Abstract
The TCF7L2 transcription factor (TF) is a member of Wnt signalling pathway, and may influence transcription of several genes by binding to distinct regulatory regions. Genome-wide studies have identified thousands of TCF7L2 binding sites and have revealed some associated TF partners. However, there is still a large uncharted region in the hierarchical regulatory network for TCF7L2 and the partner TFs in MCF7 cells. We analysed ChIP-seq data by searching for motifs in the enriched peak region based on TF-specific position weight matrix (PWM). We found association of FOXO1 and CAD with up-regulated genes, AP2α, PBF and AP1 with down-regulated genes. TCF7L2 and GATA3 were found to be associated with both up and down-regulated genes. Our study uncovers new TCF7L2 associated regulatory networks by mining ChIP-seq data in MCF7 cell, which may contribute to further study of the mechanisms related to Wnt pathway in breast cancer or other diseases.
Keywords: motif, hierarchical regulatory networks, MCF7, TCF7L2, TF, transcription factor
1 Introduction
The TCF7L2 transcription factor (TF) is an important downstream regulator in WNT signalling pathway (El-Tanani et al., 2008; Shitashige et al., 2008; Grove, 2011; Segditsas and Tomlinson, 2006) and mediates many target genes via its interaction with CTNNB1 (beta-catenin). Several studies have found that TCF7L2 can be either an activator or a repressor, depending on the availability of CTNNB1 in the nucleus (Shitashige et al., 2008; Clevers, 2006; Daniels and Weis, 2005). It is now clear that TCF7L2 is linked to a variety of human diseases including different types of cancers, such as colon, liver, breast, and pancreatic cancers (Poy et al., 2001; Prokunina-Olsson et al., 2009; El-Tanani et al., 2008; Roose and Clevers, 1999; Slattery et al., 2008; Hazra et al., 2008; Van De Wetering et al., 2002). In addition, alternative splicing variants in TCF7L2 genes are thought to be the most critical risk factors for type 2 diabetes (Cauchi and Froguel, 2008; Voight et al., 2010; Grant et al., 2006; Weedon, 2007). However, the underlying mechanisms and functional role of TCF7L2 in these diseases remains unclear. Our previous study (Frietze et al., 2012) have conducted ChIP-seq of TCF7L2 in six cancer cell types and found that TCF7L2 regulates its downstream target genes in a cell-type-specific manner, with a different set of target genes being turned on or off in each cell type. ChIP-seq data analysis revealed that TCF7L2 co-localises with a pioneer factor GATA3 in MCF7 cells. The motif analysis showed that the TCF7L2 motif is enriched in most TCF7L2 binding sites but is not enriched in the sites bound by both GATA3 and TCF7L2 (Frietze et al., 2012). This indicates that GATA3 might tether TCF7L2 to the genome at these sites. Indeed, by comparing siTCF7L2 to siControl RNA-seq data we found that TCF7L2 represses transcription when tethered to the genome via GATA3.
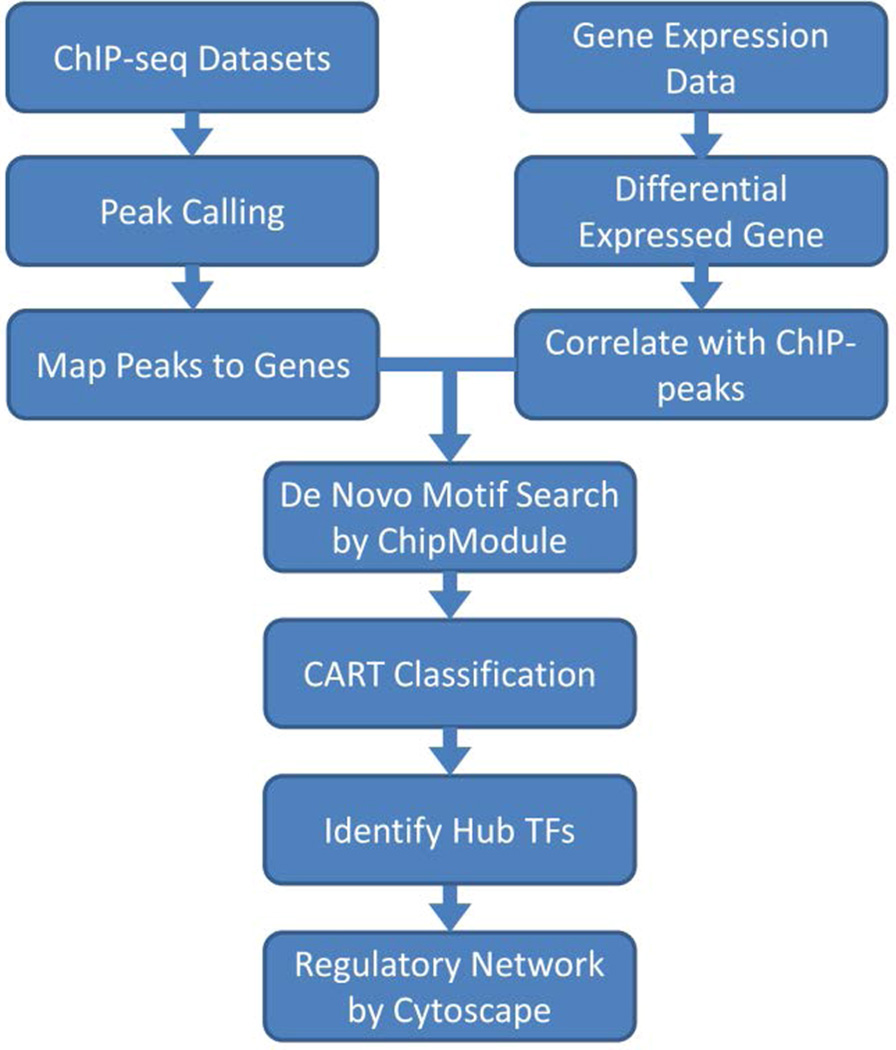
Despite we have identified thousands of TCF7L2 target genes and its partnering with GATA3 in MCF7 cell, no work has yet been done in studying the transcriptional regulatory network involving both TCF7L2 and other TFs. Given the nature of the transcriptional regulation is usually via a hierarchical architecture, it is necessary to identify other partnering factors with TCF7L2 and dissect the TCF7L2 regulated network. In this study, we performed a computational analysis using an analytical framework (Figure 1) modified from our previous approach (Gu et al., 2010), to investigate the hierarchical regulatory information for TCF7L2 in MCF7.
Figure 1.
Analytical framework of the computational analysis (see online version for colours)
Flow chart of the framework from raw ChIP-seq and gene expression data to the regulatory network.
2 Materials and methods
2.1 ChIP-seq data processing
In our previous study (Frietze et al., 2012), we have performed two experiment replicates for ChIP-seq in MCF7 cells. Since the reproducibility is very high between the replicates, we decided to combine the reads from both replicates and called TCF7L2 peaks by BELT program (http://compbio.uthscsa.edu/W-ChIPeaks/) (Lan et al., 2011). All peaks from amplified regions or gene desert were removed from the final list of total 30,119 TCF7L2 binding sites.
2.2 Correlating with gene expression
We then mapped the identified binding peaks to the regulatory regions of known genes. There are five regions, which are 5’ TSS (±1 kb around 5’TSS), 5’ Proximal (1–10 kb upstream of 5’ TSS), 5’ Distal (10–100 kb upstream of 5’ TSS), genebody (1 kb downstream 5’ TSS to the stop codon), 3’ Core (1 kb downstream of the stop codon), 3’ Proximal (1–10 kb downstream of the stop codon), 3’ Distal (10–100 kb downstream of the stop codon). We selected the genes that have TCF7L2 binding peaks in these five regions, and inspected their gene expression.
2.3 TCF7L2 regulatory network analysis
We applied the de novo motif discovery approach ChIPMotifs (Jin et al., 2006, 2009) to identify the known or novel TF partners of TCF7L2. ChIPMotifs is an online tool for searching the most significant motifs in given region on the genome based on the PWM of known factor motifs in TRANSFAC (Wingender et al., 2000) and JASPAR (Sandelin et al., 2004) databases. ChIPMotifs was applied separately on the identified binding peaks that are associated with either up- or down-regulated genes, to identify cis-regulatory modules for human TFs based on the PWMs, where stringent thresholds, core score = 1 and PWM score = 0.95, were used. The detailed equation and procedure is described in Gu et al. (2010).
Among all the motifs that were discovered, certain TFs were found at a higher frequency. These TFs, including TCF7L2, were selected as predictor variables to construct a classification model for separating the specific TF set enriched in up and down-regulated genes. The model was built using classification and regression tree, on the commercially available CART software (Salford Systems, San Diego, CA). The default setting of CART was applied. 10-fold validation mode was selected when building the model.
Then, hub TFs were selected from the variable table output by CART, corresponding gene were identified, and relationships between these TFs and genes were visualised through a TCF7L2 regulatory network with Cytoscape (Shannon et al., 2003).
3 Results and discussion
3.1 Regulation of TCF7L2 target genes in MCF7 cells
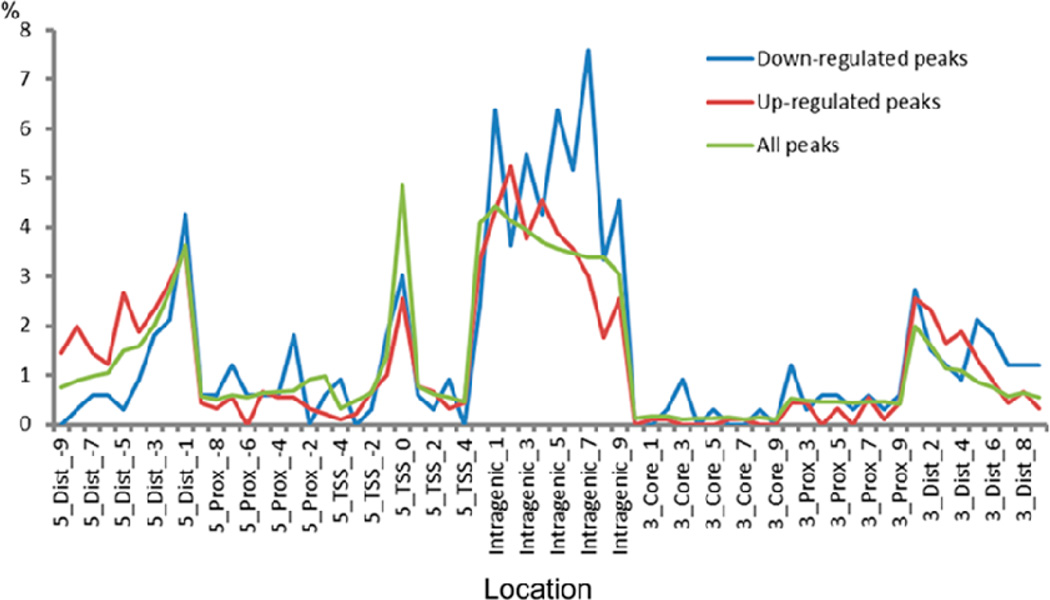
We identified TCF7L2 binding sites through peak calling and located these identified TCF7L2 binding peaks to five regulatory regions of known annotated genes. Definition of the five regulatory regions in described in the Method section and the result of the location analysis is shown in Figure 2.
Figure 2.
Location analysis of TCF7L2 binding sites (see online version for colours)
Shown is the percentage of TCF7L2 binding sites in each genomic region (according to hg19 RefSeq gene annotations). The regions are 5’ Distal (5_Dist_xxx as in the x-axis labels), 5’ Proximal (5_Prox_xxx), 5’ TSS (5_TSS_xxx), genebody (Intragenic_xxx), 3’ Core (3_Core_xxx), 3’ Proximal (3_Prox_xxx), and 3’ Distal (3_Dist_xxx).
In order to understand how the regulatory network is mediated by TCF7L2 in MCF7 cells, we integrated the ChIP-seq data with gene expression data by RNA-seq analysis of MCF7 cells before and after knockdown of TCF7L2. In our previous study (Frietze et al., 2012), we found that the expressions of 469 genes were significantly changed compared to cells treated with siRNA for TCF7L2. These included 188 down-regulated genes and 281 up-regulated genes. After performing the correlation of binding sites with annotated genes, 235 (50%) of these genes, including 60 down-regulated and 175 up-regulated genes, respectively, were identified to have at least one TCF7L2 binding peak. We considered all binding peaks except those in the gene desert regions (larger than 100 kb away from a TSS) and amplified genome regions. As a result, we found a total of 329 TCF7L2 binding peaks associated with 60 down-regulated genes and 899 TCF7L2 binding peaks associated with the 175 up-regulated genes.
For TCF7L2 binding sites associated with down-regulated genes, a relatively small number of peaks were located in known 5’ transcriptional start sit (TSS) regions (8%), while a big portion were located within gene bodies (50%). However, although comparable percentage of binding sites associated with up-regulated genes fell into the known 5’TSS region (7%), only 36% of the binding sites were located within gene bodies of up-regulated genes, which is different from that in the down-regulated genes. Overall, 37% of TCF7L2 binding peaks were within gene bodies, while 10% of TCF7L2 binding peaks were in known 5’TSS regions. Our location analysis showed that the majority of TCF7L2 binding peaks are outside of proximal promoter regions, suggesting that most of the TCF7L2-associated genes may be regulated by the long-distance interaction between TCF7L2-bound distal enhancer and proximal promoter regions. Nevertheless, 15% of TCF7L2 binding sites (21% of down-regulated and 15% of up-regulated TCF7L2 binding sites) are located in the 3’ ends (including 3’ Core, 3’ Proximal and 3’ Distal regions). This finding might imply that some alternative transcripts are transcribed in the 3’ end. This could also simply due to loops formed with the promoters.
3.2 De novo identification of TCF7L2 binding motifs and its partner factors in MCF7
It is well established that TFs control gene expression through interacting with the DNA, but how exactly a single TF functions at the gene regulatory region is still far from entirely revealed (Ji and Sharrocks, 2015; Li et al., 2015). In fact, each TF can recruit and interact with a set of co-regulatory proteins or partner factors, and the consequent regulatory complex will bind to DNA and control gene expression. This probably explains our observation that the majority of TCF7L2 direct targets are not activated or repressed, as some important TCF7L2 partner TFs might be absent.
We did de novo search on TCF7L2 binding motifs and its partner TFs, in the TCF7L2 binding sites identified as above. Then, we created a motif matrix with each element indicating the existence (or absence) of one particular TF in the regulatory region of one particular gene. Feeding this matrix into classification algorithm, we found several TFs significantly associated with the differentially expressed genes. FOXO1 and CAD were identified in up-regulated genes, while AP2α, PBF and AP1 were identified in down-regulated genes. Besides, it is also found that two TF motifs, TCF7L2 and GATA3 are associated with both up and down-regulated genes.
3.3 Regulatory network for TCF7L2 in MCF7 cells
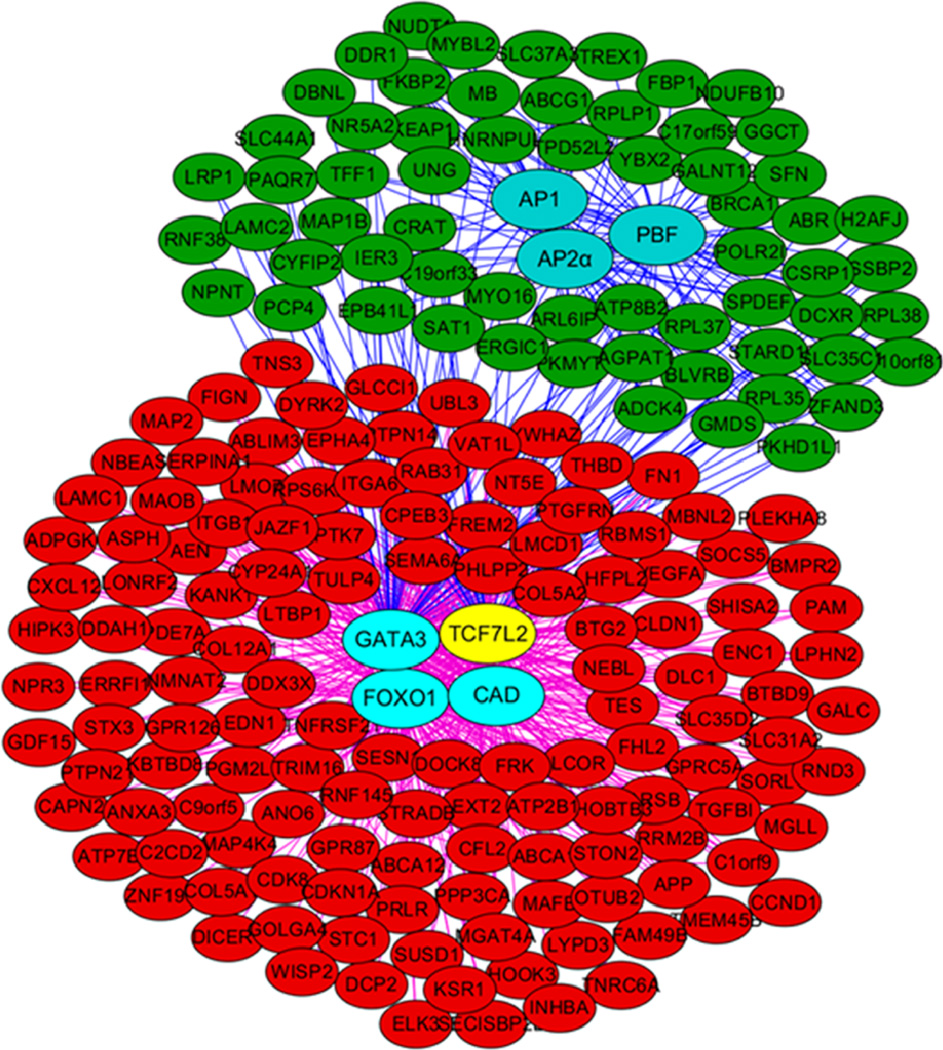
The resulted regulatory networks were thus constructed and topologically visualised using Cytoscape (Shannon et al., 2003) software platform. In the network, all the genes/TFs were represented as a node (red nodes as up-regulated genes, green nodes as down-regulated genes, and blue nodes as hub TFs, with the exception of TCF7L2 which is in yellow colour), and all the connections were represented as an edge between two nodes (Figure 3, Table S1).
Figure 3.
The regulatory network for TCF7L2 in MCF7 (see online version for colours)
Regulatory networks topologically visualised using Cytoscape. Up-regulated genes and down-regulated genes were indicated by nodes in red and green, respectively. Nodes in cyan represent hub TFs while TCF7L2 is indicated with yellow node. Connections between hub TFs and down-regulated genes were indicated with blue edges and connection between hub TFs and up-regulated genes were indicated with purple edges.
4 Conclusion
In this study, we applied computational approaches to investigate the transcriptional regulation by TCF7L2 in MCF7 cells. We found that in addition to GATA3, other TFs had binding sites co-enriched with TCF7L2 binding sites, which indicated a close collaboration between TCF7L2 and those factors. Our study supports a specific regulatory network for TCF7L2 in MCF7 cells. The hierarchical regulatory network analysis revealed that GATA3 is a potential TCF7L2 partner, which is consistent to our previous reports (Frietze et al., 2012) showing that GATA3 is serving as pioneer factors that enhance the ability of TCF7L2 to access its sites in breast cells. We also identified 5 new Hub TFs in MCF7 cells, namely up-regulated FOXO1 and CAD, and down-regulated AP2α, PBF, AP1, respectively. This indicates these factors may play important role for TCF7L2 regulation in MCF7 cells. Our studies reveal new insights into TCF7L2-mediated gene regulation and suggest that cooperation with other factors dictates different roles for TCF7L2 in MCF7 cells. The computational analytical approach applied here may also provide a framework for dissecting transcriptional regulatory networks in breast cancer and other human diseases.
Supplementary Material
Acknowledgments
This work was supported by PhRMA Foundation (Grant No. GRT00026196) and Department of Molecular Medicine, University of Texas Health Science Center at San Antonio. Rui Wang was supported by a Chinese Ministry of Education Visiting Fellowship. We sincerely thank members in our team for discussion and advice on our project.
Biographies
Yao Wang is a Postdoctoral Fellow in the Department of Molecular Medicine at the University of Texas Health Science Center, San Antonio (UTHSCSA). Her research interests include developing computational pipelines to aid molecular biomarker discovery as well as understanding genetic/epigenetic regulatory mechanisms in cancers.
Rui Wang is currently a Lecturer at the School of Chemical and Environment Science Shaanxi University of Technology. Her research interests include theoretical and computational chemistry.
Victor X. Jin received his PhD in Biological/Macromolecular Chemistry in 2001 at Queen’s University, Canada. He is currently an Associate Professor at UTHSCSA. He has more than 75 peer-reviewed papers in some of the most prestigious and highly cited journals such as Nature, Nature Cell Biology, Cancer Cell, Mol. Cell, Genome Res. He is currently an Associate Editor for BMC Medical Genetics and on Editorial Board in several journals. The major focus of his group is to perform functional genomics and develop statistical methods and machine learning algorithms for high-throughput ‘omics data, to understand epigenetic regulatory mechanisms in cancers.
Contributor Information
Yao Wang, Departments of Molecular Medicine and Epidemiology and Biostatistics, University of Texas Health Science Center at San Antonio, San Antonio, TX 78229, USA, wangy14@uthscsa.edu.
Rui Wang, School of Chemical and Environment Science, Shaanxi University of Technology, Hanzhong, Shaanxi 723000, China, wangrui830413@163.com.
Victor X. Jin, Departments of Molecular Medicine and Epidemiology and Biostatistics, University of Texas Health Science Center at San Antonio, San Antonio, TX 78229, USA, jinv@uthscsa.edu
References
- Cauchi S, Froguel P. TCF7L2 genetic defect and type 2 diabetes. Current Diabetes Reports. 2008;8(2):149–155. doi: 10.1007/s11892-008-0026-x. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nature Structural and Molecular Biology. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- El-Tanani MK, Ravindranath A, O’Connell A, Johnston PG. The role of LEF/TCF factors in neoplastic transformation. Current Molecular Medicine. 2008;8(1):38–50. doi: 10.2174/156652408783565559. [DOI] [PubMed] [Google Scholar]
- Frietze S, Wang R, Yao L, Tak YG, Ye Z, Gaddis M, Jin VX. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biology. 2012;13(9):R52. doi: 10.1186/gb-2012-13-9-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Grove EA. Wnt signaling meets internal dissent. Genes and Development. 2011;25(17):1759–1762. doi: 10.1101/gad.17594311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Hsu HK, Hsu PY, Wu J, Ma Y, Parvin J, Jin VX. Inference of hierarchical regulatory network of estrogen-dependent breast cancer through ChIP-based data. BMC Systems Biology. 2010;4:170. doi: 10.1186/1752-0509-4-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A, Fuchs CS, Chan AT, Giovannucci EL, Hunter DJ. Association of the TCF7L2 polymorphism with colorectal cancer and adenoma risk. Cancer Causes and Control. 2008;19(9):975–980. doi: 10.1007/s10552-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Sharrocks AD. Changing partners: transcription factors form different complexes on and off chromatin. Mol. Syst. Biol. 2015;11(1):782. doi: 10.15252/msb.20145936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VX, Apostolos J, Nagisetty NSVR, Farnham PJ. W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data. Bioinformatics. 2009;25(23):3191–3193. doi: 10.1093/bioinformatics/btp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VX, Rabinovich A, Squazzo SL, Green R, Farnham PJ. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data – a case study using E2F1. Genome Research. 2006;16(12):1585–1595. doi: 10.1101/gr.5520206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Bonneville R, Apostolos J, Wu W, Jin VX. W-ChIPeaks: a comprehensive web application tool for processing ChIP-chip and ChIP-seq data. Bioinformatics. 2011;27(3):428–430. doi: 10.1093/bioinformatics/btq669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang W, Wang J, Malovannaya A, Xi Y, Li W, Chen J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015;11(1):775. doi: 10.15252/msb.20145504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-beta-catenin complex. Nature Structural Biology. 2001;8(12):1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, Hall JL. Tissue-specific alternative splicing of TCF7L2. Human Molecular Genetics. 2009;18(20):3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J, Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochimica et Biophysica Acta – Reviews on Cancer. 1999;1424(2):23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Research. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25(57):7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Science. 2008;99(4):631–637. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery ML, Folsom AR, Wolff R, Herrick J, Caan BJ, Potter JD. Transcription factor 7-like 2 polymorphism and colon cancer. Cancer Epidemiology Biomarkers and Prevention. 2008;17(4):978–982. doi: 10.1158/1055-9965.EPI-07-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wetering M, Sancho E, Verweij C, De Lau W, Oving I, Hurlstone A, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Grarup N. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN. The importance of TCF7L2. Diabetic Medicine. 2007;24(10):1062–1066. doi: 10.1111/j.1464-5491.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Research. 2000;28(1):316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.