Abstract
In eubacteria, the tmRNA system frees ribosomes that stall during protein synthesis and adds an ssrA tag to the incompletely translated polypeptide to target it for degradation. The AAA+ ClpXP protease degrades most ssrA-tagged proteins in the E. coli cytoplasm and was recently shown to degrade an ssrA-tagged protein in the inner membrane. However, we find that tmRNA-mediated tagging of E. coli ProW1-182, a different inner-membrane protein, results in degradation by the membrane-tethered AAA+ FtsH protease. ClpXP played no role in degradation of ProW1-182 in vivo. These studies suggest that a complex distribution of proteolytic labor maintains protein quality control in the inner membrane.
Graphical Abstract
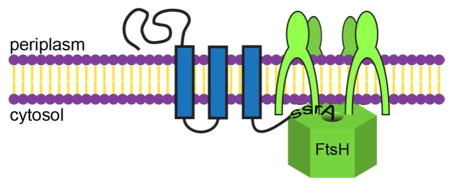
Ribosomes that stall during protein synthesis are a common source of defective intracellular polypeptides.1 In addition to producing an incomplete polypeptide, stalling depletes the pool of ribosomes available for translation and inhibits normal growth. In Escherichia coli and most eubacteria, the hybrid transfer-messenger RNA (tmRNA) system rescues stalled ribosomes and simultaneously appends a short sequence of C-terminal amino acids, called the ssrA tag, that targets the incomplete protein for degradation by ATP-fueled AAA+ proteases in the cytoplasm and by energy-independent proteases in the periplasm.2,3 For example, ~10% of E. coli translation events appear to terminate with tmR-NA rescue and ssrA tagging,1,4 but our understanding of how these ssrA-tagged aberrant proteins are degraded in different cellular compartments is incomplete, especially for membrane proteins.
E. coli contains five AAA+ proteases: ClpXP, ClpAP, FtsH, HslUV, and Lon, each consisting of a multimeric AAA+ ring that binds, unfolds, and translocates protein substrates into a degradation chamber formed by attached domains or separate peptidase subunits.5 Cytoplasmic ssrA-tagged proteins are principally degraded by the ClpXP protease, with some help from ClpAP.2,6,7 FtsH, which is anchored to the cytoplasmic face of the inner membrane, degrades soluble ssrA-tagged proteins in vitro and has been shown to degrade several integral membrane proteins in vivo.8–11 Thus, it has been widely assumed that FtsH is also responsible for degrading ssrA-tagged membrane proteins.12 Challenging this assumption, however, a recent study showed that ClpXP degrades ssrA-tagged AcrB, an E. coli inner-membrane protein.13 Here, we investigate a different ssrA-tagged integral membrane protein and find that its degradation requires FtsH but not ClpXP or ClpAP in vivo. Thus, which AAA+ protease degrades which ssrA-tagged inner-membrane protein depends on factors that are currently poorly defined.
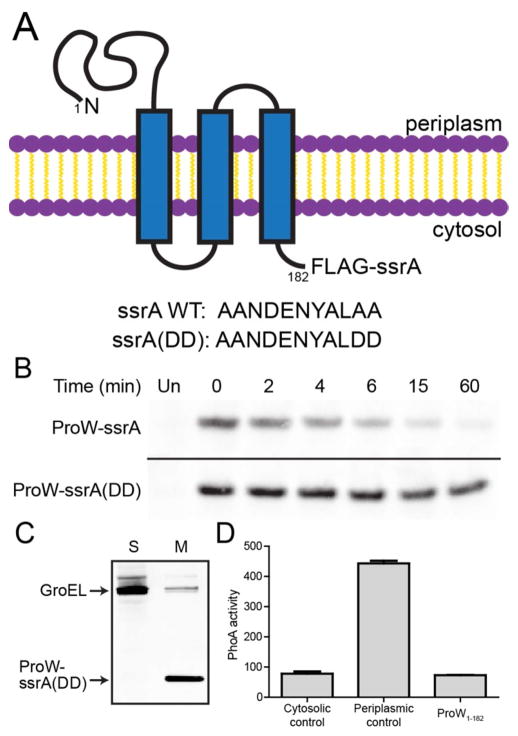
E. coli ProW is a multiple-pass inner-membrane protein.14 For our studies, we used a truncated variant (ProW1-182) with an N-terminal periplasmic domain, three transmembrane segments, and a C-terminus (with two added lysines) predicted to be cytosolic.15 ProW1-182 was fused to a FLAG tag and an ssrA tag (ProW1-182-FLAG-ssrA; Fig. 1A) and cloned under control of an IPTG-inducible Ptrc promoter. We also generated an otherwise identical construct in which the last two residues of the ssrA tag were replaced with aspartic acids (ssrA(DD)), as this mutant tag is not recognized by cellular proteases.3 In 35S pulse-chase experiments (Table 1 and Fig. 1B), ProW1-182-FLAG-ssrA was degraded quickly (t1/2 ~5 min), whereas ProW1-182-FLAG-ssrA(DD) was not (t1/2 > 60 min). The stability of ProW1-182-FLAG-ssrA-(DD) allowed us to show that it purified with cellular membranes but was undetectable in the soluble cytosolic fraction in fractionation experiments (Fig. 1C). As expected from results using a very similar construct,14 the activity of PhoA fusions to ProW1-182-FLAG-ssrA confirmed that its C-terminus is cytoplasmic (Fig. 1D).
Figure 1.
An ssrA-tagged model membrane protein is degraded in E. coli. (A) Topology of ProW1-182-FLAG-ssrA (top) and amino-acid sequences of wild-type and mutant ssrA tags (bottom). (B) Expression of ProW1-182-FLAG-ssrA or ProW1-182-FLAG-ssrA(DD) in E. coli X90 was induced, pulsed with 35S-labeled methionine and cysteine, and chased with unlabeled amino acids. Samples taken at different times were immunoprecipitated and analyzed by SDS-PAGE and autoradiography. “Un” indicates an uninduced control sample. (C) E. coli cells expressing ProW1-182-FLAG-ssrA(DD) were lysed and fractionated into soluble (S) and membrane (M) fractions, which were analyzed by SDS-PAGE and immunoblotted using antibodies against GroEL (a cytosolic protein) and the FLAG tag. (D) Activity (in Miller units) of C-terminal FLAG-ssrA-PhoA fusions of a cytosolic protein (λ repressor), a periplasmic protein (cytochrome b562), and ProW1-182.
Table 1.
Half-lives of ProW constructs determined from two independent experiments. n.d. – not determined.
| Half-life (min) | ||
|---|---|---|
| X90 | X90 ΔssrA::Cam | |
| ProW1-182-FLAG-ssrA | 7, 4 | n.d. |
| ProW1-182-FLAG-ssrA(DD) | >60, >60 | n.d. |
| ProW1-182-FLAG-trpAt | 3, 2 | ~60, ~60 |
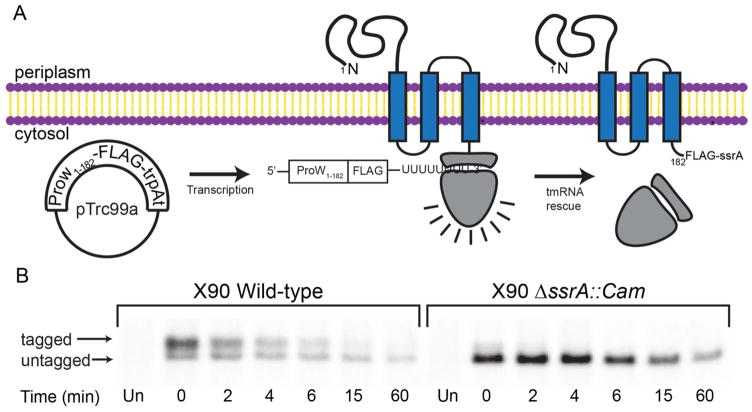
tmRNA-mediated addition of the ssrA tag in vivo can be induced using a non-stop mRNA that causes ribosome stalling at the 3′ end.3 When ProW1-182-FLAG was expressed from a gene with a strong transcriptional terminator and no stop codon (Fig. 2A), we observed ~80% ssrA-tagged ProW1-182-FLAG (higher molecular-weight band) and ~20% untagged ProW1-182-FLAG (lower molecular-weight band) at time zero in a pulse-chase experiment (Fig. 2B; left panel). Importantly, the tagged species was degraded rapidly (t1/2 ~3 min) compared to the untagged species (t1/2 ~60 min) (Table 1). As expected, the tagged species was not observed in a pulse-chase experiment performed in ΔssrA cells lacking tmRNA (Fig. 2B; right panel). Thus, whether ProW1-182-FLAG contains a genetically encoded ssrA tag or acquires an ssrA tag as a consequence of ribosome stalling and tmRNA rescue, the tagged protein is rapidly degraded in E. coli.
Figure 2.
tmRNA-mediated tagging of ProW1-182 results in rapid degradation. (A) A non-stop mRNA encoding ProW1-182-FLAG results in ribosomal stalling and ssrA tagging. (B) A plasmid encoding ProW1-182-FLAG-trpAt was transformed into wild-type E. coli X90 or an otherwise isogenic ΔssrA::Cam strain, and the half-life of the tagged and untagged proteins was measured by pulse-chase experiments as described in Fig. 1B.
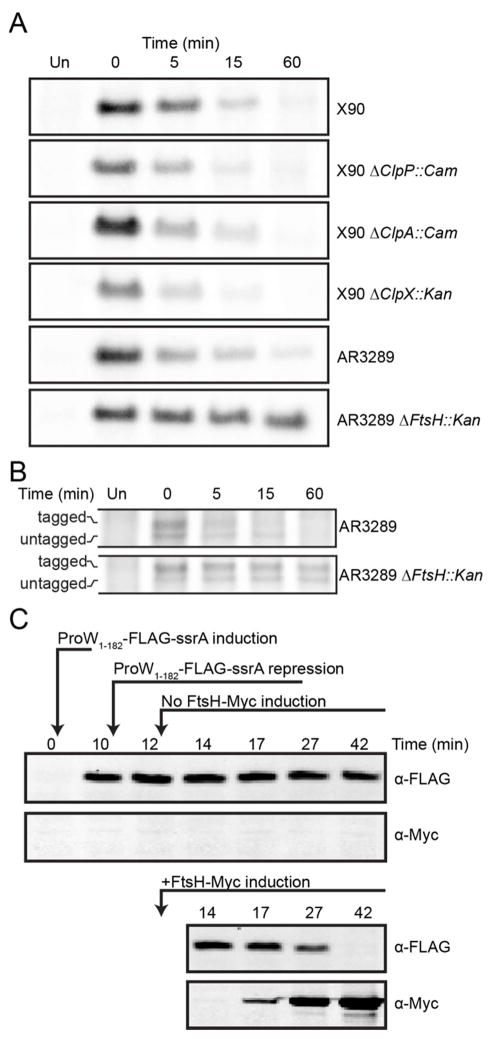
To test the importance of different E. coli AAA+ proteases in intracellular degradation of ProW1-182-FLAG-ssrA, we performed pulse-chase experiments (Fig. 3A) in cells lacking ClpP (the pep-tidase component of ClpXP and ClpAP), ClpX (the AAA+ un-foldase of ClpXP), ClpA (the AAA+ unfoldase of ClpAP), or FtsH (fused AAA+ unfoldase and peptidase). ProW1-182-FLAG-ssrA was degraded at similar rates in a wild-type strain and otherwise isogenic strains lacking ClpP, ClpX, or ClpA. Thus, ClpXP and ClpAP do not contribute to intracellular degradation of ProW1-182-FLAG-ssrA. Strikingly, however, deletion of FtsH resulted in much slower degradation of ProW1-182-FLAG-ssrA (t1/2 > 60 min) compared to an otherwise isogenic wild-type strain (t1/2 ~5 min). We also used the non-stop stalling construct to induce ssrA tagging of ProW1-182-FLAG in cells without FtsH (Fig. 3B). The ssrA-tagged species was observed in the wild-type and ΔFtsH::Kan strains but persisted only in the latter strain (t1/2 ≥ 60 min). Taken together, these results show that rapid degradation of the ssrA-tagged ProW1-182 inner-membrane protein depends upon FtsH.
Figure 3.
FtsH degrades ProW1-182-FLAG-ssrA. (A) Pulse-chase experiments demonstrate that ProW1-182-FLAG-ssrA has a longer half-life in cells lacking FtsH compared to wild-type cells or those lacking ClpP, ClpX, or ClpA (see Fig. 1B legend for experimental details). (B) Auto-radiograms of whole-cell lysates from pulse-chase experiments show that endogenously ssrA-tagged ProW1-182-FLAG-trpAt is also stabilized in a strain lacking FtsH. (C) Transient induction of plasmid-encoded ProW1-182-FLAG-ssrA in E. coli AR3289 ΔftsH::Kan resulted in little degradation (top panels) unless expression of plasmid-encoded Myc-tagged FtsH was subsequently induced (bottom panels). In all panels, ProW1-182-FLAG-ssrA expression was induced with IPTG for 10 min, and then pGAL was added to repress further expression. In the bottom panels, expression of Myc-tagged FtsH was induced by addition of ATc two min after addition of pGAL. Samples were separated by SDS-PAGE and immunoblotted with anti-FLAG and anti-Myc tag antibodies.
Insertion of proteins into the inner membrane occurs rapidly, on time scales comparable to their rates of biosynthesis.16 For ProW1-182-FLAG-ssrA, synthesis should take ~15 s. To test if FtsH degrades ProW1-182-FLAG-ssrA after membrane insertion, we transformed E. coli ΔftsH::Kan with one plasmid encoding ProW1-182-FLAG-ssrA under IPTG control and a second plasmid encoding myc-FtsH under anhydrotetracyline (ATc) control. After briefly inducing synthesis of ProW1-182-FLAG-ssrA from Ptrc with IPTG, we added phenyl-D-galactopyranoside (pGAL) to repress further synthesis. Two min after pGAL addition, one aliquot of cells was treated with ATc to induce myc-FtsH expression and a control aliquot was mock treated. As shown in the upper panel of Fig. 3C, when FtsH synthesis was not induced, the level of intracellular ProW1-182-FLAG-ssrA remained relatively constant from the time of pGAL addition until ~40 min. By contrast, ProW1-182-FLAG-ssrA was completely degraded after 40 min in cells in which myc-FtsH synthesis was induced. We conclude that FtsH-mediated degradation of ProW1-182-FLAG-ssrA can occur after this protein has been inserted into the inner membrane.
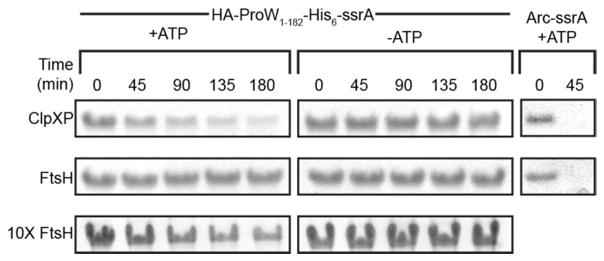
To study degradation in vitro, we purified His6-tagged variants of FtsH and ClpXP and first verified that they efficiently degraded Arc-ssrA (Fig. 4A, right lanes), a soluble model substrate previously shown to be degraded by both proteases.9,17 Next, we tested FtsH and ClpXP degradation of purified HA-ProW1-182-His6-ssrA. We used this substrate because it was easier to purify than ProW1-182-FLAG-ssrA. FtsH and ClpXP both degraded HA-ProW1-182-His6-ssrA, but roughly 10X more FtsH than ClpXP was required to achieve a comparable rate of degradation (Fig. 4).
Figure 4.
In vitro degradation of a model membrane protein. Purified HA-ProW1-182-His6-ssrA was degraded by purified ClpXP (top panel) or FtsH (middle and bottom panels) in ATP-dependent reactions. Arc-ssrA was also degraded by both proteases. Reactions were initiated by addition of 4 mM ATP (with regeneration system), quenched at different times, and then analyzed by SDS-PAGE and staining with Coomassie Blue.
Degradation of ssrA-tagged ProW1-182 in vivo requires FtsH but it is not affected by deletion of ClpXP or ClpAP. Degradation of ssrA-tagged ProW1-182 in vitro was observed with both FtsH and ClpXP, but ClpXP degradation was ~10-fold more efficient. Several factors could be responsible for these differences. For example, the detergent-solubilized environment of the substrate in vitro might interfere with FtsH degradation, FtsH and the substrate might need to be present in the same membrane for efficient recognition and degradation, and/or an additional cellular component or components required for efficient FtsH degradation might be missing in our reconstituted assay. The fact that ClpXP degrades ssrA-tagged ProW1-182 in vitro but not in vivo is likely to be a consequence of the substrate being embedded in the inner-membrane in vivo, which could reduce accessibility of the ssrA tag and/or create an energetic barrier that is too high for ClpX to pull ssrA-tagged ProW1-182 out of the membrane. Further studies will be required to distinguish between these mechanistic possibilities.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by NIH grant R01 AI-016892 (R.T.S.) and by a Ruth L. Kirschstein National Research Service Award (F32GM116241) (S.B.H.).
We are grateful for E. coli strains provided by Teru Ogura. We thank Karl Schmitz, Juhee Park, and Ohad Iosefson for helpful suggestions.
Footnotes
Author Contributions
S.B.H. designed and performed all experiments. S.B.H. and R.T.S. contributed to data analysis and writing the manuscript.
The authors declare no competing financial interests.
- Methods and materials (PDF)
References
- 1.Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 4.Sin C, Chiarugi D, Valleriani A. Quantitative assessment of ribosome drop-off in E. coli. Nucleic Acids Res. 2016;44:2528–2537. doi: 10.1093/nar/gkw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 6.Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 7.Lies M, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiyama Y, Kihara A, Ito K. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 1996;399:26–28. doi: 10.1016/s0014-5793(96)01283-5. [DOI] [PubMed] [Google Scholar]
- 9.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 10.Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci U S A. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kihara A, Akiyama Y, Ito K. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J Mol Biol. 1998;279:175–188. doi: 10.1006/jmbi.1998.1781. [DOI] [PubMed] [Google Scholar]
- 12.Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai Q, Wang Z, Webb SR, Dutch RE, Wei Y. The ssrA-Tag Facilitated Degradation of an Integral Membrane Protein. Biochemistry. 2016;55:2301–2304. doi: 10.1021/acs.biochem.6b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitley P, Zander T, Ehrmann M, Haardt M, Bremer E, von Heijne G. Sec-independent translocation of a 100-residue periplasmic N-terminal tail in the E. coli inner membrane protein proW. EMBO J. 1994;13:4653–4661. doi: 10.1002/j.1460-2075.1994.tb06788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K, Sato T, Yura T. Synthesis and assembly of the membrane proteins in E. coli. Cell. 1977;11:551–559. doi: 10.1016/0092-8674(77)90073-3. [DOI] [PubMed] [Google Scholar]
- 17.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.