Summary
The cyp125 gene of Rhodococcus jostii RHA1 was previously found to be highly upregulated during growth on cholesterol and the orthologue in Mycobacterium tuberculosis (rv3545c) has been implicated in pathogenesis. Here we show that cyp125 is essential for R. jostii RHA1 to grow on 3-hydroxysterols such as cholesterol, but not on 3-oxo sterol derivatives, and that CYP125 performs an obligate first step in cholesterol degradation. The involvement of cyp125 in sterol side-chain degradation was confirmed by disrupting the homologous gene in Rhodococcus rhodochrous RG32, a strain that selectively degrades the cholesterol side-chain. The RG32Ωcyp125 mutant failed to transform the side-chain of cholesterol, but degraded that of 5-cholestene-26-oic acid-3β-ol, a cholesterol catabolite. Spectral analysis revealed that while purified ferric CYP125RHA1 was < 10% in the low-spin state, cholesterol (KDapp = 0.20 ± 0.08 μM), 5α-cholestanol (KDapp = 0.15 ± 0.03 μM) and 4-cholestene-3-one (KDapp = 0.20 ± 0.03 μM) further reduced the low spin character of the haem iron consistent with substrate binding. Our data indicate that CYP125 is involved in steroid C26-carboxylic acid formation, catalysing the oxidation of C26 either to the corresponding carboxylic acid or to an intermediate state.
Introduction
Cytochromes P450 (P450s) are a widely distributed class of haem-containing monooxygenases that are present in all domains of life. Their essential roles in diverse metabolic pathways have also generated considerable interest for their use as biocatalysts (Julsing et al., 2008). Genome sequence data analysis has revealed that actinobacteria posses a remarkable number of genes encoding P450s compared with other prokaryotes (Lamb et al., 2006; McLean et al., 2006). For example, Rhodococcus jostii RHA1 harbours 29 genes predicted to encode P450s (McLeod et al., 2006). While the biological function of most of these monooxygenases is still unknown, several of them have been implicated in sterol/steroid catabolism.
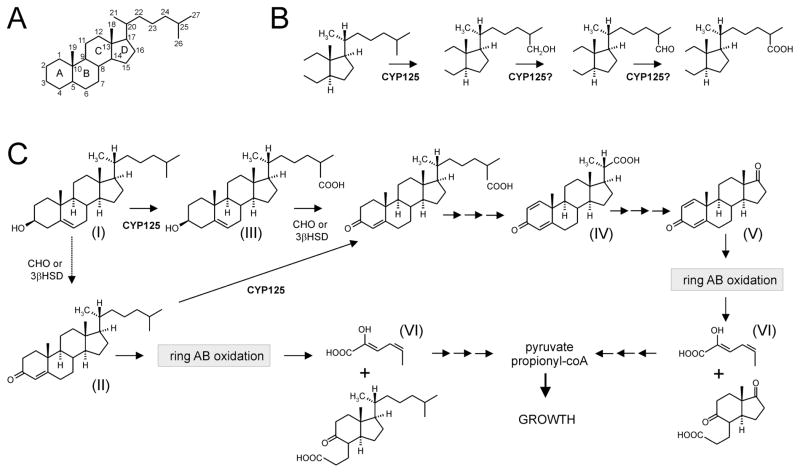
The microbial degradation of cholesterol (5-cholestene-3β-ol; Fig. 1, compound I) involves two processes: sterol side-chain elimination and steroid ring opening (Van der Geize and Dijkhuizen, 2004). The order of these two processes in vivo is unknown and may vary between microorganisms. Generally, oxidation of the cholesterol 3β-hydroxyl moiety and isomerization of Δ5 into Δ4 is thought to initiate sterol degradation (Sojo et al., 1997; Chen et al., 2006; Chiang et al., 2008). This transformation is catalysed by either cholesterol oxidase (CHO; MacLachlan et al., 2000) or 3β-hydroxysteroid dehydrogenase (3β-HSD; Yang et al., 2007) and results in the formation of 4-cholestene-3-one (Fig. 1, compound II). Further degradation of 4-cholestene-3-one proceeds via hydroxylation at C26 to initiate side-chain degradation or oxidation of rings A and B analogous to ring degradation of 4-androstene-3,17-dione, resulting in the formation of 2-hydroxyhexa-2,4-diene-oic acid (Fig. 1, compound VI; Van der Geize et al., 2007). Microbial sterol side-chain degradation has been studied at the biochemical level in more detail in Nocardia species and Mycobacterium sp. strains NRRL B-3683 and NRRL B-3805 (Sih et al., 1968a,b; Marsheck et al., 1972; Fujimoto et al., 1982a,b). The latter two are capable of selectively degrading the 17-alkyl side-chains of cholesterol and phytosterols. Microbial cholesterol side-chain degradation is initiated by C26 hydroxylation followed by further oxidation to the sterol C26-oic acid (Fig. 1, compound III). Subsequent degradation occurs via a mechanism similar to β-oxidation of fatty acids that leads to the formation of a steroid C22-oic acid intermediate (Fig. 1, compound IV) with the concomitant release of propionyl-CoA and acetyl-CoA. The remaining C3 side-chain is released as propionyl-CoA via a different mechanism (Sih et al., 1967; 1968b).
Fig. 1. The initial steps of aerobic cholesterol degradation in bacteria (Sih et al., 1968a,b; Szentirmai, 1990; van der Geize et al., 2007).
A. Steroid nomenclature.
B. CYP125 is involved in steroid C26 hydroxylation. Subsequent oxidation leads to a C26-oic acid metabolite.
C. Sterol degradation proceeds via steroid ring oxidation and side-chain degradation (upper route). The exact order of side-chain degradation and ring oxidation in vivo is unknown and may vary between microorganisms. In R. jostii RHA1, ring oxidation is not initiated until sometime after the side-chain attack by CYP125 (dotted arrow). The depicted metabolites are: (I) 5-cholestene-3β-ol (cholesterol), (II) 4-cholestene-3-one, (III) 5-cholestene-26-oic acid-3β-ol, (IV) 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (Δ1,4-BNC), (V) 1,4-androstadiene-3,17-dione and (VI) 2-hydroxyhexa-2,4-diene-oic acid. R. rhodochrous mutant strain RG32 (see text) converts compound I into compounds IV and V by selective side-chain degradation. Abbreviations: CYP125, steroid 26-monooxygenase; CHO, cholesterol oxidase; 3βHSD, 3β-hydroxysteroid dehydrogenase (Yang et al., 2007).
Rhodococcus rhodochrous DSM43269 (synonym IFO3338) is able to selectively degrade the sterol side-chain in the presence of iron chelators, which inhibit 3-ketosteroid 9α-hydroxylase (KSH) activity (Arima et al., 1978). This phenotype was replicated in a stable multiple gene deletion mutant strain of R. rhodochrous DSM43269 (strain RG32) lacking KSH activity (M.H. Wilbrink, L. Dijkhuizen and R. van der Geize, unpublished). Mutant strain RG32 is completely blocked in steroid ring degradation and capable of selective sterol side-chain degradation, thereby accumulating 1,4-androstadiene-3,17-dione (ADD) (Fig. 1, compound V) and 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (Δ1,4-BNC) (Fig. 1, compound IV) from sterols. The strain RG32 phenotype thus allows us to specifically analyse sterol side-chain degradation.
To date, genes involved in sterol side-chain degradation have not been identified. Using transcriptomic analysis, we recently identified a cholesterol catabolic gene cluster in R. jostii RHA1 that includes two P450-encoding genes (Van der Geize et al., 2007). Interestingly, ro04679 (cyp125RHA1) was one of the most highly upregulated genes within this cluster during growth on cholesterol, suggesting an important role for this enzyme in cholesterol catabolism. In the RHA1 genome, cyp125 is located proximal to genes predicted to encode β-oxidation enzymes, and suggested to be involved in degradation of the alkyl side-chain of cholesterol (Van der Geize et al., 2007). Moreover, cyp125 is located within the ro04482-ro04705 gene cluster encompassing the mce4 genes, which encode the uptake system for cholesterol and related steroids with unsubstituted alkyl side-chains (Mohn et al., 2008).
Here we report the molecular characterization of CYP125 as a steroid 26-monooxygenase. The cyp125 gene was inactivated in each of R. jostii RHA1 and R. rhodochrous RG32 and the effect on cholesterol catabolism was elucidated. CYP125RHA1 was heterologously expressed and purified, and its binding to cholesterol and its analogues was investigated. This study provides novel insights into bacterial steroid degradation, revealing that degradation in R. jostii RHA1 is initiated by side-chain oxidation, not oxidation of the rings.
Results
CYP125 possesses conserved amino acid residues for interaction with sterols
Bioinformatic analysis revealed that CYP125RHA1 has high amino acid sequence identity with P450s from other actinobacteria, including Nocardia farcinica strain IFM10152 [Nfa5180, 79% (Ishikawa et al., 2004)] and Mycobacterium tuberculosis strain H37Rv [Rv3545c, 69% (Cole et al., 1998; Camus et al., 2002)]. These proteins belong to the uncharacterized CYP125 family (subfamily A) of P450 enzymes (Nelson et al., 1996), in which CYP125RHA1 has been assigned CYP125A14P (http://drnelson.utmem.edu/biblioE.html#125). These monooxygenases presumably transform lipid-like compounds, as the CYP125 family includes many actinobacterial proteins associated with lipid degradation (Ventura et al., 2007).
Bioinformatic analysis further revealed that the annotated sequence of CYP125RHA1 was about 50 residues longer than that of the annotated orthologues. Careful analysis of the cyp125RHA1 nucleotide sequence indicated that the start codon most likely is located 159 nucleotides downstream from that in the original annotation, and is preceded by a Shine–Dalgarno sequence (aggag). Thus, cyp125RHA1 is a gene of 1257 nucleotides, encoding a protein of 418 amino acids with a calculated molecular mass of 47.2 kDa. The re-annotated sequence of cyp125RHA1 (RHA1 genome co-ordinates 4930900 … 4932156) was used in this study.
Amino acid sequence alignments revealed that the actinobacterial CYP125s share the conserved motifs characteristic for the P450 super-family, as well as key residues of cholesterol-transforming eukaryotic P450s (Fig. S1). The latter belong to various families, including: CYP3A4, which performs 4β-hydroxylation of cholesterol; CYP11A1, which transforms cholesterol to pregnenolone via C20–C22 bond-cleavage; CYP27A1, which hydroxylates cholesterol at C27; and CYP46A1, which transforms cholesterol to 24S-hydroxycholesterol (Mast et al., 2006; Pikuleva, 2006; Storbeck et al., 2007). The presence of these conserved residues in CYPs125 and in P450 enzymes known to interact with sterols suggests that sterols are substrates for CYP125.
CYP125 is essential for growth on 3-hydroxy-sterols
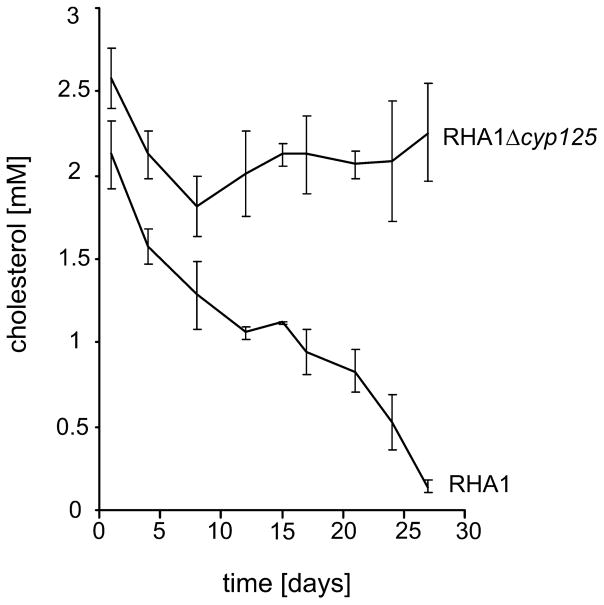
To elucidate the role of cyp125 in sterol/steroid catabolism, an unmarked single gene deletion mutant strain, RHA1Δcyp125, was constructed. Growth experiments in mineral medium (MM) supplemented with cholesterol revealed that the RHA1Δcyp125 strain was unable to grow on cholesterol (Table 1). To confirm that the observed phenotype was solely due to inactivation of cyp125, a complementation experiment was performed in which cyp125RHA1 was supplied in trans. The complemented strain, RHA1Δcyp125+pTip-QC1cyp125RHA1, displayed a restored wild-type growth phenotype in MM supplemented with cholesterol (Table 1). Wild-type RHA1 and RHA1Δcyp125 were subsequently grown in mineral liquid media on a range of other sterols, steroids and their metabolites as sole carbon and energy sources (Table S1). RHA1 grew readily on all tested compounds. By contrast, RHA1Δcyp125 failed to grow on epicholesterol, 5α-cholestanol and on the plant sterol mixture β-sitosterol/β-sitostanol/campesterol. Remarkably, growth of strain RHA1Δcyp125 on 3-ketone oxidized derivatives of two of these sterols, 4-cholestene-3-one and 5α-cholestane-3-one, was unimpaired, likely due to degradation of the steroid ring structure (Table S1, Fig. 1). We thus conclude that CYP125 is essential for 3-hydroxy-sterol degradation. The phenotype of RHA1Δcyp125 was investigated further by growing the mutant in mineral liquid media supplemented with cholesterol and an additional non-repressing carbon source (i.e. pyruvate or glycerol). In contrast to the wild-type strain, RHA1Δcyp125 did not significantly transform cholesterol under these conditions (Fig. 2).
Table 1.
Growth in mineral media supplemented with cholesterol (2.5 mM) as sole carbon and energy source of wild-type strain RHA1, mutant strain RHA1Δcyp125, complemented mutant strain RHA1Δcyp125 (RHA1Δcyp125+pTip-QC1cyp125), and RHA1Δcyp125 mutant strain harbouring null vector (RHA1Δcyp125+pTip-QC1) after 10 days of growth.
| Strain | Protein content (mg l−1) | Residual cholesterol (%) |
|---|---|---|
| RHA1 | 49 ± 6 | 56 ± 5 |
| RHA1Δcyp125 | 5 ± 3 | 112 ± 4 |
| RHA1Δcyp125+pTip-QC1cyp125 | 57 ± 4 | 55 ± 4 |
| RHA1Δcyp125+pTip-QC1 | 3 ± 3 | 116 ± 16 |
| Control (medium + cholesterol) | 0 | 100 ± 13 |
Non-inoculated mineral medium with cholesterol was included as a negative control. Values represent mean ± standard deviation (n = 3).
Fig. 2.
Cholesterol degradation by cell cultures of strains RHA1 and RHA1Δcyp125 grown in mineral liquid media supplemented with pyruvate (20 mM) and cholesterol (2.5 mM). The data represent averages of triplicates. Error bars indicate standard deviations.
To further investigate the initial cholesterol-transforming enzymes of RHA1, we assayed pyruvate-grown cultures of wild-type RHA1 and mutant strain RHA1Δcyp125 that had been induced with cholesterol for total 3β-hydroxysteroid oxidation activity (Yang et al., 2007). These studies comprised assays for extracellular and intracellular activities arising from CHO and 3β-HSD. When cholesterol was used as a substrate in these assays, no activity was detected in either supernatants or cell lysates of these cultures, consistent with the lack of transformation of cholesterol by RHA1Δcyp125. By contrast, 3β-hydroxysteroid oxidation activity was detected in lysates of cholesterol-induced cells of RHA1 (0.27 μM min−1 mg−1) and RHA1Δcyp125 (0.76 μM min−1 mg−1) when 5-pregnene-3β-ol-20-one was used as a substrate in the assay. Overall, these data indicate that CYP125 is essential for cholesterol degradation by RHA1, and that it catalyses an obligate first reaction in the cholesterol catabolic pathway.
CYP125 has a role in sterol side-chain degradation
We hypothesized that CYP125RHA1 might have a specific role in sterol side-chain degradation. To substantiate this hypothesis, we used R. rhodochrous RG32, a mutant of R. rhodochrous DSM43269 which only degrades the side-chain of cholesterol, transforming it to ADD (Fig. 1, compound V) and Δ1,4-BNC (Fig. 1, compound IV) (Fig. 3A). First, we cloned cyp125 from R. rhodochrous DSM43269 by screening a genomic library of this strain with degenerate PCR primers based on conserved amino acid sequences found in actinobacterial CYP125s. A positive clone, containing 8.7 kb of insert DNA, was obtained, sequenced and analysed. The insert carried cyp125DSM43269, encoding a protein sharing 76% amino acid sequence identity with CYP125RHA1 (Fig. S1). Moreover, the cyp125 locus is similarly organized in R. jostii RHA1 and R. rhodochrous DSM43269. More specifically, the genes immediately downstream of cyp125 in DSM43269 encode proteins sharing 56%, 74% and 86% amino acid sequence identity to those encoded by ro04676, ro04677 and ro04678, respectively, in RHA1. Upstream of cyp125DSM43269, orthologues of ro04654 (82% identity) and ro04653 (82% identity) were located, as well as genes encoding hypothetical proteins that have no counterparts in RHA1.
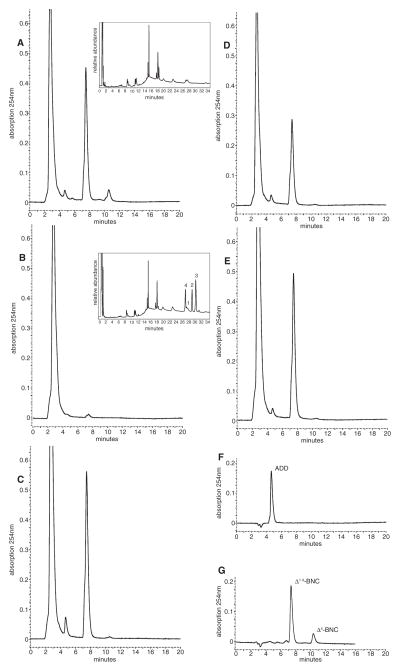
Fig. 3.
HPLC profiles of whole-cell biotransformations of cholesterol by cell cultures of (A) R. rhodochrous strain RG32 showing the formation of 1,4-androstadiene-3,17-dione (ADD) and 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (Δ1,4-BNC) via selective sterol side-chain degradation, (B) mutant strain RG32Ωcyp125 and (C) cyp125DSM43269 complemented mutant strain RG32Ωcyp125. HPLC profiles of whole-cell biotransformations of 5-cholenic acid-3β-ol (D) and 5-cholestene-26-oic acid-3β-ol (E) by cell cultures of R. rhodochrous mutant strain RG32Ωcyp125 are also shown. Profiles of authentic ADD (50 μM, F) and Δ1,4-BNC (G), obtained by incubating authentic 3-oxo-23,24-bisnorchol-4-en-22-oic acid (50 μM, Δ4-BNC) with purified Δ1-KSTD1 (Knol et al., 2008), are included as reference samples. Insets: GC profiles showing the accumulation of 4-cholestene-3-one (2), 1,4-cholestadiene-3-one (3) and 5α-cholestane-3-one (4) from cholesterol (1) by R. rhodochrous mutant strain RG32Ωcyp125, but not strain RG32.
We then specifically disrupted cyp125 in RG32, yielding mutant strain RG32Ωcyp125. Whole-cell biotransformations of 3-hydroxy-sterols by RG32Ωcyp125 revealed that the mutant was blocked in the ability to degrade sterol side-chains (Fig. 3). Cell cultures of RG32Ωcyp125 incubated with cholesterol showed no formation of ADD or Δ1,4-BNC (Fig. 3B). Similar results were obtained when RG32Ωcyp125 cell cultures were incubated with 5α-cholestanol and β-sitosterol (data not shown). Contrary to RHA1Δcyp125, cholesterol was rapidly converted by RG32Ωcyp125 to 4-cholestene-3-one and 1,4-cholestadiene-3-one, which accumulated in the medium (Fig. 3B inset). Indeed, cholesterol-induced cells of RG32 and RG32Ωcyp125 contained high levels of 3β-hydroxysteroid total oxidation activity using cholesterol as a substrate (0.34 and 0.73 μM min−1 mg−1 respectively). By contrast, no extracellular activity was detected in either strain. Reintroduction of cyp125DSM43269 into RG32Ωcyp125 under its native promoter fully restored the ability of the strain to degrade the cholesterol side-chain (Fig. 3C). This excludes the possibility that side-chain degradation in RG32Ωcyp125 was blocked by polar effects rather than by disruption of cyp125 directly.
CYP125DSM43269 is involved in formation of the sterol C26-oic acid intermediate
We then tested the ability of mutant strain RG32Ωcyp125 to convert each of two predicted sterol side-chain degradation pathway intermediates: 5-cholestene-26-oic acid-3β-ol (Fig. 1, compound III) and the C24-oic intermediate 5-cholenic acid-3β-ol. Whole cell biotransformations performed with cultures of mutant strain RG32Ωcyp125 resulted in conversion of both 5-cholenic acid-3β-ol and 5-cholestene-26-oic acid-3β-ol to ADD and Δ1,4-BNC (Fig. 3D and E). As predicted, RHA1Δcyp125 was able to grow on both of these compounds (Table S1). Both diastereomers of 5-cholestene-26-oic acid-3β-ol appeared to be metabolized, because 75 mol% of the added substrate was converted into ADD and Δ1,4-BNC. These results show that CYP125 is essential for the conversion of cholesterol into the C26-oic acid catabolite during sterol side-chain degradation by both RG32 and RHA1.
Production and purification of CYP125RHA1
To biochemically characterize CYP125RHA1, we homologously produced and purified recombinant CYP125RHA1 with a 6-histidine tag. Expression of cyp125RHA1 was first attempted in Escherichia coli BL21(DE3) using T7 promoter-based expression vectors and conditions known to promote expression of P450 proteins, such as the addition of δ-aminolevulinic acid, FeCl3, trace elements and thiamine (Parikh et al., 1997; Keizers et al., 2004). However, CYP125RHA1 was not produced in significant amounts in E. coli. By contrast, cyp125RHA1 was well expressed in R. jostii RHA1 using the pTip-QC1 vector (Nakashima and Tamura, 2004). Addition of δ-aminolevulinic acid and other additives, usually necessary to promote expression of properly folded and soluble P450 proteins in E. coli, was not needed for homologous production of CYP125RHA1 in R. jostii RHA1.
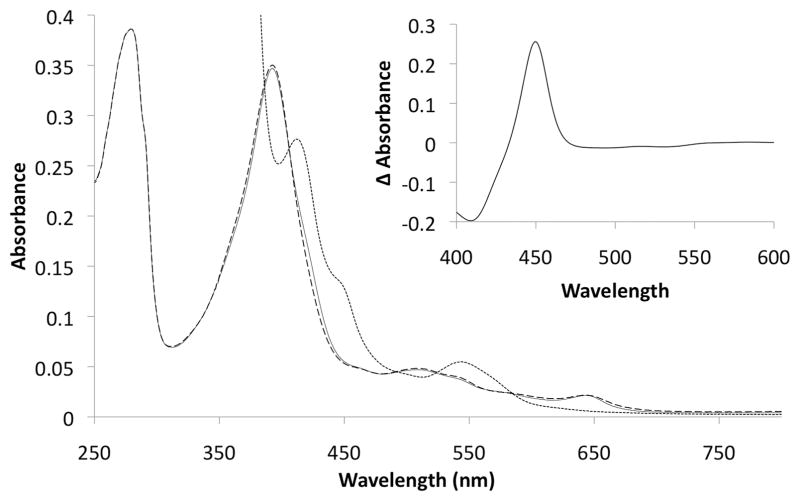
CYP125RHA1 was purified using Ni2+-NTA affinity chromatography and was determined by SDS-PAGE analysis to be in excess of 95% pure. The CO-difference spectrum of purified CYP125RHA1 displayed a maximum at 451 nm (Fig. 4, inset), indicating the haem iron thiolate ligation remained intact throughout the protein’s purification. The absorption spectrum of the purified ferric CYP125RHA1 had a maximum at 392 nm and a shoulder at 422 nm (Fig. 4). Based on analysis of this spectrum (see Experimental procedures), the preparation is estimated to contain ~93% high spin state haem iron.
Fig. 4.
The absorption spectrum of CYP125RHA1 in the oxidized state as isolated (solid line) and incubated with 10 μM cholesterol in oxidized (dashed line) and reduced (dotted line) states. The inset shows the reduced CO-difference spectrum of the enzyme incubated with 10 μM cholesterol. The sample contained 2.9 μM purified CYP125RHA1, 0.1 M potassium phosphate buffer, pH 7.0, 25°C; cholesterol was added from a 1 mM stock solubilized in 10% 2-hydroxypropyl-β-cyclodextrin.
Spectroscopic analysis of sterol binding
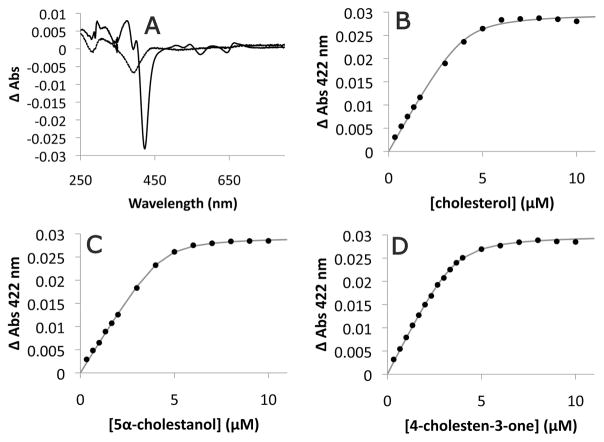
Spectroscopic assays were performed with purified CYP125RHA1 to investigate its binding to sterols. Following the addition of cholesterol (Fig. 5A) or 5α-cholestanol (data not shown) in a solution of 10% 2-hydroxypropyl-β-cyclodextrin, CYP125RHA1 exhibited a spectral change with a pronounced trough at 422 nm and a peak at 392 nm, consistent with the decrease in the low-spin character of the haem iron associated with substrate binding. The difference spectrum also exhibited a perturbation at 395 nm in comparison with the typical type I binding spectrum. A perturbation at the same wavelength was observed upon addition of 5-cholestene-26-oic acid-3β-ol in 10% 2-hydroxypropyl-β-cyclodextrin, although the acid elicited no underlying type I spectral change at concentrations up to 20 μM (Fig. 5A). Cholesterol also induced a type I binding spectrum when added in the presence of other solubilizing agents, such as Triton WR1339 and dimethylsulphoxide. However, the spectral shifts were much weaker than in the presence of 2-hydroxypropyl-β-cyclodextrin (data not shown).
Fig. 5. Binding of steroids to purified CYP125RHA1.
A. Spectral responses of 3.7 μM purified CYP125RHA1 induced by 10 μM cholesterol (solid line) and 10 μM 5-cholestene-26-oic acid-3β-ol (dashed line). The dependence of the absorbance change of CYP125RHA1 at 422 nm on (B) cholesterol, (C) 5α-cholestanol and (D) 4-cholestene-3-one concentration. The best fit of Eq. 1 to the data as determined using R is represented as a grey line with fitted parameters KD = 0.20 ± 0.08 μM, ΔAmax = 0.0298 ± 0.0006, and [E] = 4.0 ± 0.2 μM for cholesterol; KD = 0.15 ± 0.03 μM, ΔAmax = 0.0293 ± 0.0002, and [E] = 4.3 ± 0.1 μM for 5α-cholestanol; and KD = 0.20 ± 0.03 μM, ΔAmax = 0.0300 ± 0.0002, and [E] = 3.6 ± 0.1 μM for 4-cholestene-3-one. Steroids were prepared as stock solutions in 10% 2-hydroxypropyl-β-cyclodextrin which alone did not induce a CYP125RHA1 spectral response.
Using Eq. 1, apparent KD values for cholesterol, 5α-cholestane-3β-ol, and 4-cholestene-3-one were evaluated to be 0.20 ± 0.08 μM, 0.15 ± 0.03 μM and 0.20 ± 0.03 μM respectively. The concentrations of enzyme calculated using this equation (4.0, 4.3 and 3.6 μM respectively) were within 15% of the enzyme concentration calculated using the extinction coefficient for the reduced CO-difference spectrum of ε450–490 = 91 mM−1 cm−1 (3.7 μM), although this extinction coefficient has not been independently verified for this isozyme. The high quality fit of the equation to the binding data (Fig. 5B–D) supports a binding stoichiometry of 1:1 and suggests that CYP125RHA1 does not harbour a ligand as isolated despite the proportion of high-spin iron. Finally, CYP125RHA1 exhibited maxima at 451 nm in CO-difference spectra taken after each binding experiment, indicating that the haemthiolate ligation remained intact.
Discussion
The current study presents several lines of evidence identifying CYP125 as a steroid 26-monooxygenase that catalyses the initial step in microbial sterol side-chain degradation (Fig. 1). First, a cyp125 deletion mutant of R. jostii RHA1 was unable to grow on or transform several 3-hydroxysterols with relatively long unactivated aliphatic side-chains. Second, a cyp125 disruption mutant of R. rhodochrous RG32 was completely blocked in cholesterol side-chain degradation. However, this mutant was still able to degrade the side-chain of 5-cholestene-26-oic acid-3β-ol (Fig. 1, compound III), an expected intermediate of cholesterol side-chain degradation. Mutant RG32Ωcyp125 thus is unable to form the sterol C26-oic acid intermediate, strongly indicating that CYP125DSM43269 catalyses the oxidation of the sterol at C26. Finally, CYP125RHA1 bound cholesterol, 5α-cholestane-3β-ol and 4-cholestene-3-one in a manner typical of P450 substrates: each compound induced a transition in the spin state of the haem iron and each bound with apparent submicromolar dissociation constants. The conclusion that CYP125 is a steroid 26-monooxygenase extends previous studies in which an NADH-dependent mixed function oxidase system was reported to be responsible for the first step in the mycobacterial sterol side-chain degradation pathway (Szentirmai, 1990) catalysing sterol C26-oxidation (Ambrus et al., 1995).
Our data indicate that the oxidation of C26 is an essential first step of cholesterol degradation in R. jostii RHA1. The RHA1Δcyp125 mutant not only failed to detectably transform cholesterol, but grew on 3-oxo steroids, such as 4-cholestene-3-one and 5α-cholestane-3-one, as effectively as the wild-type strain. This indicates that in R. jostii RHA1, C26-oxidation precedes oxidation of the 3β-hydroxyl moiety (Fig. 1). RHA1Δcyp125 is likely able to grow on 3-oxo steroids by degrading steroid A and B rings, resulting in the formation of 2-hydroxyhexa-2,4-diene-oic acid that is further metabolized to form pyruvate and propionyl-CoA for growth (Fig. 1; van der Geize et al., 2007). Previously, it was suggested that the microbial catabolism of cholesterol was initiated by ring oxidation (Sojo et al., 1997; Chen et al., 2006; Chiang et al., 2008). Indeed, R. rhodochrous RG32Ωcyp125 is capable of performing ring oxidation in the absence of CYP125, illustrating that the order of ring oxidation and sterol side-chain oxidation may vary between different species of bacteria. Consistent with the conclusion that CYP125RHA1 initiates cholesterol degradation, genes encoding putative CHOs in RHA1 (ro03863, ro04305, ro06201) were not upregulated during growth on cholesterol and are located outside of the cholesterol catabolic gene cluster (McLeod et al., 2006; Van der Geize et al., 2007). Although 3β-HSD has not been definitively identified in RHA1, ro04707 encodes a protein sharing 43% amino acid similarity with 3β-HSD of M. tuberculosis (Rv1106c) and is located proximal to the genes encoding the Mce4 steroid transporter (Mohn et al., 2008). Indeed, ro04707 was upregulated in cholesterol-grown RHA1 cells (Van der Geize et al., 2007). While no cholesterol-transforming 3β-HSD activity was detected in RHA1, a 3β-HSD was expressed that transformed 5-pregnene-3β-ol-20-one, a 3β-hydroxysteroid with a shortened C21 side-chain. 3β-HSDRHA1 thus appears to have a high substrate specificity for side-chain-degraded cholesterol metabolites. This is similar to 3β-HSD of M. tuberculosis (Rv1106c) which had threefold higher activity towards 5-pregnene-3β-ol-20-one compared with cholesterol (Yang et al., 2007). It is possible that in RHA1, side-chain and ring degradation occur concurrently after C26 and C3-ol have been oxidized.
It is unclear whether CYP125 catalyses the oxidation of C26 to the corresponding carboxylic acid or only to an intermediate state. Various P450s have been reported to catalyse multistep oxidations (Helliwell et al., 1999; 2001; Ro et al., 2006), including a P450 from Pseudomonas putida PpG777 which catalyses two sequential oxidations of linalool, to 8-hydroxylinalool and 8-oxolinalool respectively (Ropp et al., 1993). It was proposed that a second oxygenation step results in a transient gem-diol adduct that spontaneously dehydrates to a more stable carbonyl compound (Ullah et al., 1990). Interestingly, CYP125RHA1 displays significant amino acid sequence identity (32%) with linalool 8-monooxygenase and thus might well catalyse the complete oxidation of the aliphatic sterol side-chain into the sterol 26-oic acid intermediate via a similar mechanism. Attempts to reconstitute the activity of CYP125RHA1 have so far proved unsuccessful despite using a variety of electron donors, including the spinach ferredoxin and ferredoxin-reductase electron transport chain and the peroxide shunt using cumene hydroxyperoxide (Hrycay et al., 1975). The physiological reductase of CYP125RHA1 has not been identified yet.
Mycobacterium tuberculosis contains a CYP125 encoded by rv3545c located within the recently described igr operon (Chang et al., 2007; 2009). The bioinformatic data strongly suggest that the CYP125s of RHA1 and M. tuberculosis perform the same function: they are reciprocal best hits with 69% amino acid sequence identity that both occur in the cholesterol catabolic gene cluster (Van der Geize et al., 2007). However, the recently reported phenotype of an Δigr mutant indicates that Rv3545c is not a steroid 26-hydroxylase (Chang et al., 2009): the mutant appeared to partially degrade cholesterol and transform the cholesterol labelled with 14C at C26 into mycobacterial lipids. Additional studies are clearly required to definitively establish the role of CYP125 in M. tuberculosis. Indeed, while it is unclear if cholesterol degradation in mycobacteria occurs in the same manner as in R. jostii RHA1, two studies suggest that it does. First, Mycobacterium sp. NRRL B-3683, a mutant strain blocked in steroid ring degradation and able to selectively degrade the sterol side-chain, displayed a clear preference for substrates possessing a 3β-hydroxy-Δ5 ring configuration compared with the 3-keto-Δ4 configuration (Marsheck et al., 1972). Second, 3β-HSD of M. tuberculosis (Rv1106c) showed threefold higher activity towards 5-pregnene-3β-ol-20-one, a sterol with a C21 side-chain, compared with cholesterol, suggesting that sterols with shortened side-chains are preferred substrates of 3β-HSD (Yang et al., 2007). Regardless of the precise function of CYP125 in M. tuberculosis, its gene is upregulated during growth of M. tuberculosis in macrophages (Kendall et al., 2004) and CYP125H37Rv is more resistant to nitric oxide than other P450s of H37Rv (Ouellet et al., 2009). Moreover, the gene appears to be important for infection in mice (Chang et al., 2007; 2009). CYP125 may thus be an interesting target for the development of novel antituberculosis drugs.
Experimental procedures
Bacterial strains, plasmids and chemicals
Plasmids and bacterial strains used are listed in Table S2. 5-Cholestene-3β-ol, 5α-cholestane-3β-ol, 4-cholestene-3-one and 5-cholestene-24β-ethyl-3β-ol (75%) were obtained from Sigma-Aldrich. 5α-Cholestane-3-one was obtained from Acros Organics. 5-Pregnene-3β-ol-20-one was obtained from ICN Biomedicals. 5-Cholestene-3α-ol, 23,24-bisnor-5-cholenic acid-3β-ol, 5-cholenic acid 3β-ol and 1-(5α)-androstene-3,17-dione were obtained from Steraloids. 4-Androstene-3,17-dione and 9,17-dioxo-1,2,3,4,10, 19-hexanorandrostane-5-oic acid were provided by Schering-Plough (Oss, the Netherlands).
Construction of R. jostii RHA1Δcyp125
A cyp125 unmarked single gene deletion mutant of R. jostii RHA1 was constructed using the sacB counter-selection system (Van der Geize et al., 2001). Genomic DNA of R. jostii RHA1 was isolated as described (Van der Geize et al., 2000). Mutagenic plasmid pDELcyp125RHA1 was constructed for cyp125 deletion, as follows. The upstream region of cyp125 was amplified by PCR using forward primer 5′-tcgac atccacttgatgaaggagaccg-3′ and reverse primer 5′-gcgACTAG Tcactgctgtctcctgccctaagc-3′, containing a SpeI restriction site (shown in capital letters). The resulting 1421 bp amplicon was cloned into SmaI-digested pK18mobsacB, resulting in pK18mobsacBUPcyp125. A 1451 bp amplicon of the cyp125 downstream flanking region including the cyp125 stop codon was obtained using forward primer 5′-cgcACTAGTtgaccccctg attcagcggcgtcgg-3′ (SpeI restriction site) and reverse primer 5′-cgcAAGCTTgaacgaggacggcaagatcacgtccc-3′ (HindIII restriction site). This amplicon was digested with SpeI/HindIII and ligated into SpeI/HindIII linearized pK18mobsacB UPcyp125, resulting in pDELcyp125RHA1. Deletion of cyp125 from RHA1 was confirmed by PCR using forward 5′-gcctcga cgattactggtgtgc-3′ and reverse primer 5′-cctcggacagaa ggagaacagc-3′.
Functional complementation of mutant strain RHA1Δcyp125 was performed by electrotransformation (Van der Geize et al., 2000) of RHA1Δcyp125 cells with expression plasmid pTip-QC1cyp125RHA1 (see below).
Growth of R. jostii RHA1 and mutant RHA1Δcyp125 strain on sterols/steroids
Pre-cultures of wild-type strain RHA1 and mutant strain RHA1Δcyp125 were grown for 3 days at 30°C with shaking (220 r.p.m.) in MM (Masai et al., 1995) supplemented with pyruvate (20 mM) and used to inoculate MM liquid media (1:50) supplemented with various sterols/steroids (1 g l−1; Table S1) as sole carbon and energy source. Biomass production of R. jostii RHA1 cell cultures incubated with cholesterol were quantified by total protein content determination of sonicated cells (10 cycles of 30 s at 8 μm) using the Bradford protein assay (Bio-Rad, Hercules, CA) with BSA as protein standard.
Biotransformation of cholesterol by R. jostii RHA1 and mutant RHA1Δcyp125
For biotransformation of cholesterol, precultures of RHA1 and RHA1Δcyp125 were grown in MM supplemented with pyruvate (20 mM) for 3 days at 30°C with shaking (220 r.p. m.). The precultures were used to inoculate MM liquid media (1:50) containing pyruvate (20 mM) and cholesterol (2.5 mM).
Determination of intracellular and extracellular total 3β-hydroxysteroid oxidation activity
Total 3β-hydroxysteroid oxidation activity was determined by high-performance liquid chromatography (HPLC) analysis essentially as described by Yang et al. (2007). Cell cultures of RHA1 and RHA1Δcyp125 were grown in MM supplemented with pyruvate (20 mM) for 3 days to an OD600 of 3. Cell cultures of RG32 and RG32Ωcyp125 were grown overnight in Luria–Bertani (LB) medium. Grown cultures were induced for 16 h by adding 0.5 mM cholesterol from a 10 mM stock prepared in isopropanol. The cell cultures (50 ml) were pelleted and the resulting supernatants were filter-sterilized and used for assaying extracellular cholesterol oxidation. The cell pellets were washed two times with 50 mM phosphate buffer (pH 7) supplemented with 5% (v/v) isopropanol and resuspended in 2 ml of the same buffer. Cell lysates were prepared by bead-beating. Cell lysates were centrifuged to remove cell debris. The 3β-hydroxysteroid oxidation assay was performed in 100 mM triethanolamide hydrochloride buffer (pH 8.5) supplemented with 0.05% (v/v) Triton X-100, 3.5 mM NAD+ and either 200 μM cholesterol or 200 μM 5-pregnene-3β-ol-20-one and incubated at 30°C for several hours (Yang et al., 2007). 4-Cholestene-3-one and 4-progestene-3-one formation was quantified by HPLC-UV254nm using calibration curves.
Steroid analysis
Steroid content of the cell cultures was analysed by HPLC and gas chromatography (GC). Culture samples (0.5 ml) were mixed with 2 ml of 80% methanol in water solution and filtered (0.2 μm) prior to analysis by HPLC-UV254nm. HPLC was performed on an Alltima C18 column (250 × 4.6 mm; Alltech, Deerfield, USA, 35°C) using a mobile phase consisting of methanol: water (80:20) supplemented with 1% formic acid at a flow rate of 1 ml min−1. For analysis of 4-cholestene-3-one and 1,4-cholestadiene-3-one a mobile phase consisting of acetonitrile: tetrahydrofuran (75:25) at a flow rate of 2 ml min−1 was used. Samples (0.5 ml) for GC analysis were mixed with 10% H2SO4 (10 μl) and ethyl acetate (2 ml) and the upper organic layer was subjected to GC. GC was performed on a (5% phenylo)-95% methoxypoly siloxane Heliflex AT-5 ms column (30 m × 0.25 mm, ID × 0.25 μm; Alltech, Deerfield, USA) with FID-40 detection at 300°C.
Production of CYP125RHA1
The cyp125RHA1 gene was amplified by PCR on genomic DNA of RHA1 with forward primer 5′-CATATGgcgcagcccaat cttccagaggg-3′, containing an NdeI restriction site, and reverse primer 5′-GGATCCtcagtgtctgaccgggcaaccg-3′, containing a BamHI restriction site, such that the recombinant protein contains a 6-histidine tag. PCR was performed in a reaction mixture (25 μl) consisting of Tris-HCl (10 mM, pH 8), polymerase buffer, dNTP (0.2 mM), primers (0.8 μM) and Vent polymerase (0.1 U, New England Biolabs, Ipswich, MA) under the following conditions: 5 min 95°C, 30 cycles of 45 s 95°C, 45 s 65°C, 2 min 72°C, followed by 5 min at 72°C. A band of the expected size for cyp125RHA1 (1266 bp) was purified from agarose gel using GenElute Gel Extraction Kit (Sigma-Aldrich, Steinheim, Germany) and cloned into SmaI-digested pBlueScript KS(II) (Stratagene, La Jolla, CA, USA). The resulting plasmid was digested with NdeI and BamHI and the DNA fragment containing cyp125RHA1 was ligated into NdeI/BamHI-digested digested pTip-QC1.
CYP125RHA1 was homologously produced in R. jostii RHA1 using expression plasmid pTip-QC1cyp125RHA1. Cells were cultured in LB broth in the presence of 25 μg ml−1 chloramphenicol. R. jostii RHA1 cells were transformed with pTip-QC1cyp125RHA1 by electroporation and grown on LB-agar plates containing 25 μg ml−1 chloramphenicol for 2 days, after which a single colony was used to inoculate 50 ml of liquid medium which was incubated at 30°C (200 r. p.m.). When OD600 reached ~1.0 (~2–3 days) 2 l of medium inoculated with 20 ml of this preculture was incubated at 30°C. When the culture reached an OD600 of 0.6, thiostrepton was added to a final concentration of 50 μg ml−1 and the cells were incubated for a further 20 h before harvesting by centrifugation (4600 g, 4°C, 10 min) and subsequent washing with 0.1 M potassium phosphate buffer, pH 8.0. Cell pellets were flash frozen in liquid nitrogen and stored at −80°C until use.
Purification of CYP125RHA1
The cell pellets were suspended in potassium phosphate buffer (pH 7.4) (Lussenburg et al., 2005) containing DNase I (Roche diagnostics, IN). Cells were disrupted by bead beating and debris was removed by centrifugation at 10 000 g for 45 min at 4°C. The clear supernatant was passed through a syringe-driven 0.45 μm filter. Cell free extracts were loaded on a NTA column (Qiagen) equilibrated with 0.1 mM potassium phosphate, pH 7.4. The protein was washed with Buffer A containing 0.5 M NaCl and a brown fraction eluted with buffer A further supplemented with 50 mM L-histidine. The protein was exchanged into 0.1 M potassium phosphate, pH 7.4, concentrated to 20 mg ml−1, flash frozen as beads in liquid nitrogen and stored at −80°C. P450 protein concentrations were calculated from the reduced CO-bound difference spectrum using the extinction coefficient ε450−490 = 91 mM−1 cm−1 (Omura and Sato, 1964).
Spectroscopic analysis
UV-vis absorption spectra were recorded using a Cary 5000 spectrophotometer equipped with a thermojacketed cuvette holder (Varian, Walnut Creek, CA). The CO-bound form of CYP125RHA1 was generated by first incubating samples with ~8 mM sodium dithionite for 10 min then slowly bubbling them with CO for 30 s. The proportion of purified protein containing high-spin ferric haem iron was estimated by comparing the spectra of CYP125RHA1 to linear combinations of the spectra of CYP125RHA1 in high and low spin states (Jung et al., 1991; Jefcoate, 1978) generated by adding 0.5% Triton X-100 and 40% methanol, respectively, to the sample. The same values were obtained when using substrate-free cytochrome P450cam from P. putida as a low spin standard. Substrate-induced spectral responses were recorded in 0.1 mM KPi, pH 7.0 by titrating solutions of CYP125RHA1 with 1.0 mM stock solutions of cholesterol, 5α-cholestane-3β-ol, and 4-cholesten-3-one in 10% 2-hydroxypropyl-β-cyclodextrin (Sigma). Equilibrium dissociation constants were calculated using Eq. 1.
| (1) |
In this equation, ΔA is the change in absorbance observed in the sample, [S]T is the total ligand concentration, [E]T is the total enzyme concentration, KD is the equilibrium dissociation constant, and ΔAMax is the change in absorbance at infinite ligand concentration. A non-linear least-squares fit of the equation to the data was obtained using the program R (http://www.R-project.org).
Construction of R. rhodochrous RG32Ωcyp125
The cyp125 orthologue in R. rhodochrous DSM43269 (cyp125DSM43269) was identified using degenerate cyp125 primers (forward 5′ (a/g)ac(a/c/g/t)gc(a/c/g/t)cc(a/c/g/t)at (a/c/t)tggtggaa and reverse 5′-gg(a/g)tt(c/t)tc(a/g)aa(a/c/g/t) gc(a/g)tc(c/t)tc(a/g)) based on the deduced amino acid sequences T33APIWWN39 and D329EDAFENP336 from CYP125RHA1; these sequences are highly conserved in Nfa5180 and Rv3545c from N. farcinica IFM10152 and M. tuberculosis H37Rv respectively. A genomic library of R. rhodochrous DSM43269 in pRESQ (Petrusma et al., 2009; Table S2) was screened by PCR using these degenerate primers. A single clone (pRESQ4679) was identified containing an 8.7 kb DNA insert. Nucleotide sequencing confirmed the presence of full-length (1254 bp) cyp125 (DDBJ/EMBL/Genbank Accession No. FJ824698).
The cyp125 gene was disrupted in R. rhodochrous strain RG32 essentially as described (Van der Geize et al., 2000). An internal cyp125DSM43269 fragment of 811 bp was amplified by PCR using forward primer 5′-gcacgaggaggtccgtgaggtc and reverse primer 5′-cgttgttggccgaggcgtacag and ligated into EcoRV-digested pK18mobsacB, yielding pΩcyp125. This construct was used to transform E. coli S17-1 and was subsequently mobilized to mutant strain RG32 by conjugational transfer (Van der Geize et al., 2001). Transconjugants were checked by PCR to confirm the cyp125 gene disruption using forward primer 5′-acgcagccaccgatgacctgtt, annealing to a sequence upstream of cyp125DSM43269, and reverse primer 5′-ctgcgtgcaatccatcttgttc, which is reverse complementary to part of the aphII gene of pK18mobsacB. A PCR product of the expected size (1903 bp) confirmed insertion of the disruption plasmid pΩcyp125 at the correct genomic locus.
Functional complementation RG32Ωcyp125
The intact cyp125 DSM43269 gene and its flanking regions were isolated from DraIII/BspHI-digested pRESQ4679. A DNA fragment of 2.3 kb harbouring cyp125 was treated with T4 DNA polymerase and blunt-ligated into EcoRV-digested shuttle vector pRRE1 (see below), resulting in pCOMPcyp125DSM43269 that was used to transform electro-competent cells of RG32Ωcyp125 as described (Fernandes et al., 2001). E. coli-Rhodococcus shuttle vector pRRE1 was constructed as follows. The repA and repB genes from R. rhodochrous DSM43269 endogenous plasmid pRC4 (GenBank/EMBL/DDJB accession number AB040101) were amplified from genomic DNA of strain DSM43269 using forward primer 5′-cgatggcaagccaccgcgaagc and reverse primer 5′-atcggacagaagctgactaagg. This amplicon (2.5 kb) was ligated into SmaI-digested pK18mobsacB. A 2.6 kb EcoRI/XbaI DNA fragment of the latter construct was subsequently treated with Klenow fragment and blunt-ligated into PsiI-digested pBs-Apra-ori (Van der Geize et al., 2008), resulting in pRRE1.
Whole-cell steroid biotransformations with RG32 and RG32Ωcyp125
Cell cultures of parent strain R. rhodochrous RG32, mutant strain RG32Ωcyp125 and the cyp125DSM43269 complemented mutant strain were grown overnight in liquid LB medium, supplemented with kanamycin 25 μg ml−1 when appropriate, at 30°C with shaking (200 r.p.m.) until OD600 ~4 was reached. Sterols were added to the cell cultures at a final concentration of 0.5 mM from a 25 mM stock solution dissolved in acetone. Bioconversions were followed for 3 days of incubation at 30°C with shaking (200 r.p.m.). Accumulation of ADD and Δ1,4-BNC was analysed by HPLC-UV254nm as described above in Steroid analysis.
Chemical synthesis of 5-cholestene-26-oic acid-3β-ol
Synthesis of 5-cholestene-26-oic acid -3β-ol was carried out using a modification of the method described by Williams et al. (2002) with diosgenin as starting material (Fig. S2). In the first step, the 3-hydroxy group was protected as a methyl ether, using NaH and MeI and a reaction time of 24 h. The resulting 3-methyl ether (product 1) was isolated in near 100% yield after precipitation from water. Next, the ether rings were reductively ring-opened under Clemmensen conditions by treatment with Zn/HCl in ethanol at reflux temperature. After removal of the salts, extractive work up and a precipitation from acetone/water, the 16, 27-dihydroxylated product (product 2) was obtained in near 100% yield. A regio selective protection of the primary alcohol at C27 was carried out by reaction with tert-butyldimethylsilyl chloride and imidazole in N,N-dimethylformamide with 97% yield (product 3). For removal of the 16-hydroxy group, the Barton deoxygenation conditions were chosen. The C16-hydroxy group was transformed in the corresponding thiocarbonate with CS2 under the influence of NaH. The intermediate thiocarbonate anion was quenched with methyl iodide. Next, a radical reduction reaction was carried out using Bu3SnH and AIBN. After purification by silica gel column chromatography, the tert-butyldimethylsilyl-protected 3-methyl ether form of 27-hydroxycholesterol (product 4) could be isolated in near 100% yield with an estimated 1H-NMR purity of > 80%. The tert-butyldimethylsilyl ether was removed under standard conditions using tetrabutylammonium fluoride in tetrahydrofuran, and silica gel column chromatography was used to purify product 5 with a yield of 73%. Oxidation to the 26-oic acid was carried out under Jones’ condition, using a mixture of sulphuric acid and chromine trioxide. 5-Cholestene-26-oic acid-3β-ol-3-methyl ether (product 6) was obtained by column chromatography purification in 89% yield with an estimated 1H-NMR purity of 80%. The final step in the synthesis was the removal of the 3-methyl ether by treatment with TFA in DCM at room temperature for 2 days. After aqueous work up, the trifluoroethanol ester was saponified with K2CO3 in methanol and purified by silica gel column chromatography, generating 5-cholestene-26-oic acid-3β-ol (product 7) in a low yield of 15% with a 1H NMR purity of approximately 95% and consisting of a 1:1 mixture of diastereomers at C26. Apparently, during the strong acidic conditions used for the removal of the 3-methyl ether, enolization and protonation at C26 had occurred giving rise to a 1:1 mixture of stereo-isomers. The structure was confirmed by mass spectrometry (Fig. S3).
Supplementary Material
Acknowledgments
We are grateful to N. P. E. Vermeulen, J. Commandeur and B. van Vugt-Lussenburg (VU Amsterdam) for technical advice and support. We thank T. Tiemersma-Wegman (University of Groningen) for technical assistance in GC analyses. We thank T. Tamura for kindly providing pTip-QC1. This work was supported by grants from the Integration of Biosynthesis and Organic Synthesis program of Advanced Chemical Technologies for Sustainability and the Canadian Institutes for Health Research (to L.D.E.). J.C. is the recipient of studentships from NSERC of Canada and MSFHR.
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ambrus G, Jekkel A, Ilkoy E, Horvath G, Bocskei Z. Novel 26-oxygenated products in microbial degradation of ergosterol. Steroids. 1995;60:626–629. doi: 10.1016/0039-128x(95)00078-5. [DOI] [PubMed] [Google Scholar]
- Arima K, Nakamatsu T, Beppu T. Microbial production of 3-oxobisnorchola-1,4-dien-22-oic acid. Agric Biol Chem. 1978;42:411–416. [Google Scholar]
- Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- Chang JC, Harik NS, Liao RP, Sherman DR. Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. J Infect Dis. 2007;196:788–795. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- Chang JC, Miner MD, Pandey AK, Gill WP, Harik NS, Sassetti CM, Sherman DR. igr genes and Mycobacterium tuberculosis cholesterol metabolism. J Bacteriol. 2009;191:5232–5239. doi: 10.1128/JB.00452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Huan HH, Cheng TF, Tang TY, Liu WH. Expression of a cholesterol oxidase gene from Arthrobacter simplex in Escherichia coli and Pichia pastoris. Enzyme Microb Technol. 2006;39:258–262. [Google Scholar]
- Chiang YR, Ismail W, Heintz D, Schaeffer C, Van Dorsselaer A, Fuchs G. Study of anoxic and oxic cholesterol metabolism by Sterolibacterium denitrificans. J Bacteriol. 2008;190:905–914. doi: 10.1128/JB.01525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Fernandes PJ, Powell JA, Archer JA. Construction of Rhodococcus random mutagenesis libraries using Tn5 transposition complexes. Microbiology. 2001;147:2529–2536. doi: 10.1099/00221287-147-9-2529. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Chen CS, Szeleczky Z, DiTullio D, Sih CJ. Microbial degradation of the phytosterol side chain. 1. Enzymatic conversion of 3-oxo-24-ethylcholest-4-en-26-oic acid into 3-oxochol-4-en-24-oic acid and androst-4-ene-3,17-dione. J Am Chem Soc. 1982a;104:4718–4720. [Google Scholar]
- Fujimoto Y, Chen CS, Gopalan AS, Sih CJ. Microbial degradation of the phytosterol side chain. 2. Incorporation of NaH14CO3 onto the C-28 position. J Am Chem Soc. 1982b;104:4720–4722. [Google Scholar]
- Helliwell CA, Poole A, Peacock WJ, Dennis ES. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycay EG, Gustafsson JA, Ingelman-Sundberg M, Ernster L. Sodium periodate, sodium chloride, organic hydroperoxides, and H2O2 as hydroxylating agents in steroid hydroxylation reactions catalyzed by partially purified cytochrome P-450. Biochem Biophys Res Commun. 1975;66:209–216. doi: 10.1016/s0006-291x(75)80315-9. [DOI] [PubMed] [Google Scholar]
- Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, et al. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci USA. 2004;101:14925–14930. doi: 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate CR. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- Julsing MK, Cornelissen S, Bühler B, Schmid A. Heme-iron oxygenases: powerful industrial biocatalysts? Curr Opin Chem Biol. 2008;12:177–186. doi: 10.1016/j.cbpa.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Jung C, Ristau O, Rein H. The high-spin/low-spin equilibrium in cytochrome P-450 – a new method for determination of the high-spin content. Biochim Biophys Acta. 1991;1076:130–136. doi: 10.1016/0167-4838(91)90229-s. [DOI] [PubMed] [Google Scholar]
- Keizers PH, Lussenburg BM, de Graaf C, Mentink LM, Vermeulen NP, Commandeur JN. Influence of phenylalanine 120 on cytochrome P450 2D6 catalytic selectivity and regiospecificity: crucial role in 7-methoxy-4-(aminomethyl) -coumarin metabolism. Biochem Pharmacol. 2004;68:2263–2271. doi: 10.1016/j.bcp.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Kendall SL, Rison SCG, Movahedzadeh F, Frita R, Stoker NG. What do microarrays really tell us about M. tuberculosis? Trends Microbiol. 2004;12:537–544. doi: 10.1016/j.tim.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Knol J, Bodewits K, Hessels GI, Dijkhuizen L, van der Geize R. 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J. 2008;410:339–346. doi: 10.1042/BJ20071130. [DOI] [PubMed] [Google Scholar]
- Lamb DC, Guengerich FP, Kelly SL, Waterman MR. Exploiting Streptomyces coelicolor A3(2) P450s as a model for application in drug discovery. Expert Opin Drug Metab Toxicol. 2006;2:27–40. doi: 10.1517/17425255.2.1.27. [DOI] [PubMed] [Google Scholar]
- Lussenburg BM, Babel LC, Vermeulen NP, Commandeur JN. Evaluation of alkoxyresorufins as fluorescent substrates for cytochrome P450 BM3 and site-directed mutants. Anal Biochem. 2005;341:148–155. doi: 10.1016/j.ab.2005.02.025. [DOI] [PubMed] [Google Scholar]
- MacLachlan J, Wotherspoon AT, Ansell RO, Brooks CJ. Cholesterol oxidase: sources, physical properties and analytical applications. J Steroid Biochem Mol Biol. 2000;72:169–195. doi: 10.1016/s0960-0760(00)00044-3. [DOI] [PubMed] [Google Scholar]
- McLean KJ, Clift D, Lewis DG, Sabri M, Balding PR, Sutcliffe MJ, et al. The preponderance of P450s in the Mycobacterium tuberculosis genome. Trends Microbiol. 2006;14:220–228. doi: 10.1016/j.tim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsheck WJ, Kraychy S, Muir RD. Microbial degradation of sterols. Appl Microbiol. 1972;23:72–77. doi: 10.1128/am.23.1.72-77.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E, Yamada A, Healy JM, Hatta T, Kimbara K, Fukuda M, et al. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:2079–2085. doi: 10.1128/aem.61.6.2079-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N, Murtazina D, Liu H, Graham SE, Bjorkhem I, Halpert JR, et al. Distinct binding of cholesterol and 5beta-cholestane-3alpha,7alpha,12alpha-triol to cytochrome P450 27A1: evidence from modeling and site-directed mutagenesis studies. Biochemistry. 2006;45:4396–4404. doi: 10.1021/bi052654w. [DOI] [PubMed] [Google Scholar]
- Masuda S, Prosser DE, Guo YD, Kaufmann M, Jones G. Generation of a homology model for the human cytochrome P450, CYP24A1, and the testing of putative substrate binding residues by site-directed mutagenesis and enzyme activity studies. Arch Biochem Biophys. 2007;460:177–191. doi: 10.1016/j.abb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Mohn WW, Van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, et al. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Tamura T. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl Environ Microbiol. 2004;70:5557–5568. doi: 10.1128/AEM.70.9.5557-5568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- Ouellet H, Lang J, Couture M, Ortiz de Montellano PR. Reaction of Mycobacterium tuberculosis cytochrome P450 enzymes with nitric oxide. Biochemistry. 2009;48:863–872. doi: 10.1021/bi801595t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Gillam EM, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol. 1997;15:784–788. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- Petrusma M, Dijkhuizen L, Van der Geize R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9alpha-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl Environ Microbiol. 2009;75:5300–5307. doi: 10.1128/AEM.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuleva IA. Cholesterol-metabolizing cytochromes P450. Drug Metab Dispos. 2006;34:513–520. doi: 10.1124/dmd.105.008789. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Ropp JD, Gunsalus IC, Sligar SG. Cloning and expression of a member of a new cytochrome P-450 family: cytochrome P-450lin (CYP111) from Pseudomonas incognita. J Bacteriol. 1993;175:6028–6037. doi: 10.1128/jb.175.18.6028-6037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sih CJ, Tai HH, Tsong YY. The mechanism of microbial conversion of cholesterol into 17-keto steroids. J Am Chem Soc. 1967;89:1957–1958. doi: 10.1021/ja00984a039. [DOI] [PubMed] [Google Scholar]
- Sih CJ, Tai HH, Tsong YY, Lee SS, Coombe RG. Mechanisms of steroid oxidation by microorganisms. XIV. Pathway of cholesterol side-chain degradation. Biochemistry. 1968b;7:808–818. doi: 10.1021/bi00842a039. [DOI] [PubMed] [Google Scholar]
- Sih CJ, Wang KC, Tai HH. Mechanism of steroid oxidation by microorganisms XIII. C22 acid intermediates in the degradation of the cholesterol side chain. Biochemistry. 1968a;7:796–807. doi: 10.1021/bi00842a038. [DOI] [PubMed] [Google Scholar]
- Sojo M, Bru R, Lopez-Molina D, Garcia-Carmona F, Arguelles JC. Cell-linked and extracellular cholesterol oxidase activities from Rhodococcus erythropolis. Isolation and physiological characterization. Appl Microbiol Biotechnol. 1997;47:583–589. doi: 10.1007/s002530050977. [DOI] [PubMed] [Google Scholar]
- Storbeck KH, Swart P, Swart AC. Cytochrome P450 side-chain cleavage: insights gained from homology modeling. Mol Cell Endocrinol. 2007;265–266:65–70. doi: 10.1016/j.mce.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Szentirmai A. Microbial physiology of sidechain degradation of sterols. J Ind Microbiol. 1990;6:101–116. [Google Scholar]
- Ullah AJ, Murray RI, Bhattacharyya PK, Wagner GC, Gunsalus IC. Protein components of a cytochrome P-450 linalool 8-methyl hydroxylase. J Biol Chem. 1990;265:1345–1351. [PubMed] [Google Scholar]
- Van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Van der Geize R, Hessels GI, van Gerwen R, Vrijbloed JW, van der Meijden P, Dijkhuizen L. Targeted disruption of the kstD gene encoding a 3-ketosteroid Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl Environ Microbiol. 2000;66:2029–2036. doi: 10.1128/aem.66.5.2029-2036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase. Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol Lett. 2001;205:197–202. doi: 10.1016/s0378-1097(01)00464-5. [DOI] [PubMed] [Google Scholar]
- Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Geize R, De Jong W, Hessels GI, Grommen AW, Jacobs AA, Dijkhuizen L. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 2008;36:e151. doi: 10.1093/nar/gkn811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JR, Chai D, Wright D. Synthesis of (25R) -26-hydroxycholesterol. Steroids. 2002;67:1041–1044. doi: 10.1016/s0039-128x(02)00060-0. [DOI] [PubMed] [Google Scholar]
- Yang X, Dubnau E, Smith I, Sampson NS. Rv1106c from Mycobacterium tuberculosis is a 3beta-hydroxysteroid dehydrogenase. Biochemistry. 2007;46:9058–9067. doi: 10.1021/bi700688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.