresmann, and C. Ehresmann. 1998. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374). Biochemistry 37:6077-6085. [DOI] [PubMed] [Google Scholar]
- 17.Doria-Rose, N. A., and V. M. Vogt. 1998. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 72:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza, V., J. Melamed, D. Habib, K. Pullen, K. Wallace, and M. F. Summers. 2001. Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: implications for genome recognition. J. Mol. Biol. 314:217-232. [DOI] [PubMed] [Google Scholar]
- 19.Ellis, R. W., D. DeFeo, J. M. Maryak, H. A. Young, T. Y. Shih, E. H. Chang, D. R. Lowy, and E. M. Scolnick. 1980. Dual evolutionary origin for the rat genetic sequences of Harvey murine sarcoma virus. J. Virol. 36:408-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, Y. X., W. Fu, A. J. Winter, J. G. Levin, and A. Rein. 1995. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J. Virol. 69:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, Y. X., D. L. Hatfield, A. Rein, and J. G. Levin. 1989. Translational readthrough of the murine leukemia virus gag gene amber codon does not require virus-induced alteration of tRNA. J. Virol. 63:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher, J., and S. P. Goff. 1998. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology 244:133-145. [DOI] [PubMed] [Google Scholar]
- 24.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu, W., and W. S. Hu. 2003. Functional replacement of nucleocapsid flanking regions by heterologous counterparts with divergent primary sequences: effects of chimeric nucleocapsid on the retroviral replication cycle. J. Virol. 77:754-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girard, P. M., B. Bonnet-Mathoniere, D. Muriaux, and J. Paoletti. 1995. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry 34:9785-9794. [DOI] [PubMed] [Google Scholar]
- 28.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 30.Haddrick, M., A. L. Lear, A. J. Cann, and S. Heaphy. 1996. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J. Mol. Biol. 259:58-68. [DOI] [PubMed] [Google Scholar]
- 31.Hoglund, S., A. Ohagen, J. Goncalves, A. T. Panganiban, and D. Gabuzda. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Izmailova, E., and A. Aldovini. 2002. Functional analysis of the murine sarcoma virus RNA packaging sequence. J. Virol. 76:4643-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khandjian, E. W., and C. Meric. 1986. A procedure for Northern blot analysis of native RNA. Anal. Biochem. 159:227-232. [DOI] [PubMed] [Google Scholar]
- 34.Kim, C. H., and I. Tinoco, Jr. 2000. A retroviral RNA kissing complex containing only two G · C base pairs. Proc. Natl. Acad. Sci. USA 97:9396-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolb, F. A., E. Westhof, C. Ehresmann, B. Ehresmann, E. G. Wagner, and P. Romby. 2001. Bulged residues promote the progression of a loop-loop interaction to a stable and inhibitory antisense-target RNA complex. Nucleic Acids Res. 29:3145-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konings, D. A., M. A. Nash, J. V. Maizel, and R. B. Arlinghaus. 1992. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J. Virol. 66:632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kung, H. J., S. Hu, W. Bender, J. M. Bailey, N. Davidson, M. O. Nicolson, and R. M. McAllister. 1976. RD-114, baboon, and woolly monkey viral RNA's compared in size and structure. Cell 7:609-620. [DOI] [PubMed] [Google Scholar]
- 38.Laughrea, M., L. Jette, J. Mak, L. Kleiman, C. Liang, and M. A. Wainberg. 1997. Mutations in the kissing loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 71:3397-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin, J. G., and M. J. Rosenak. 1976. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 73:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ly, H., and T. G. Parslow. 2002. Bipartite signal for genomic RNA dimerization in Moloney murine leukemia virus. J. Virol. 76:3135-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, A. D., D. G. Miller, J. V. Garcia, and C. M. Lynch. 1993. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 217:581-599. [DOI] [PubMed] [Google Scholar]
- 43.Mougel, M., and E. Barklis. 1997. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 71:8061-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mougel, M., Y. Zhang, and E. Barklis. 1996. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 70:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop- loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 46.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murti, K. G., M. Bondurant, and A. Tereba. 1981. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 37:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oroudjev, E. M., P. C. Kang, and L. A. Kohlstaedt. 1999. An additional dimer linkage structure in Moloney murine leukemia virus RNA. J. Mol. Biol. 291:603-613. [DOI] [PubMed] [Google Scholar]
- 49.Ortiz-Conde, B. A., and S. H. Hughes. 1999. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J. Virol. 73:7165-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paillart, J. C., L. Berthoux, M. Ottmann, J. L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prats, A. C., C. Roy, P. A. Wang, M. Erard, V. Housset, C. Gabus, C. Paoletti, and J. L. Darlix. 1990. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 64:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasmussen, S. V., J. G. Mikkelsen, and F. S. Pedersen. 2002. Modulation of homo- and heterodimerization of Harvey sarcoma virus RNA by GACG tetraloops and point mutations in palindromic sequences. J. Mol. Biol. 323:613-628. [DOI] [PubMed] [Google Scholar]
- 53.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 54.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 56.Rist, M. J., and J. P. Marino. 2002. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry 41:14762-14770. [DOI] [PubMed] [Google Scholar]
- 57.Roy, C., N. Tounekti, M. Mougel, J. L. Darlix, C. Paoletti, C. Ehresmann, B. Ehresmann, and J. Paoletti. 1990. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 18:7287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakuragi, J., S. Ueda, A. Iwamoto, and T. Shioda. 2003. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 77:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 60.Stewart, L., G. Schatz, and V. M. Vogt. 1990. Properties of avian retrovirus particles defective in viral protease. J. Virol. 64:5076-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 63.Tchenio, T., and T. Heidmann. 1995. The dimerization/packaging sequence is dispensable for both the formation of high-molecular-weight RNA complexes within retroviral particles and the synthesis of proviruses of normal structure. J. Virol. 69:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torrent, C., T. Bordet, and J. L. Darlix. 1994. Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus. J. Mol. Biol. 240:434-444. [DOI] [PubMed] [Google Scholar]
- 65.Velu, T. J., W. C. Vass, D. R. Lowy, and P. E. Tambourin. 1989. Harvey murine sarcoma virus: influences of coding and noncoding sequences on cell transformation in vitro and oncogenicity in vivo. J. Virol. 63:1384-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, S., and H. M. Temin. 1994. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 13:713-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]