Abstract
This paper presents a review of the importance and role of precision medicine and molecular imaging technologies in cancer diagnosis with therapeutics and diagnostics purposes. Precision medicine is progressively becoming a hot topic in all disciplines related to biomedical investigation and has the capacity to become the paradigm for clinical practice. The future of medicine lies in early diagnosis and individually appropriate treatments, a concept that has been named precision medicine, i.e. delivering the right treatment to the right patient at the right time. Molecular imaging is quickly being recognized as a tool with the potential to ameliorate every aspect of cancer treatment. On the other hand, emerging high-throughput technologies such as omics techniques and systems approaches have generated a paradigm shift for biological systems in advanced life science research. In this review, we describe the precision medicine, difference between precision medicine and personalized medicine, precision medicine initiative, systems biology/medicine approaches (such as genomics, radiogenomics, transcriptomics, proteomics, and metabolomics), P4 medicine, relationship between systems biology/medicine approaches and precision medicine, and molecular imaging modalities and their utility in cancer treatment and diagnosis. Accordingly, the precision medicine and molecular imaging will enable us to accelerate and improve cancer management in future medicine.
Keywords: Precision medicine, molecular imaging, precision medicine initiative, systems biology, metabolomics-based systems medicine, cancer theranostics, radio-omics, P4 medicine
Introduction
President Obama announced in his State of the Union Address on January 20, 2015, “I’m launching a new Precision Medicine Initiative to bring us closer to curing diseases like cancer and diabetes-and to give all of us access to the personalized information we need to keep ourselves and our families healthier” [1,2]. Now the President has declared a research initiative that aims to accelerate progress toward a new age of precision medicine [1]. Precision (or personalized) medicine is being strongly supported as a solution to selectively target cancer cells and minimize damage to normal tissue [3]. The goal of this initiative is to acquire momentum of precision medicine [1]. It also aims to identify diagnostic and treatment models whose central focus is on the individual variability of disease presentation [4]. As an inestimable tool in precision medicine, theranostics, which is a portmanteau term of “therapeutics” and “diagnostics”, was coined by John Funkhouser in 2002. It can be defined as a diagnostic methodology for individually tailored therapeutic intervention, and it personalizes healthcare practices to an individual patient by removing inessential treatments for whom a standard therapy is not suitable and/or by optimizing a therapeutic plan for a particular patient [5]. The rapid growth of precision medicine requires dedicated future leaders with a strong foundation in advanced genomic medicine, including molecular diagnostic techniques such as next generation sequencing and whole genome/exome sequencing interpretation. These leaders should integrate personalized medicine into healthcare, and they must attain many additional management and teaching skills [6]. The electronic medical records (EMRs) and genomic research effort work to advance personalized and precision medicine. In fact, the eMERGE network launched in 2007 is an NIH-funded consortium devoted to genomic discovery and implementation investigation by leveraging biorepositories connected to EMRs [7]. The precision medicine requires information technology (IT) infrastructure for the storage, maintenance, and transfer of large amounts of individual genomic data. With such a biobank IT, the researchers enable the management of genetic data for individual patients and other research [8]. The largest precision medicine project in Germany was launched in 2013, entitled the German National Cohort (GNC). This project is a nationwide, long-term study with an overall duration of 25-30 years and a €210 million budget for the first 10 years. Despite these projects, precision medicine in Germany is still significantly smaller than in the United States [9].
Molecular imaging is quickly acquiring recognition as a tool that has the capacity to improve every aspect of cancer treatments. Molecular imaging in oncology has been described as in vivo characterization and the measurement of key biomolecules and molecularly based events that are fundamental to the malignant state [10]. While molecular imaging is defined by the Society of Nuclear Medicine as the “visualization, characterization, and measurement of biological processes at the molecular and cellular levels in humans and other living systems” [11], the burgeoning demand among physicians, patients, and communities for personalized care is increasing the importance of molecular imaging and forming the development of biomedical imaging as a whole [10]. Molecular imaging is developing to include a variety of imaging techniques to enable in vivo monitoring of cellular and molecular processes [11]. Although molecular imaging has existed for decades, the rapid progress of molecular and cell biology, imaging technology, and imaging probe evolvement have highly raised its power and potential. Essential to the development and translation of molecular imaging is interdisciplinary collaboration across many fields, including radiology, nuclear medicine, pharmacology, chemistry, molecular and cell biology, physics, mathematics, and engineering. In fact, these include positron emission tomography (PET), single photon emission computed tomography (SPECT), computed tomography (CT), MRI, MRSI, ultrasound, and optical imaging [10]. Hence, imaging techniques and system biology approaches can be applied as two high-throughput methods to cancer therapy and diagnosis in clinical experiments.
Cancer is one of the widespread causes of death all over the world. Cancer research has focused on the identification of molecular differences between cancerous and healthy cells [12]. There is much evidence that the interaction and network between genes and proteins plays an important role in the research of cancer molecular mechanisms. It is essential and significant to present a new initiative of precision medicine in cancer investigation by integrating systems biology, clinical science, omics technology, bioinformatics, and mathematical science to improve diagnosis, therapies, and prognosis of diseases [13]. Cancer is disease established on the malfunction of system characteristics in biology. Therefore, it has been recognized as a systems biology disease [12]. Advances in science and technology have provided approaches toward the diagnosis, therapies, and prognosis of cancer such as molecular imaging and systems biology as new targeted methods in clinical science. Systems biology has appeared during the two recent decades as a powerful new pattern for investigation in life science [14]. Our understanding of cancer initiation and progress has been furthered by means of high-throughput discovery technologies, such as next generation sequencing and the core omics including genomics, radiogenomics, transcriptomics, proteomics, and metabolomics [15]. Indeed, those are systems biology approaches for better comprehending of cancer systems biology. This review’s purpose is to highlight the role and importance of precision medicine and molecular imaging technologies accompanying systems biology/medicine approaches to cancer therapeutics and diagnosis to create new targeted approaches in future medicine.
Precision medicine
According to the National Institutes of Health (NIH), precision medicine is “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person” [2,16,17]. This approach will permit physicians and investigators to predict more precisely which treatment and prevention strategies for a special disease will work in which groups of people. It is in conflict to a “one-size-fits-all” approach in which disease treatment and prevention strategies are developed for the average person, with less consideration for the differences between individuals. Although the term “precision medicine” is relatively new, the concept has been a part of healthcare for many years. For instance, a person who requires a blood transfusion is not given blood from a randomly selected donor; instead, the donor’s blood type is matched to the recipient to decrease the risk of complications [1,2]. Although examples can be found in several areas of medicine, the role of precision medicine in day-to-day healthcare is relatively limited. Scientists hope that this approach will expand to many areas of health in the coming years [16]. Indeed, precision medicine is now widely utilized only in oncology [4], particularly for treatment the of melanoma, metastatic lung, breast, and brain cancer and leukemia [18]. Radioiodine theranostics is a typical example of precision medicine and has been utilized widely for the management of differentiated thyroid cancer [5]. Generally, precision medicine has not yet become a medical standard for many conditions in spite of its high expectations in the United States, Germany, and other countries [9].
Difference between precision medicine and personalized medicine
Between the terms “precision medicine” and “personalized medicine”, there is a lot of overlap. According to the National Research Council (United States), “personalized medicine” is an older term with a meaning similar to “precision medicine”. There was concern that the term “personalized” could be misconceived to imply that treatments and preventions are being advanced uniquely for each individual; in precision medicine, the focus is on identifying which approaches will be effective for which patients based on genetic, environmental, and lifestyle factors [2,9,19]. The Council therefore preferred the term “precision medicine” to “personalized medicine”. However, some people still utilize the two terms interchangeably [16].
Precision medicine initiative
In early 2015, President Obama announced a strong conviction that science and investigation offer great potential for focusing on bringing precision medicine to many facets of healthcare [1,9,20]. The President’s budget for fiscal year 2016 comprised of $216 million in funding for the initiatives of the NIH, the National Cancer Institute (NCI, the NIH institute focused on cancer research), and the Food and Drug Administration (FDA) [9]. The Precision Medicine Initiative has both short-term and long-term purposes. The short-term objectives include enlarging precision medicine in the field of cancer research. Investigators at the NCI hope to utilize this approach to find novel, more effective cares for different types of cancer based on increased knowledge of the biology and genetics of the disease. The long-term objectives of the Precision Medicine Initiative focus on bringing precision medicine to all aspects of health and healthcare on a large scale as well as generate knowledge applicable to the whole range of health and disease [1]. For this goal, the NIH schemes to start a study including a group (cohort) of at least one million volunteers from around the United States. Participants will provide genetic data, biological samples, lifestyle, environment, and other information about their health. This data will be utilized by investigators to examine a large range of diseases, with the aims of better predicting disease risk, comprehending how diseases happen, and finding improved diagnosis and treatment strategies [16].
Systems biology as an efficient tool in precision medicine
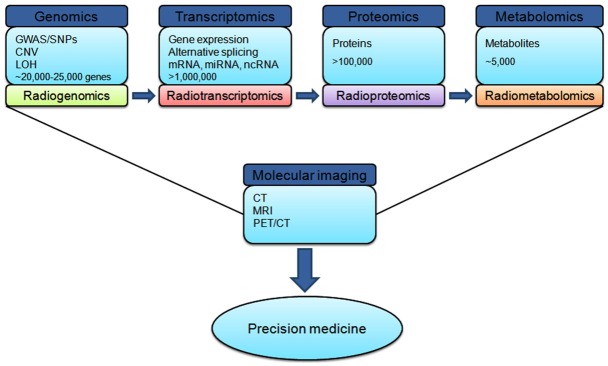
The expression “systems biology” has appeared during recent years to explain the frontier of interdisciplinary investigations in biology [21]. The modern biology is supported by the progress in high-throughput experimental techniques such as genomics, transcriptomics, proteomics, metabolomics, phenomics, and other omics technologies that may generate “big data science”. Systems biology has been described as the study of biological components and their interactions using models and/or networks to integrate genes, metabolites, proteins, regulatory elements and other biological components via high-throughput technologies such as DNA-microarray, Fluorescence microscopy, Illumina sequencer, GC-MS, LC-MS, MALDI-TOF, HPLC, FT-IR, mass spectrometry, HPLC-MS, NMR, and so on [22,23]. Through systems biology, we can find out more puzzling images regarding the fundamental components of biology including the genome, transcriptome, proteome, metabolome, membrane systems, organelles [24,25]. The big data biology, network biology, and other new branches of modern biology have emerged as a result of the combination of versatile knowledge (Figure 1). For instance, understanding of systems-level of the cell or cellular components and subprocesses will be facilitated by network biology. The aim of systems biology is to comprehend the functions and mechanisms in the different levels of organisms or cells [26]. Today, systems biology is encountering the challenges of analyzing huge biological networks and big molecular biological data.
Figure 1.
A general hierarchical diagram of the systems biology approaches integrated to the molecular imaging techniques toward diagnosis and prognosis a typical cancer disease.
Systems approaches
It is necessary to integrate experimental and computational research in order to understand complex biological systems. In other words, it is a systems biology approach [27]. Nowadays, the holistic systems biology approaches have created a new outlook in biology, medicine, and pharmacological sciences [28,29]. Systems biology has provided the base for encoding the structure, variation, and function of the human genome and relating them to health and disease states by high-throughput technologies for DNA sequencing and for analyses of transcriptomes, proteomes, and metabolomes [29]. Many biological processes have already applied omics approaches to lead a large number of genes potentially included in corresponding modules. Development and implementation of computational methods was an important step of progress by which genes or proteins that conduct similarly under different experimental situations [30]. In this section, we discuss on the principal “omics” techniques including genomics, transcriptomics, proteomics, and metabolomics as the most frequently approaches applied in systems medicine research. An overview of the omics technologies for systems medicine and precision medicine has also been presented (Table 1).
Table 1.
An overview of the common omics techniques for systems medicine and precision medicine
| Technique | Molecules of interest | Description | Temporal variance | Affect by disease status | High-throughput instruments |
|---|---|---|---|---|---|
| Genomics | DNA | Evaluation of variability in DNA sequence in the genome | None | No | Illumina sequencing, Pyrosequencer (GS-FLX Titanium), Roche 454 [37,115] |
| Transcriptomics | RNA | Evaluation of variability in composition and abundance of the transcriptome | High | Yes | Affx arrays (Illumina sequencing, Roche 454) [37,115] |
| Proteomics | Protein | Evaluation of variability in composition and abundance of the proteome | High | Yes | MS, 2DE, iTRAQ [47,115,116] |
| Metabolomics | Small molecules (metabolites) | Evaluation of variability in composition and abundance of the metabolome | High | Yes | NMR, DE-NMR, LC-MS, GC-MS, MALDI, MS [47,115] |
Genomics and transcriptomics
Evolution in high-throughput technologies has produced a paradigm shift for biological systems in advanced life science investigations. The systems biology by means of methodically organizing the genomics, functional genomics, and proteomics data attempts to provide a systems-level comprehension of the biological phenomena [31]. In 1986, the term “genomics” was coined by Thomas Roderick for first time. Genomics expression was quickly applied as the novel journal name intended to sustain the new areas and discipline of genome mapping and sequencing [21,32]. Indeed, this was a big and fantastic transformation in molecular biology through the evolution of dramatically efficient approaches for DNA sequencing [33]. Genomics is the branch of science that studies the genome, or the genetic material or blue print of a plant, animal, human, microorganisms, and/or other species in order to understand the functions, gene interactions, and regulation of gene networks with each other and the environment [34]. Nevertheless, the focus of genomics research is mostly on the investigation of sequencing (sequence genomics), functions (functional genomics), structures, and interactions of genes. An example of a huge international genomics project was the Human Genome Project that was successfully completed in 2003 [21]. One of the first omics technologies that had thereafter developed was transcriptomics [35]. Transcriptomics is defined as the study of transcriptome under particular circumstances or in a certain cell by high-throughput technologies such as RNA-seq and microarray analysis [36]. The most important purposes of transcriptomics are: to make an itemized list of all species’ transcripts, including mRNAs, non-coding RNAs, and small RNAs; to appoint the transcriptional structure of genes in terms of their beginning sites, 5’ and 3’ ends, splicing models, and other post-transcriptional alterations; and to quantify the modifying expression levels of each transcript during evolution and under various situations. In order to conclude and quantify the transcriptome, various state of the art technologies have been created such as hybridization or sequence-based approaches [37].
Radiogenomics
Radiogenomics is the newest member of omics family. The concept of radiogenomics is the study of the correlation between cancer imaging features and gene expression (Imaging Genomics) and genotypic variations observed in response to radiation therapy (Radiation Genomics) [38]. The combination of imaging tools with molecular techniques (such as functional genomics assays) offers the potential for the quick clinical translation of powerful high-throughput technology [39]. Radiogenomics may create imaging biomarkers that can recognize the genomics of a disease, particularly cancer, without the use of a biopsy [40]. Numerous techniques are applied to reveal correlations between MRI, CT, and PET imaging features and the genomics of disease (such as large-scale MRI microRNA-mRNA correlative study in glioblastoma, liver cancer genome from non-invasive imaging features, and link image characteristics of non-small cell lung nodules in CT scans) to predict survival using gene expression data [39]. In addition, functional CT in oncology and cardiovascular imaging is a key player in the age of precision medicine and radiogenomics [41].
Proteomics
Proteomics, the study of proteomes on a large scale, promises to transform biology and medicine [42,43]. The study of proteomes in the large scale is defined as proteomics. A proteome is defined as the set of proteins that is generated in a system, organism, or other biological entity. For example, we can refer to the proteome of an organ, such as the liver, or of a species, such as Homo sapiens. The proteome changes from cell to cell at any given time; thus, it is not constant [44]. The term “complete proteome” is applied in the field of mass spectrometry (MS)-based proteomics, pointing toward the presently unachieved aim realizing all the proteins of a given species [45]. To investigate proteomes, several high-throughput techniques have been developed, among them mass spectrometry (MS)-based techniques, such as tandem-MS, and gel-based techniques, such as differential in-gel electrophoresis, which are the most utilized in proteomics. Via these high-performance devices, we can produce enormous amounts of data. Therefore, we require databases for registering, storing, and maintaining these huge amounts of data so that investigators can create links between their outcomes and the available knowledge [44,46].
Metabolomics
Metabolomics enables the integration and merging of biological data from different levels, revealing communication and connectivity inside a system as a key for precision medicine and phenotyping [47]. Metabolomics is an omics technique in systems medicine that is used in the global quantitative evaluation of endogenous metabolites in a biological system [48]. Metabolomics is progressing, with powerful technology enabling the assay and detection of the huge numbers of metabolites in tissues and biofluids [49,50]. Currently, metabolomics research can provide applicable and useful information regarding health and disease status. Metabolomics techniques such as NMR and mass spectroscopy are applied to detect and treat various cancers [49]. In recent years, the universal metabolic profiling of diseases has become feasible via the use of high-throughput analytical devices [51].
Systems approaches in cancer management
At present, researchers utilize a number of complementary omics techniques to investigate an extensive range of diseases, including cancer [52]. Major attempts have been made to apply systems biology approaches to oncology [22,53,54]. High-throughput and sophisticated omics technologies, such as genomics, radiogenomics, proteomics, and metabolomics, have provided a foundation for a new kind of oncological investigation [55]. These advances in experimental systems biology, along with novel analytical techniques and quantitative imaging software tools, are helping to create a more perfect image of many cancers connected to signaling routes [22,55,56]. Recently, metabolomics approaches have been applied as an important tool for detection, prognosis, biomarker discovery, and the design of therapeutics [57]. Denkert et al. utilized gas chromatography/time of flight mass spectrometry (GC-TOF-MS) to analyze ovarian cancer metabolism in both invasive carcinomas and borderline tumors [58,59]. In recent studies, Japanese investigators have diagnosed gastroenterological cancer via the metabolomic discovery of a pancreatic cancer biomarker [60]. Wikoff et al. have succeeded in discovering blood-based biomarkers relevant to lung cancer screening and early detection via metabolomic analysis approaches [61]. In the field of thyroid cancer investigation, there are abundant studies demonstrating the successful application of omics technologies, such as genomics, transcriptomics, and proteomics. Metabolomics tools have been utilized in identifying a great number of metabolites in thyroid cancer in recent years [62]. Systems biology/medicine approaches also improve the understanding, treatment, and clinical management of neuroendocrine prostate cancer [63]. A schematic illustration of the metabolomics-based systems medicine and molecular imaging tools used in cancer diagnosis studies is presented in Figure 2.
Figure 2.
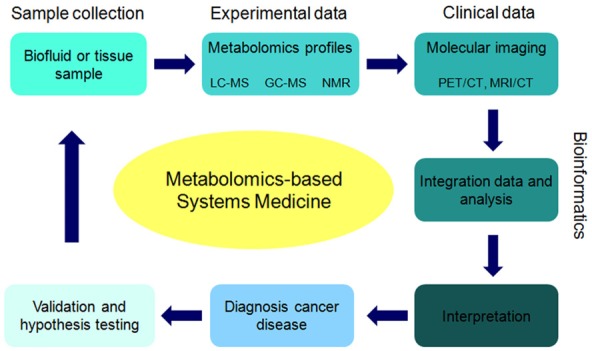
A schematic illustration of metabolomics-based systems medicine and molecular imaging tools into cancer diagnosis studies.
P4 medicine
Medicine is now experiencing a major revolution that will change the current healthcare system from reactive to proactive in every way [19,39,64]. In fact, systems biology and the digital revolution are transforming healthcare into a proactive type of P4 medicine, meaning that it is predictive, preventive, personalized, and participatory [64,65]. However, the term of “personalized medicine” is an expression for a revolution in medicine that will be predictive, preventive, personalized, and participatory. This aspect of medicine was nominated P4 medicine [19,66]. P4 medicine comprises of predictive, preventive, personalized, and participatory medicine [2,64,65,67]. The essence of P4 medicine is the quantification of healthiness or wellness and the clarification of disease [68]. P4 medicine will 1) create better healthcare, 2) decrease the cost of healthcare, and 3) motivate innovation and the creation of new corporations [68]. Meanwhile, systems biology is a holistic, universal, and integrative approach to biology. More specifically, systems medicine is a systems approach to health and disease. Systems biology provides the strategies, instruments, and computational and analytical capabilities needed to analyze huge amounts of information [19,29,69]. These strategies and tools can be utilized in order to combat diseases [70]. Here, we will debate precision medicine in the form of molecular imaging: 1) Personalized: P4 medicine will be ‘personalized’ because it will be established based on the genetic and epigenetic data of each person. Individualized or precision medicine may assist in data mining regarding quantitative biology and anatomy, in targeted imaging/targeted therapy, and also in the real-time monitoring of treatment response. Molecular imaging can provide special molecular profiles and assist in selecting the most efficient treatment with the minimum toxicity on a personalized basis [39,71-73]. Imaging agents that have both diagnostic and therapeutic abilities, or ‘theragnostics’, will probably be more cost-effective and popular [39,74]. The great advantage of molecular imaging in personalized medicine is its ability to merge physiologic and metabolic data with clinical phenotypes and provide inestimable information concerning treatments. 2) Predictive: generally, one human being differs from another by less than 1% in terms of their hereditary structure. These genetic differences cause physical differences, such as the tendency to develop various diseases [39,75]. Medicine will be ‘predictive’ because this individual data will allow physicians to estimate the risk of particular diseases in each person [39,73]. Imaging modalities will play a meaningful role as non-invasive screening methods that are both sensitive and accurate predictors of disease [39,76]. Investigations have shown that only PET influenced management decisions in 38% of cancer cases [39,77]. 3) Preventive: medicine will be ‘preventive’ because the prediction of risk will permit the utilization of prophylactic procedures (way of life or therapeutic) to reduce this risk. Molecular/genetic screening and intervention (often directed by imaging) are the most effective methods of disease management [39,73]. Theragnostic agents can also be used. 4) Participatory: medicine will be ‘participatory’ due to the fact that most of these prophylactic manipulations will require the participation of the patient. This consists of a domain of participatory activities, such data sharing, training patients and physicians, and also counseling patients regarding individual options connected to sickness and well-being. The increasing utilization of social networks by patients, and also the activities of patients’ associations, are examples of participatory actions [39,73]. It is also anticipated that “technology singularity” and “exponential medicine”, as new approaches, will shift the paradigm of medical philosophy and generate a considerable influence on the healthcare system and patient-physician relationship [78]. Here, a schematic representation of the procedure for precision medicine (predictive, preventive, personalized and participatory) and two principal objectives of the precision medicine are demonstrated (Figure 3).
Figure 3.

A schematic representation of the procedure of precision medicine (predictive, preventive, personalized and participatory) and two principal objectives of precision medicine as future of medicine comprising: 1) Quantifying wellness, 2) Demystifying disease.
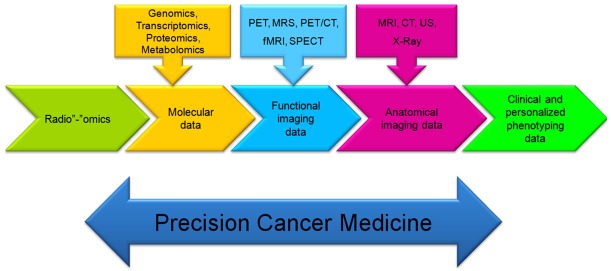
Linkage of the systems approaches and precision medicine
Systems biology is growing quickly, and the term “precision medicine” has also emerged as a significant revolution in modern medicine [29,64]. Furthermore, the biology and disease are unbelievable complicated [19,79]. The precision medicine has extremely developed by advance of high-throughput technologies and systems approaches. Indeed, the field of precision medicine is changing the paradigm of future health care from conventional medicine practice (diagnosis and treatment) to predictive and preventative medicine and personalized health supervising [80,81]. Progress of high-throughput technologies like high-throughput sequencing and mass spectrometry has made capable researchers, scientists and clinicians to study genomes, transcriptomes, proteomes, metabolomes, and other omics data in unexampled detail. The integrated ‘omics’ data can be led to a universal profiling of health and disease, and provide novel approaches for P4 medicine and health monitoring [64,82-85]. The power of systems biology in personalized medicine lies in disease risk estimation, personalized health monitoring and preventative medicine more significantly and it is not only for disease mechanism explanation. Systems biology provides forceful tools to supervise molecular profiles and discern delicate changes that can demonstrate network disturbance. Physicians and pathologists are energetically combining systems biology and medicine tools to attain molecular disease diagnosis [86,87]. Regarding to undergoing period in medicine, we require contributing between systems biology and molecular imaging in the future medicine. Therefore, we will be able to estimate and monitor of personalized disease risk and health care with applying integrative omics techniques and molecular imaging technologies. We may connect systems biology and systems medicine via the implementation of omics technologies, which are the most efficient tools available in modern biology and precision medicine for disease diagnosis (Figure 4).
Figure 4.

The components that will permit to precision medicine for discovering the biological complexity. These emerging technologies such as systems biology approaches and molecular imaging techniques will enable the implementation of precision medicine.
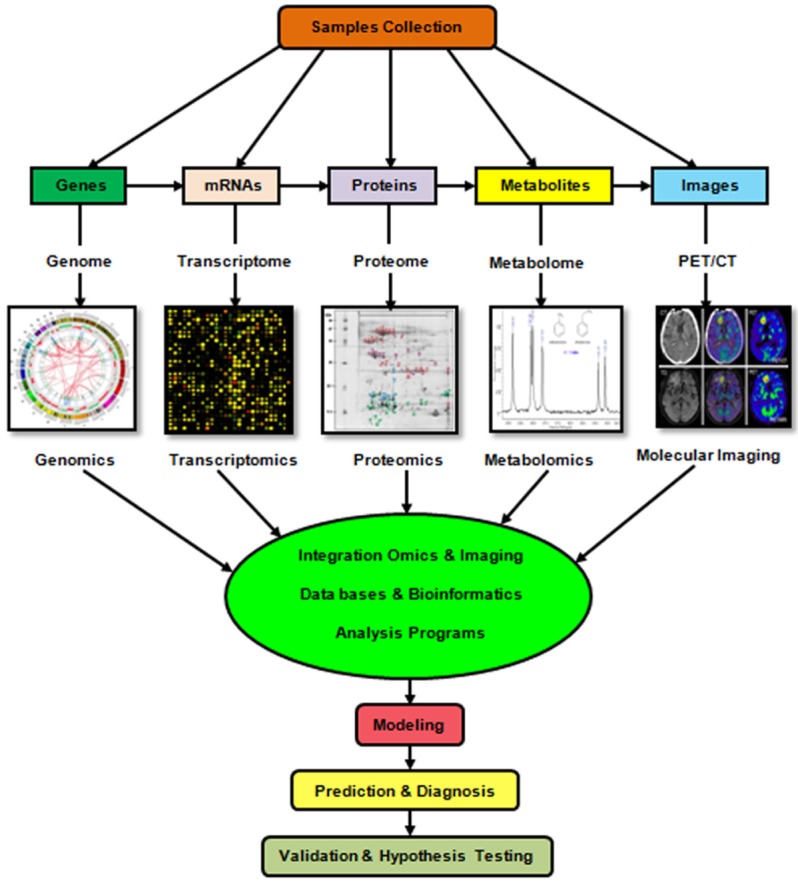
Relationship of imaging and systems approaches in precision medicine
Molecular imaging and systems biology approaches play an important role in developing precision medicine in the cancer management [88]. The evolvement of high-throughput technologies able to comprehensively evaluate DNA, RNA, protein, and metabolites in tumors management [89]. High-throughput experiments such as genomics, radiogenomics, transcriptomics, radiotranscriptomics, proteomics, radioproteomics, metabolomics and radiometabolomics provide a basis for new kinds of oncology investigation. These progresses in systems biology linked with modern analytical and also quantitative imaging software tools are assisting to create a more complete picture of many cancer related signaling pathways and personalized cancer management [22,90,91]. The omics technologies are major basis of systems biology. Recently, the radiogenomics/radiomics and radiotranscriptomics technology has emerged as a new area of omics in molecular imaging field [36,88]. In fact, novel imaging strategies in the era of precision medicine require the focus on improved characterization of disease by phenotyping disease specific imaging characteristics within the concept of genetic patient information in order to non-invasively identify patient subpopulations that benefit from similar treatments or vice versa [41]. On the other side, radiology requires “Big-Data” analysis that is feasible by bioinformatics and systems biology approaches. Since, the radiogenomics generates large amount of genotyping data by imaging from gene expression profile and genotyping variation [88]. The precision medicine requires the individual genetic data to assess many characteristics in cancer detection and diagnosis by researchers. Molecular imaging contributes with systems biology methods (such as computational and modeling) to analyze and interpret the image results. Indeed, the synergy of imaging, genomics, transcriptomics and other omics data can be constructed a bridge between systems biology approach and molecular imaging leads to precision medicine goals [36,92-94] (Figure 5). These progressions provide opportunities for the advancement of personalized oncology in which cancer detection, diagnosis and therapy are tailored to each individual’s molecular profile.
Figure 5.
From Radiogenomics to Radiometabolomics. We present a transition from the integration of systems biology with molecular imaging techniques to precision medicine.
Molecular imaging technologies
In total, Imaging is an essential part in research, trials and practice in field of oncology. There has been a vast growth in the number and type of imaging technologies and their uses in past three decades, although some issues remain. At present, molecular imaging is an emerging subject that integrates advanced imaging technology with cellular and molecular biology [95]. The molecular imaging term can be defined as visual representation, characterization, measurement and quantification of biological processes at the molecular, cellular and sub-cellular within entire living organisms using specific and appropriate imaging probes [11,96,97]. Molecular imaging technologies play a crucial role in earlier detection, precise diagnosis of diseases like cancerous tumors, and drug development and discovery [95,98-100] (Figure 6). Molecular imaging needs to high resolution and high sensitive tools to detect and realize specific imaging agents that connect the imaging signal with molecular occurrence [95,101]. There are copious classes of molecular imaging agents comprising small molecules, peptides, aptamers, high-molecular-weight antibodies, engineered protein fragments, and various nanoparticles [102]. At present, only some of evolving of molecular imaging technologies is used in clinical and preclinical stage e.g. Positron-emission tomography (PET), single-photon-emission CT (SPECT), fluorescence reflectance imaging, fluorescence-mediated tomography (FMT), fibre-optic microscopy, optical frequency-domain imaging, bioluminescence imaging, laser-scanning confocal microscopy and multiphoton microscopy [95,98,102-105]. Table 2 summarizes the characterizations and features of the some molecular imaging modalities with extraction and adaption of available references and literatures that have been published by outstanding scientists in the molecular imaging field [22,96,102,105-108]. Molecular imaging modalities, imaging agents, and applications are well written by James ML and et al. [102].
Figure 6.
A typical representative of the PET/MRI imaging benefits in Precision medicine cancer care. Staging PET/MRI scan of a 56-year-old woman with ovarian cancer. The MRI images (A and B) show multiple lesions abutting the liver posteriorly (long arrow), involving the porta hepatis (short arrow) and seeding the peritoneum (arrowheads). A round, well defined lesion with same features is also visualized in segment IV of the liver (dotted arrow). On PET/MRI images (C and D) the lesions earlier described, and others not so evident, are depicted by high FDG uptake confirming their malignant nature. Maximum intensity projection of the whole body (E) reveals several lesions both in the chest and abdomen. Reproduced from [117].
Table 2.
Comparison of emerging molecular imaging technologies for precision medicine
| Imaging Modalities | Spatial Resolution | Imaging Time | Depth of Penetration | Imaging Agents and Molecular Probes | Key Utilization | |
|---|---|---|---|---|---|---|
| Multi-photon Microscopy | 15-1000 nm | Secs | - | Fluorescent proteins, dyes, rhodamine amide, quantum dots | Visualization of cell structures | |
| Atomic Force Microscopy | 10-20 nm | Mins | - | Intermolecular forces | Mapping cell surface | |
| Electron Microscopy | 0.2-3 nm | Secs | - | Cyrofixation | Discerning protein structure | |
| Ultrasound (US) | Superficial applications | 50 μm, 0.01-0.1 mm | Secs | mm-cm | Microbubbles, nanoparticles | Vascular imaging |
| Deeper applications | 1-2 mm | |||||
| CT | Preclinical | 12-50 μm, 50-200 μm | Mins | Limitless | Iodine | Lung and bone tumor imaging |
| Clinical | 0.5-1 mm | |||||
| MRI | Preclinical | 4-100 μm, 25-100 μm | Mins-Hrs | Limitless | Gadolinium, dysprosium, iron oxide particles | Anatomical imaging |
| Clinical | ~1 mm | Functional | ||||
| fMRI | ~1 nm | Secs-Mins | - | Oxygenated hemoglobin | Functional imaging of brain activity | |
| (HbO2) deoxygenated hemoglobin (Hb) | ||||||
| MRS | ~2 nm | Secs | - | N-acetylaspartate (NAA), creatine, choline, citrate | Detection of metabolites | |
| PET | Preclinical | 1-2 mm | Mins | Limitless | Fluorodeoxyglucose (FDG), 18F, 11C, 15O | Metabolic imaging |
| Clinical | 5-7 mm | |||||
| SPECT | Preclinical | 1-2 mm | Mins | Limitless | Tc-99m, In-111, I-131-labeled compounds, Ga-67, Tl-201, Peptide | Cardiovascular imaging, diagnosis of bone metastasis |
| Clinical | 8-10 mm | |||||
| Optical fluorescence imaging (OFI) | 2-3 mm | Secs-Mins | < 1 cm | Peptide, integrins, matrix metallo-proteinases, caspases | Characterization of cancer | |
| Optical bioluminecence imaging (OBI) | 3-5 mm | Secs-Mins | 1-2 cm | Luciferins, coelenterazines, luminal, Peptide | Gene expression, cell & bacterial tracking | |
| Surface-enhanced raman scattering (SERS) imaging | mm | Min-days | ~5 mm | EGFR-SERS, EGFR affibody-gold-silica nanoparticles | Oligonucleotide targeting | |
| Photoacoustic imaging (PAI) | ~10 μm to 1 mm | Secs-Mins | 6 mm to 5 cm | EGFR Antibody-gold nanoparticles [Nle4, d-Phe7]- -melanocyte-stimulating hormone-gold, hormone-gold nanocages | Functional imaging of blood oxygenation | |
| Intravital microscopy (IVM) | 1 μm | Secs-days | < 400-800 μm, ~700 μm | Photoproteins, fluorochromes | In development (endoscopy, skin) | |
| Fluorescence reflectance imaging (FRI) | 1 mm | Secs-Mins | < 1 cm | Photoproteins, fluorochromes | Rapid screening of molecular events in surface-based disease | |
| Fluorescence mediated tomography (FMT) | 1 mm | Mins | 2-3 cm | Near Infrared, fluorochromes | Quantitative imaging of targeted or “smart” fluorochrome reporters | |
Molecular imaging in cancer management
Modern molecular imaging technologies have the potential to meaningfully increase the diagnostic and therapeutic approaches for cancer treatment [109]. Molecular imaging based personalized therapy has been a fascinating concept for individualized therapeutic strategy, which is able to obtain the highest efficacy and decrease adverse effects in specific patients [5,110-112]. Applications of molecular imaging can be assisted to detect malignant cells at cellular levels in the early stage formation of cancer [10,113]. Molecular imaging techniques also may consider tumor characterization and cancer diagnosis without any invasive operations like biopsy or even surgery [109,114]. Nevertheless, molecular imaging modalities and its probes are being evolved and enhanced more and more for realizing types of cancer in frame work of non-invasive procedures (Figure 7).
Figure 7.
Schematic overview of precision medicine strategies exploiting the tumor microenvironment. Reproduced from [3] with permission of John Wiley and Sons License.
Conclusion and future perspective
Cancer is a devastating disease that modifies the metabolism of a cell and the encircling milieu. Systems biology and molecular imaging approaches are being employed to better comprehend these alterations in cancer diagnosis and therapy in the era of precision medicine. In order to successful curing of patients, early detection, precision staging and remove of cancer tissue are very important. To detect the cancer types, we require to precision medicine tools such as systems approaches (genomics, radiogenomics, transcriptomics, proteomics, and metabolomics) and molecular imaging techniques. Molecular imaging technologies have key role for enhancing cancer diagnosis and treatment by extending imaging modalities to a functional and cellular level as well as improving intraoperative imagining of diseased and healthy tissues for surgeons. Though, medicine is now undergoing a major revolution that will change the nature of healthcare from reactive to proactive. It will increasingly transition to predictive, preventive personalized and participatory medicine (precision medicine). Meanwhile, ongoing technological progresses, particularly in the field of systems biology and molecular imaging, and also interdisciplinary cooperation with miscellaneous branches of science are compulsory for the implementation of precision medicine. These technologic improvements have been rapid in the imaging technologies such as MRI, CT, US, and imaging with radionuclides, fluoroscopy and bio-luminescence. In summary, “Radio-omics”, therefore, poses as a promising contributor to the precision medicine initiative and P4 medicine, advancement both reactive and proactive measures through disease diagnosis in pre-symptomatic stages and appropriate targeting of therapeutic programs (Figure 8).
Figure 8.
An overview of the integration of radio-omics, Molecular biology information and molecular imaging data in precision cancer medicine.
Acknowledgements
We gratefully acknowledge by the Persian Gulf Nuclear Medicine Research Center affiliated to Bushehr University of Medical Science, Bushehr, Iran for providing an opportunity to prepare this manuscript.
Disclosure of conflict of interest
None.
Abbreviations
- BLI
bioluminescence imaging
- CNV
copy number variation
- CT
computed tomography
- DE-NMR
Diffusion-edited-nuclear magnetic resonance
- EMRs
electronic medical records
- eMERGE
Electronic Medical Records and Genomics
- FDA
Food and Drug Administration
- FDG
18-fluoro-deoxyglucos
- FT-IR
fourier transform-infrared spectroscopy
- FRI
fluorescence reflectance imaging
- FMT
fluorescence mediated tomography
- GC-MS
gas chromatography-mass spectrometry
- GC-TOF-MS
gas chromatography time of flight-mass spectrometry
- GWAS
genome-wide association studies
- HPLC
high performance liquid chromatography
- HPLC-MS
high performance liquid chromatography-mass spectrometry
- iTRAQ
isobaric tag for relative and absolute quantitation
- IVUS
intravascular ultrasonography
- LC-MS
liquid chromatography mass spectrometry
- LOH
loss of heterozygosity
- MD-LCMS
Multidimensional liquid chromatography mass spectroscopy
- MALDI-TOF
matrix assisted laser desorption/ionization-time of flight
- MAS-NMR
magic angle spinning-nuclear magnetic resonance
- mRNA
messenger RNA
- miRNA
micro-RNA
- MRI
magnetic resonance imaging
- MRSI
magnetic resonance spectroscopic imaging
- MS
mass spectrometry
- NCI
National Cancer Institute
- ncRNA
non-coding RNA
- NIH
National Institutes of Health
- NMR
nuclear magnetic resonance
- OI
optical imaging
- PET
positron emission tomography
- RAMAN
raman spectroscopy
- SERS
surface-enhanced raman scattering imaging
- SNPs
Single-nucleotide polymorphisms
- SPECT
single photon emission computed tomography
- TEI
transthoracic echocardiography imaging
- US
ultrasound
References
- 1.Collins FS, Varmus H. A New Initiative on Precision Medicine. N Engl J Med. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nabipour I, Assadi M. Precision medicine, an approach for development of the future medicine technologies. ISMJ. 2016;19:167–84. [Google Scholar]
- 3.Penet MF, Krishnamachary B, Chen Z, Jin J, Bhujwalla ZM. Molecular Imaging of the Tumor Microenvironment for Precision Medicine and Theranostics. Adv Cancer Res. 2014;124:235–56. doi: 10.1016/B978-0-12-411638-2.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupte AA, Hamilton DJ. Molecular Imaging and Precision Medicine. Cardiology. 2016;133:178–80. doi: 10.1159/000442044. [DOI] [PubMed] [Google Scholar]
- 5.Ahn B. Personalized Medicine Based on Theranostic Radioiodine Molecular Imaging for Differentiated Thyroid Cancer. Biomed Res Int. 2016;2016:1680464. doi: 10.1155/2016/1680464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason-Suares H, Sweetser D, Lindeman N, Morton C. Training the Future Leaders in Personalized Medicine. J Pers Med. 2016;6 doi: 10.3390/jpm6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smoller J, Karlson E, Green R, Kathiresan S, MacArthur D, Talkowski M, Murphy SN, Weiss ST. An eMERGE Clinical Center at Partners Personalized Medicine. J Pers Med. 2016;6 doi: 10.3390/jpm6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutin N, Holzbach A, Mahanta L, Aldama J, Cerretani X, Embree K, Leon I, Rathi N, Vickers M. The Information Technology Infrastructure for the Translational Genomics Core and the Partners Biobank at Partners Personalized Medicine. J Pers Med. 2016;6 doi: 10.3390/jpm6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kichko K, Marschall P, Flessa S. Personalized Medicine in the U. S. and Germany: Awareness, Acceptance, Use and Preconditions for the Wide Implementation into the Medical Standard. J Pers Med. 2016;6 doi: 10.3390/jpm6020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher MF, Hricak H, Larson SM. Molecular imaging for personalized cancer care. Mol Oncol. 2012;6:182–95. doi: 10.1016/j.molonc.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurdzeil K, Ravizzini G, Croft B, Tatum J, Choyke P, Kobayashi H. The evolving role of nuclear molecular imaging in cancer. Expert Opin Med Diagn. 2008;2:829–42. doi: 10.1517/17530059.2.7.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: A Systems Biology disease. BioSystems. 2006;83:81–90. doi: 10.1016/j.biosystems.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Wu D, Rice CM, Wang X. Cancer bioinformatics: A new approach to systems clinical medicine. BMC Bioinformatics. 2012;13:71. doi: 10.1186/1471-2105-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner HM, Mills GB, Ram PT. Cancer Systems Biology: a peek into the future of patient care? Nat Rev Clin Oncol. 2014;11:167–76. doi: 10.1038/nrclinonc.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albanese C, Rodriguez OC, VanMeter J, Fricke ST, Rood BR, Lee Y, Wang SS, Madhavan S, Gusev Y, Petricoin EF 3rd, Wang Y. Preclinical magnetic resonance imaging and systems biology in cancer research: current applications and challenges. Am J Pathol. 2013;182:312–8. doi: 10.1016/j.ajpath.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference GH, editor. Help Me Understand Genetics Precision Medicine. 2016. https://ghr.nlm.nih.gov/ Precision Medicine.
- 17.Herold CJ, Lewin JS, Wibmer AG, Thrall JH, Krestin GP, Dixon AK, Schoenberg SO, Geckle RJ, Muellner A, Hricak H. Imaging in the Age of Precision Medicine: Summary of the Proceedings of the 10th Biannual Symposium of the International Society for Strategic Studies in Radiology. Radiology. 2016;279:226–38. doi: 10.1148/radiol.2015150709. [DOI] [PubMed] [Google Scholar]
- 18.Weiss ST, Shin MS. Infrastructure for Personalized Medicine at Partners HealthCare. J Pers Med. 2016;6 doi: 10.3390/jpm6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hood L, Balling R, Auffray C. Revolutionizing medicine in the 21st century through systems approaches. Biotechnol J. 2012;7:992–1001. doi: 10.1002/biot.201100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FACT SHEET: President Obama’s Precision Medicine Initiative. 2015. https://www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative.
- 21.Likić VA, McConville MJ, Lithgow T, Bacic A. Systems biology: The next frontier for bioinformatics. Adv Bioinformatics. 2010:268925. doi: 10.1155/2010/268925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kherlopian AR, Song T, Duan Q, Neimark MA, Po MJ, Gohagan JK, Laine AF. A review of imaging techniques for systems biology. BMC Syst Biol. 2008;2:74. doi: 10.1186/1752-0509-2-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan JS, Galbraith DW, Dai SY, Griffin P, Stewart CN. Plant systems biology comes of age. Trends Plant Sci. 2008;13:165–71. doi: 10.1016/j.tplants.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Soto AM, Sonnenschein C, Maini PK, Noble D. Systems biology and cancer. Prog Biophys Mol Biol. 2011;106:337–9. doi: 10.1016/j.pbiomolbio.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone WS. The Concept of the Gene in Development and Evolution: Historical and Epistemological Perspectives. Am J Psychiatry. 2002;159:335–335. [Google Scholar]
- 26.Altaf-Ul-Amin M, Afendi FM, Kiboi SK, Kanaya S. Systems biology in the context of big data and networks. Biomed Res Int. 2014;2014:428570. doi: 10.1155/2014/428570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 28.Buriani A, Garcia-Bermejo ML, Bosisio E, Xu Q, Li H, Dong X, Simmonds MS, Carrara M, Tejedor N, Lucio-Cazana J, Hylands PJ. Omic techniques in systems biology approaches to traditional Chinese medicine research: Present and future. J Ethnopharmacol. 2012;140:535–44. doi: 10.1016/j.jep.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 29.Auffray C, Chen Z, Hood L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 2009;1:2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge H, Walhout AJ, Vidal M. Integrating “omic” information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–60. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Aggarwal K, Lee KH, Lee KH, Aggarwal K. Functional genomics and proteomics as a foundation for systems biology. Brief Funct Genomic Proteomic. 2003;2:175–84. doi: 10.1093/bfgp/2.3.175. [DOI] [PubMed] [Google Scholar]
- 32.McKusick VA, Ruddle FH. “A new discipline, a new name, a new journal”. Genomics. 1987;1:1–2. [Google Scholar]
- 33.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–4. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon S. Genomics and World Health. Trans R Soc Trop Med Hyg. 2002;96:669. doi: 10.1016/s0035-9203(02)90222-1. [DOI] [PubMed] [Google Scholar]
- 35.Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–35. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 36.Katrib A, Hsu W, Bui A, Xing Y. “Radiotranscriptomics”: A synergy of imaging and transcriptomics in clinical assessment. Quant Biol. 2016;4:1–12. doi: 10.1007/s40484-016-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenstein BS, West CM, Bentzen SM, Alsner J, Andreassen CN, Azria D, Barnett GC, Baumann M, Burnet N, Chang-Claude J, Chuang EY, Coles CE, Dekker A, De Ruyck K, De Ruysscher D, Drumea K, Dunning AM, Easton D, Eeles R, Fachal L, Gutiérrez-Enríquez S, Haustermans K, Henríquez-Hernández LA, Imai T, Jones GD, Kerns SL, Liao Z, Onel K, Ostrer H, Parliament M, Pharoah PD, Rebbeck TR, Talbot CJ, Thierens H, Vega A, Witte JS, Wong P, Zenhausern F Radiogenomics Consortium. Radiogenomics: Radiobiology enters the era of big data and team science. Int J Radiat Oncol Biol Phys. 2014;89:709–13. doi: 10.1016/j.ijrobp.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assadi M, Nabipour I. The future of molecular imaging in paradigm shift from reactive to proactive (P4) medicine. Nucl Med Commun. 2014;35:1193–6. doi: 10.1097/MNM.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 40.Das AK, Bell MH, Nirodi CS, Story MD, Minna JD. Radiogenomics-predicting tumor responses to radiotherapy in lung cancer. Semin Radiat Oncol. 2010;20:149–55. doi: 10.1016/j.semradonc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henzler T, Fink C. Functional computed tomography in oncology and cardiovascular imaging: A key player in the era of precision medicine and radiogenomics. Eur J Radiol. 2015;84:2345–6. doi: 10.1016/j.ejrad.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Weston AD, Hood L. Systems Biology, Proteomics, and the Future of Health Care: Toward Predictive, Preventative, and Personalized Medicine Introduction: Paradigm Changes in Health Care. J Proteome Res. 2004;3:179–96. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 43.Belhocine TZ, Tait JF, Vanderheyden JL, Li C, Blankenberg FG. Nuclear medicine in the era of genomics and proteomics: Lessons from annexin V. J Proteome Res. 2004;3:345–9. doi: 10.1021/pr049968a. [DOI] [PubMed] [Google Scholar]
- 44. http://www.ebi.ac.uk/training/online/course/proteomics-introduction-ebi-resources/what-proteomics.
- 45.Beck M, Claassen M, Aebersold R. Comprehensive proteomics. Curr Opin Biotechnol. 2011;22:3–8. doi: 10.1016/j.copbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Gehlenborg N, O’Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, Kohlbacher O, Neuweger H, Schneider R, Tenenbaum D, Gavin AC. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 47.van der Greef J, Hankemeier T, McBurney RN. Metabolomics-based systems biology and personalized medicine: moving towards n = 1 clinical trials? Pharmacogenomics. 2006;7:1087–94. doi: 10.2217/14622416.7.7.1087. [DOI] [PubMed] [Google Scholar]
- 48.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical Applications of Metabolomics in Oncology: A Review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beger R. A Review of Applications of Metabolomics in Cancer. Metabolites. 2013;3:552–74. doi: 10.3390/metabo3030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis GD, Asnani A, Gerszten RE. Application of Metabolomics to Cardiovascular Biomarker and Pathway Discovery. J Am Coll Cardiol. 2008;52:117–23. doi: 10.1016/j.jacc.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oskouie AA, Taheri S. Recent developments and application of metabolomics in cancer diseases. J Paramed Sci. 2015;6:116–35. [Google Scholar]
- 52.Pin E, Fredolini C, Petricoin EF. The role of proteomics in prostate cancer research: Biomarker discovery and validation. Clin Biochem. 2013;46:524–38. doi: 10.1016/j.clinbiochem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, Lander ES, Golub TR, Look AT. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 54.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–9. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 55.Cavallo F, De Giovanni C, Nanni P, Forni G, Lollini PL. 2011: the Immune Hallmarks of Cancer. Cell. 2011;60:319–26. doi: 10.1007/s00262-010-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–5. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072–7. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- 58.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Könsgen D, Dietel M, Fiehn O. Mass Spectrometry-Based Metabolic Profiling Reveals Different Metabolite Patterns in Invasive Ovarian Carcinomas and Ovarian Borderline Tumors Metabolite Patterns in Invasive Ovarian Carcinomas and Ovarian Borderline Tumors. Cancer Res. 2006;66:10795–805. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 59.Nagrath D, Caneba C, Karedath T, Bellance N. Metabolomics for mitochondrial and cancer studies. Biochim Biophys Acta. 2011;1807:650–63. doi: 10.1016/j.bbabio.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida M, Nishiumi S, Azuma T. Gastroenterological Cancer Diagnosis by Metabolomics-Discovery of Pancreatic Cancer Biomarker. Rinsho Byori. 2015;63:450–6. [PubMed] [Google Scholar]
- 61.Wikoff W, Hanash S, DeFelice B, Miyamoto S, Barnett M, Zhao Y, Goodman G, Feng Z, Gandara D, Fiehn O, Taguchi A. Diacetylspermine Is a Novel Prediagnostic Serum Biomarker for Non-Small-Cell Lung Cancer and Has Additive Performance With Pro-Surfactant Protein B. J. Clin. Oncol. 2015;33:3880–6. doi: 10.1200/JCO.2015.61.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shajahan-Haq A, Cheema M, Clarke R. Application of Metabolomics in Drug Resistant Breast Cancer Research. Metabolites. 2015;5:100–18. doi: 10.3390/metabo5010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yadav K, Khader S, Readhead B, Yadav S, Li L, Kasarksis A, Tewari AK, Dudley JT. Systems Medicine approaches to improving understanding, treatment, and clinical management of Neuroendocrine Prostate Cancer. Curr Pharm Des. 2016;22:5234–5248. doi: 10.2174/1381612822666160513145924. [DOI] [PubMed] [Google Scholar]
- 64.Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: Predictive, preventive, personalized and participatory. N Biotechnol. 2012;29:613–24. doi: 10.1016/j.nbt.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Nabipour I, Assadi M. Infrastructures for systems medicine in Iran’s health roadmap. ISMJ. 2014;17:974–92. [Google Scholar]
- 66.Nabipour I, Assadi M. The future of medicine, systems medicine, P4 medicine. Bushehr University of Medical Sciences. 2014 [Google Scholar]
- 67.Hood L. A personal journey of discovery: developing technology and changing biology. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:1–43. doi: 10.1146/annurev.anchem.1.031207.113113. [DOI] [PubMed] [Google Scholar]
- 68.Hood L. Systems Biology and P4 Medicine: Past, Present, and Future. Rambam Maimonides Med J. 2013;4:e0012. doi: 10.5041/RMMJ.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nabipour I, Assadi M. Converging technologies: shaping the future of medicine. ISMJ. 2015;17:1045–67. [Google Scholar]
- 70.Hood L, Friend S. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–7. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 71.Zhao B, Schwartz L, Larson S. Imaging surrogates of tumor response to therapy: anatomic and functional biomarkers. J Nucl Med. 2009;50:239–49. doi: 10.2967/jnumed.108.056655. [DOI] [PubMed] [Google Scholar]
- 72.Larson S, Morris M, Gunther I, Beattie B, Humm J, Akhurst T. Tumor localization of 16beta-18F-fluoro-5alpha-dihydrotestosterone versus 18F- FDG in patients with progressive, metastatic prostate cancer. J Nucl Med. 2004;45:366–73. [PubMed] [Google Scholar]
- 73.Assadi M, Nabipour I. The evolving role of molecular imaging in transforming reactive to proactive (P4) medicine: predictive, preventive, personalized and participatory. J Nucl Med. 2014;55(Suppl 1):1310. doi: 10.1097/MNM.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 74.Jokersta JV, Gambhir SS. Molecular Imaging with Theranostic Nanoparticles Jesse. Acc Chem Res. 2011;44:1050–60. doi: 10.1021/ar200106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bengoechea J. Infection systems biology: from reactive to proactive (P4) medicine. Int Microbiol. 2012;15:55–60. doi: 10.2436/20.1501.01.158. [DOI] [PubMed] [Google Scholar]
- 76.Bradley WG, Golding SG, Herold CJ, Hricak H, Krestin GP, Lewin JS, Miller JC, Ringertz HG, Thrall JH. Globalization of P4 medicine: predictive, personalized, preemptive, and participatory- summary of the proceedings of the Eighth International Symposium of the International Society for Strategic Studies in Radiology, August 27-29, 2009. Radiology. 2011;258:571–82. doi: 10.1148/radiol.10100568. [DOI] [PubMed] [Google Scholar]
- 77.Hillner BE, Siegel BA, Shields AF, Liu D, Gareen IF, Hunt E, Coleman RE. Relationship between cancer type and impact of PET and PET/CT on intended management: findings of the national oncologic PETregistry. J Nucl Med. 2008;49:1928–35. doi: 10.2967/jnumed.108.056713. [DOI] [PubMed] [Google Scholar]
- 78.Nabipour I, Assadi M. The technological singularity and exponential medicine. ISMJ. 2016;18:1287–98. [Google Scholar]
- 79.Hood L. Deciphering complexity: A personal view of systems biology and the coming of “Big” science. Genet Eng Biotechnol News. 2011;31:131. [Google Scholar]
- 80.Chen R, Snyder M. Systems biology: Personalized medicine for the future? Curr Opin Pharmacol. 2012;12:623–8. doi: 10.1016/j.coph.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nabipour I, Assadi M. Converging technologies: shaping the future of medicine. ISMJ. 2015;17:1045–67. [Google Scholar]
- 82.Wetterstrand K. DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program. 2012 [Google Scholar]
- 83.Chen R, Mias G, Li-Pook-Than J, Jiang L, Lam H, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Mol Syst Biol. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes Dev. 2010;24:423–31. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moch H, Blank P, Dietel M, Elmberger G, Kerr K, Palacios J, Penault-Llorca F, Rossi G, Szucs TD. Personalized cancer medicine and the future of pathology. Virchows Arch. 2012;460:3–8. doi: 10.1007/s00428-011-1179-6. [DOI] [PubMed] [Google Scholar]
- 87.Berman DM, Bosenberg MW, Orwant RL, Thurberg BL, Draetta GF, Fletcher CD, Loda M. Investigative pathology: leading the post-genomic revolution. Lab Invest. 2012;92:4–8. doi: 10.1038/labinvest.2011.147. [DOI] [PubMed] [Google Scholar]
- 88.Rutman AM, Kuo MD. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–41. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 89.Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: A systems biology approach. J. Clin. Oncol. 2010;28:2777–83. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Djekidel M. Radiogenomics and Radioproteomics. Omi J Radiol. 2012;2:2–4. [Google Scholar]
- 91.Ehlerding EB, Cai W. Harnessing the Power of Molecular Imaging for Precision Medicine. J Nucl Med. 2016;57:171–2. doi: 10.2967/jnumed.115.166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herold CJ, Lewin JS, Wibmer AG, Thrall JH, Krestin GP, Dixon AK, Schoenberg SO, Geckle RJ, Muellner A, Hricak H. Imaging in the Age of Precision Medicine: Summary of the Proceedings of the 10th Biannual Symposium of the International Society for Strategic Studies in Radiology. Radiology. 2016;279:226–38. doi: 10.1148/radiol.2015150709. [DOI] [PubMed] [Google Scholar]
- 93.Thrall JH. Moreton Lecture: Imaging in the Age of Precision Medicine. J Am Coll Radiol. 2015;12:1106–11. doi: 10.1016/j.jacr.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 94.Penet MF, Krishnamachary B, Chen Z, Jin J, Bhujwalla ZM. Molecular imaging of the tumor microenvironment for precision medicine and theranostics. Adv Cancer Res. 2014;124:235–56. doi: 10.1016/B978-0-12-411638-2.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen ZY, Wang YX, Lin Y, Zhang JS, Yang F, Zhou QL, Liao YY. Advance of molecular imaging technology and targeted imaging agent in imaging and therapy. Biomed Res Int. 2014;2014:819324. doi: 10.1155/2014/819324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003:545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 97.Ametamey SM, Honer M, Schubiger PA. Molecular imaging with PET. Chem Rev. 2008;108:1501–16. doi: 10.1021/cr0782426. [DOI] [PubMed] [Google Scholar]
- 98.New SE, Aikawa E. Molecular Imaging Insights into Early Inflammatory Stages of Arterial and Aortic Valve Calcification. Circ Res. 2011;108:1381–91. doi: 10.1161/CIRCRESAHA.110.234146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gomes CM, Abrunhosa AJ, Ramos P, Pauwels EK. Molecular imaging with SPECT as a tool for drug development. Adv Drug Deliv Rev. 2011;63:547–54. doi: 10.1016/j.addr.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 100.Byrne T, O’Connor E, Hall M, Murtagh J, O’Neill K, Curran KM, Mongrain K, Rousseau JA, Lecomte R, McGee S, Callanan JJ, O’Shea DF, Gallagher WM. Vascular-targeted photodynamic therapy with BF2-chelated Tetraaryl-Azadipyrromethene agents: a multi-modality molecular imaging approach to therapeutic assessment. Br J Cancer. 2009;101:1565–73. doi: 10.1038/sj.bjc.6605247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32:7127–38. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 102.James ML, Gambhir SS. A Molecular Imaging Primer: Modalities, Imaging Agents, and Applications. Physiol Rev. 2012;92:897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 103.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–9. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.MPolidais. Medical Imaging in Cancer Care: CHARTING THE PROGRESS. US Oncol Natl Electr Manuf Assoc. 2006:1–32. [Google Scholar]
- 105.Condeelis J, Weissleder R. In vivo imaging in cancer. Cold Spring Harb Perspect Biol. 2010;2:a003848. doi: 10.1101/cshperspect.a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khalil MM, Tremoleda JL, Bayomy TB, Gsell W. Molecular SPECT Imaging: An Overview. Int J Mol Imaging. 2011;2011:796025. doi: 10.1155/2011/796025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hassan M, Klaunberg BA. Overview Biomedical Applications of Fluorescence Imaging In Vivo. Comp Med. 2004;54:635–44. [PubMed] [Google Scholar]
- 108.Xie W, Schlücker S. Medical applications of surface-enhanced Raman scattering. Phys Chem Chem Phys. 2013;15:5329–44. doi: 10.1039/c3cp43858a. [DOI] [PubMed] [Google Scholar]
- 109.Hussain T, Nguyen QT. Molecular imaging for cancer diagnosis and surgery. Adv Drug Deliv Rev. 2014;66:90–100. doi: 10.1016/j.addr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Personalized medicine: Molecular imaging predicts treatment success in many cancers. ScienceDaily. 2010. www.sciencedaily.com/releases/2010/09/100901103731.htm.
- 111.American College of Surgeons National Cancer Database Public Reports. 2013. https://www.facs.org/quality%20programs/cancer/ncdb.
- 112.Scott W, Howington J, Feigenberg S, Movsas B, Pisters K. Treatment of non-small cell lung cancer stage I and stage II: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:234S–242S. doi: 10.1378/chest.07-1378. [DOI] [PubMed] [Google Scholar]
- 113.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 114.Hricak H. Oncologic imaging: a guiding hand of personalized cancer care. Radiology. 2011;259:633–40. doi: 10.1148/radiol.11110252. [DOI] [PubMed] [Google Scholar]
- 115.Vlaanderen J, Moore LE, Smith MT, Lan Q, Zhang L, Skibola F, Rothman N, Vermeulen R. Application of Omics Technologies in Occupational and Environmental Health Research; Current Status and Projections. Occup Env Med. 2010;67:136–43. doi: 10.1136/oem.2008.042788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chandramouli K, Qian PY. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009 doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Partovi S, Kohan A, Rubbert C, Vercher-Conejero JL, Gaeta C, Yuh R, Zipp L, Herrmann KA, Robbin MR, Lee Z, Muzic RF Jr, Faulhaber P, Ros PR. Clinical oncologic applications of PET/MRI: a new horizon. Am J Nucl Med Mol Imaging. 2014;4:202–12. [PMC free article] [PubMed] [Google Scholar]