Abstract
Somatic instability of CAG repeats has been associated with the clinical progression of CAG repeat diseases. Aging and DNA repair processes influence the somatic stability of CAG repeat in disease and in mouse models. However, most of the studies have focused on genetically engineered transgenic repeats and little is known about the stability of naturally polymorphic CAG repeats. To study whether age and/or DNA repair activity have an effect on the somatic stability of CAG repeats, we analyzed variations of the length of naturally polymorphic CAG repeats in the striatum of young and aged WT and ogg1 KO mice. Some multiple and long polymorphic CAG repeats were observed to have variable length in the striatum of aged mice. Interestingly, a low level of repeat variability was detected in the CAG repeat located in tbp, the only mouse polymorphic CAG repeat that is associated with a trinucleotide disease in humans, in the striatum of aged mice and not in young mice. We propose that age may have an effect on the somatic stability of polymorphic CAG repeats and that such an effect depends on intrinsic CAG repeat characteristics.
1. INTRODUCTION
Length variability of trinucleotide repeat sequences is a common form of genetic polymorphism but, for reasons yet to be understood, some polymorphic trinucleotide repeats become abnormally unstable, expand and cause neurodegeneration. At least sixteen (16) neurodegenerative disorders have been associated with trinucleotide repeat expansion, 11 of them associated with CAG repeat expansions, and Huntington’s disease (HD) is the best known neuropathology linked to increased CAG repeats in a specific gene [1]. The observation of intergenerational expansion that accompanies anticipation in families carrying CAG repeats diseases led to propose meiotic instability as an underlying mechanism for repeat expansion [2]. However, CAG repeats have also been found to expand in non-proliferative somatic cells such as neurons [3, 4] and, more specifically, in specific brain regions, and in neuron sub-populations [5, 6]. This type of expansion at the somatic level is proposed to occur in an age-related manner [4, 7] and to influence the course and severity of CAG repeat diseases [8–10]. Different from instability at the germline level, somatic CAG instability is hypothesized to involve molecular mechanisms associated with DNA repair and transcription [7, 11, 12]. Studies in HD mouse models have proposed that the activity of ogg1, a glycosylase that specifically recognizes and removes the oxidatively modified guanine base from DNA, is an initiating step in the generation of CAG expansion with age [13]. Further supporting the role of DNA repair in CAG repeat instability, components of other DNA repair systems have been involved [12, 14–16]. These studies have highlighted potential factors influencing CAG somatic instability but they use mouse models carrying transgenic CAG repeats that would be uncommonly long and highly unstable in humans. The main reason for this is that CAG repeats located in mouse homologues have different characteristics from their human counterparts and this is reflected in the fact that disease-related CAG repeats are unstable in humans but not in mice. For instance, the polymorphic CAG repeat whose expansion is responsible for HD is located in exon 1 of Huntingtin gene (HTT), normal HTT alleles have 7 to 35 repeats whereas most of the individuals affected with HD have 40 to 50 CAG repeats [17]. In contras, the HTT mouse homologue htt (htt) has 7 uninterrupted CAG repeats which have been shown to be stable [18]. Additionally, even if the CAG repeats in the mouse gene were unstable, the biological consequences appear to be minimal, since in order to observe phenotypic changes and repeat instability, the mouse model requires to carry transgenic constructs with more than 100 CAG repeats [6, 7]. There is little information about the stability of naturally polymorphic CAG repeats in mice but it could be inferred that the same factors that induce somatic instability in transgenic repeats may exert a similar destabilizing effect on them. Detecting somatic instability in naturally polymorphic repeats may help to understand why specific CAG repeats become unstable in susceptible neurons and brain regions. This in turn may help to understand conversion of polymorphic CAG repeats from normal to pre-mutation stages in human trinucleotide repeat disorders. In this study we aimed to determine age-related changes in the stability of mouse polymorphic CAG repeats in the striatum and how the activity of the DNA repair enzyme ogg1 may be involved.
2. MATERIALS AND METHODS
2.1. Animals and DNA extraction
Brain tissue from 3–4 (young) and 18–24 (old) month-old Svj129 wild-type and ogg1 KO mice (n=3) was dissected to obtain striatum. Liver and tail tissue were used as control tissue. DNA was extracted from fresh tissue using the Promega Wizard Genomic DNA kit. All extracted DNA samples were diluted in 1x TE and normalized to 50ng/μL. All animal use was conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and with protocols approved by the Institutional Animal Care and Use Committee (IACUC).
2.2. PCR and CAG repeat length measurement
2.2.1. Size screening PCR of CAG repeat regions
We used the sequence of primers previously reported which amplify the region containing CAG repeats in the 27 genes and 30 CAG repeats found to be polymorphic in mice [19]. Reaction conditions were as follows: 1X Taq polymerase buffer, 1,5 mM MgCl2, 0.125 mM dNTPs, 0.25 μM each primer, 0.025 U/μl Taq DNA polymerase (native Invitrogen Cat#18038-018) and 50 ng DNA. Thermal conditions were as follows: denaturation 4:00 at 95°C, 30 cycles of 0:30 at 95°C, annealing 0:45 at 55–57°C and elongation 1:00 at 72°C, and a final elongation for 7:00 at 72°C. Two types of analyses were done using these PCR conditions: determination of CAG repeat length by Sanger sequencing and screening of size variations in the PCR amplicons. Initially, PCR products were purified and sequenced to determine that the PCR was specific for the region of interest and to measure the exact length of the non-repeat region and CAG repeat in the Svj129 strain. For this sequencing protocol, DNA was extracted from tail tissue sample of three different 3-month-old WT mice. Repeats were defined as more than 2 consecutive CAG sequences allowing only one synonymous (CAA) interruption. CAG repeat length determined by sequencing was used as the reference for microfluidic electrophoresis repeats length estimation. Second, PCR was performed in DNA extracted from striatum/liver, young/old, and WT/ogg1 KO mice (n=3) products were resolved in 1.2% agarose for 2h at 25 mA to confirm amplification and visually estimate variations in the length of PCR amplicons. The same tail snip sample used for sequencing was run in parallel with the other samples to visually compare size differences.
2.2.2. Quantitative estimation of the CAG repeat length
Subsequent determination of CAG repeats length in striatum was done by microfluidic electrophoresis on an Agilent 2100 Bioanalyzer. Each genotype group (WT and ogg1 KO) consisted of 6 mice. The reference length obtained by sequencing for each PCR product was used to calculate the number of CAG repeats in the amplicon using the formula: # of repeats = size of PCR fragment in base pairs – size of non-repeat region in base pairs/3, as previously described [20]. Sequences were analyzed for the presence of additional repeats in the surrounding sequence using the RepeatMasker track on UCSC and for the formation of secondary structures using mFOLD [21]. Gene diversity (GD) was calculated for each repeat by the formula: h = 1 − Σmi=1 x2i where h is GD, xi is the frequency of a certain repeat length i and m is the total number of observed lengths [19, 22]. When multiple PCR amplicons were detected by the Bioanalyzer electrophoresis, GD calculation was based on the PCR fragment with the size closer to the sequenced reference sample.
2.2.3. Small pool PCR (SP-PCR)
SP-PCR was performed by a two-stage PCR as previously described [23]. For the first-stage PCR, the conditions of the size screening PCR were adjusted by increasing dNTPs to 0.8 mM and each primer to 0.5 μM. This first-stage PCR used 1 μl of DNA per 10 μl PCR reaction at a concentration of 75 genomic equivalents/μL. Thirty reactions of the same sample and 2 negative controls were run simultaneously. The PCR thermal conditions were 95°C for 4min, 4 cycles of 94°C 30sec, 62°C 30sec and 69°C 1min, 14 cycles of 94°C 30sec, 64°C 30sec touchdown 0.7°C/cycle and 69°C 1min, 15 cycles of 94°C 30sec, 54°C 30sec and 69°C 1min touchdown 0.1°C/cycle, and a final elongation 69°C 10min. For the second-stage PCR, the conditions of size screening PCR were adjusted to use 5 μl of the first-stage PCR product per 15 μl PCR reaction. Two negative template controls were run simultaneously to control for contamination. PCR amplicons were run in the Agilent 2100 Bioanalyzer and the output size were used to quantitate the CAG repeat length as it was said above.
3. RESULTS
We analyzed 30 CAG repeats located in 27 mouse genes previously reported to be polymorphic in 16 inbred mouse strains by Ogasawara et al [19]. A PCR was performed to amplify the CAG repeat and surrounding genomic region followed by purification and sequencing. This DNA sequence was used as reference to calculate the number of CAG repeats. Based on their length (n = number of CAG/CAA repeats), the 30 CAG repeats in the genes analyzed were classified into three groups: short (<10n), intermediate (between 10n and 20n) and long (> 20n) repeats (Table 1). Most of the studied polymorphic repeats (18) were short. Four polymorphic CAG repeats were classified as intermediate and other five were long. Eighteen repeats were unique but there were also 12 repeats that were located near one or 2 more repeats in the same gene and they were classified as multiple. Additionally, depending on the presence or absence of synonymous CAA interruptions in the CAG repeat sequence and the location of these interruptions in the repeat sequence, repeats can be further classified uninterrupted, terminally interrupted, and internally interrupted. In table 1, a consolidated classification is presented for all studied repeats which takes into account the repeat length, presence of CAA interruptions, location of CAA interruptions: SU= short uninterrupted, STI= short terminally interrupted, SSI= short internally interrupted, IU: intermediate uninterrupted, ISI: intermediate internally interrupted, LSI: long internally interrupted, ITI: intermediate terminally interrupted.
Table 1. Analyzed mouse genes with polymorphic CAG repeats.
List of mouse genes carrying polymorphic CAG repeats analyzed and comparison with their corresponding human homologue.
| Repeat class | # | Official full gene name/Gene accesed sequence | MOUSE | HUMAN | |||
|---|---|---|---|---|---|---|---|
| Gene symbola | Repeat type (N + type)b | Human homologyc | Gene symbola | Repeat type (N + type)b | |||
| SHORT CAG REPEATS | 1 | Ankyrin repeat domain 24 / AK019475 | ankrd24 | 5 SU | 70.9 | ANKRD24 | 2 SU |
| 2 | Aryl hydrocarbon receptor nuclear translocator / U10325 | arnt | 5 SU | 95.7 | ARNT | 5 SSI | |
| 3 | Nik related kinase / AB020741 | nrk | 6 SU | 52.2 | NRK | - | |
| 4 | Bone morphogenetic protein 6 / X80992 | bmp6 | 7 SU | 94.7 | BMP6 | 6 SU | |
| 5 | Interleukin 2 / X01772 | il2 | 8 SU | 60.9 | IL2 | 1 | |
| 6 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) / X04435 | nr3c1 | 8 SU | 92.9 | NR3C 1 | 2 SU | |
| 7 | AT rich interactive domain 3B (BRIGHT-like) / AF116847 | arid3b | 9 SU | 90.7 | ARID3B | 11 ISI | |
| 8 | Clock homolog / NM_007715 | clock | 9 STI | 97.8 | CLOCK | 6 SU | |
| 9 | CXXC finger protein 1 / AK010337 | cxxc1 | 9 STI | 98.2 | CXXC1 | 6 SU | |
| INTERMEDIATE CAG REPEATS | 10 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosamide alpha-2,6-sialyltransferase 5 / AB028840 | st6galnac5 | 10 IU | 91.4 | ST6GALNAC5 | 12 ISI |
| 11 | Chromogranin A (parathyroid secretory protein 1) / M64278 | chga | 11 IU | 76.6 | CHGA | 2 SU | |
| 12 | TATA box binding protein / NM_013684.3 | tbp | 13 II | 99.1 | TBP | 25–42 LSI | |
| 13 | PAX interacting (with transcription-activation domain) protein 1 / AF104261 | paxip1 | 13 II | 91.2 | PAXIP1 | 7§ SU | |
| LONG CAG REPEATS | 14 | Transcription factor 20 (AR21) / AY007594 | tcf20 | 20 LI | 93.8 | TCF20 | 5 SU |
| 15 | Polyhomeotic-like protein 1 isoform a (Drosophila) / NM_007905 | phc1 | 24 LI | 95.4 | PHC1 | 15 ISI | |
| 16 | mastermind-like domain containing 1 / AF125313 | mamld1 | 25 LI | 73 | MAMLD1 | 11 ISI | |
| 17 | Nuclear receptor coactivator 6 / AF216186 | ncoa6 | 31 LI | 94 | NCOA6 | 25 LSI | |
| 18 | Hyperpolarization activated cyclic nucleotide-gated potassium channel 1 / AF028737 | hcn1 | 37 LI | 91.9 | HCN1 | 3 SNI | |
| MULTIPLE CAG REPEATS | 19 | Bromodomain containing 4 / AF273217 | brd4_a | 8 SU | 97.4 | BRD4_a | 5 SNI |
| 20 | Bromodomain containing 4 / AF273217 | brd4_b | 7 SU | 97.4 | BRD4_b | 8 SSI | |
| 21 | Involucrin / L28819 | ivl_a | 19 IU | 54 | IVL_a | 2 SU | |
| 22 | Involucrin / L28819 | ivl_b | 6 SU | 54 | IVL_b | - | |
| 23 | Transducer of ERBB2,1 / D78382 | tob1_a | 7 SU | 88 | TOB1_a | 5 SNI | |
| 24 | Transducer of ERBB2,1 / D78382 | tob1_b | 6 SU | 88 | TOB1_b | 6 SSI | |
| 25 | Transducer of ERBB2,1 / D78382 | tob1_c | 10 TI | 88 | TOB1_c | - | |
| 26 | TSC22 domain family, member 1 / AF201285 | tsc22d1_a | 7 STI | 91.6 | TSC22D1_a | 2 SU | |
| 27 | TSC22 domain family, member 1 / AF201285 | tsc22d1_b | 5 SU | 91.6 | TSC22D1_b | 2 SU | |
| 28 | Zinc finger homeobox 3 / D26046 | zfhx3_a | 13 ITI | 96.1 | ZFHX3_a | 19 ISI | |
| 29 | Zinc finger homeobox 3 / D26046 | zfhx3_b | 6 SU | 96.1 | ZFHX3 | 4 SU | |
| 30 | Zinc finger homeobox 3 / D26046 | zfhx3_c | 9 SU | 96.1 | ZFHX3_c | 4 SU | |
The official gene symbol is followed by _ to denote that multiple repeats are found in the same gene.
N as determined by sequencing and type of repeat is denoted according to these abbreviations. SU: short uninterrupted, STI: short terminally interrupted by a synonymous CAA codon, SSI: short internally interrupted by a synonymous CAA codon, IU: intermediate uninterrupted, ISI: intermediate internally interrupted by a synonymous CAA codon, LSI: long internally interrupted by a synonymous CAA codon, ITI: intermediate terminally interrupted by a synonymous CAA codon.
HomoloGene highest identity percentage by Pairwise Alignment Score at the DNA level.
In order to estimate the conservation of mouse CAG polymorphic repeats in humans, we obtained the Pairwise Alignment Score by comparing the mouse gene to the corresponding human homologue using the HomoloGene tool from NCBI. All the studied genes have a human ortholog and the sequence homology of the repeat region in the human genes is variable (Table 1). As for the studied CAG repeat regions in each gene, seven of these mouse polymorphic repeats are not found in the human ortholog (ANKRD24, NRK, IL2, NR3C1, CHGA, IVL and TSC22D1) and only one, TBP, has been linked to a CAG repeat disease in humans as the cause of Spinocerebellar ataxia type 17 (MIM 607136).
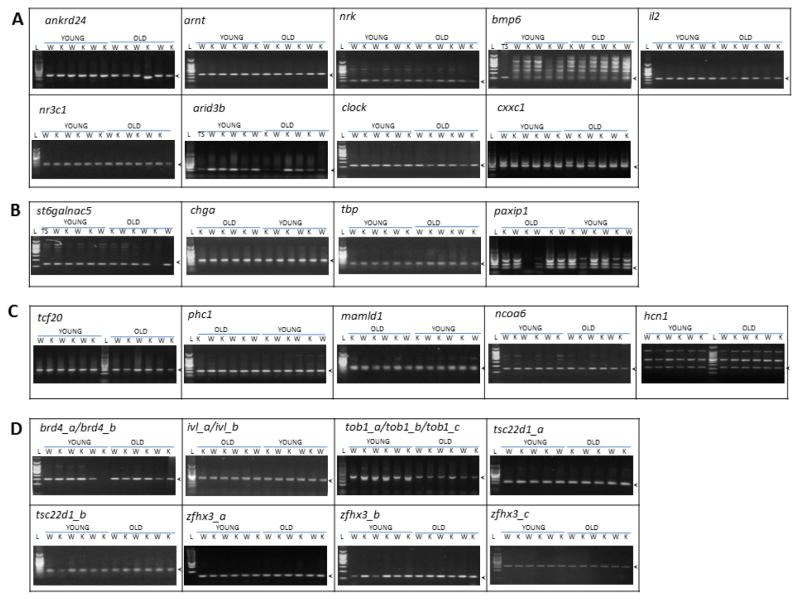
Variability of the CAG repeat length was first qualitatively evaluated by performing electrophoresis of the amplified PCR fragments containing each repeat in young vs. old, caudate vs. liver, and WT vs. ogg1 KO mice. A side-by-side comparison of the amplicon size of WT and ogg1 KO mice revealed that there are some samples with different sizes (Figure 1A ankrd24 lane 11); but such differences were not consistently observed in a specific age, tissue or ogg1 genotype group. We also observed a variable presence of unspecific bands, which may indicate repeat contractions or expansions in some samples and tissues. Some examples of this variability in the caudate are shown in Figure 1. In bmp6, tail snip samples (Figure 1A lane 1) that were treated following the same protocols didn’t show a ladder pattern observed in caudate. In cxxc1, two WT old mice (Figure 1B lanes 8 and 10) had unspecific bands with sizes comparable higher than others. Lastly, in tob1, unspecific bands of lower size are observed only in 4 young mice (Figure 1D lanes 3,4,5 and 7). In general, the length of the CAG repeat amplicons was qualitatively equal regardless of age, tissue or ogg1 activity.
Figure 1. Assessment of length of polymorphic CAG repeats in relation to age- and ogg1 activity.
Representative agarose gel electrophoresis of the CAG repeat –containing region of analyzed mouse polymorphic CAG repeats in striatum of young (3mo) and old (18–24mo). Arrow heads indicate the reference size as indicated by the length of the PCR product from tail snip (TS). A. short unique CAG repeats. B. Intermediate unique CAG repeats. C. Long unique CAG repeats. D. Multiple CAG repeats. (L: MW ladder, W: wild type, K: ogg1-ko)
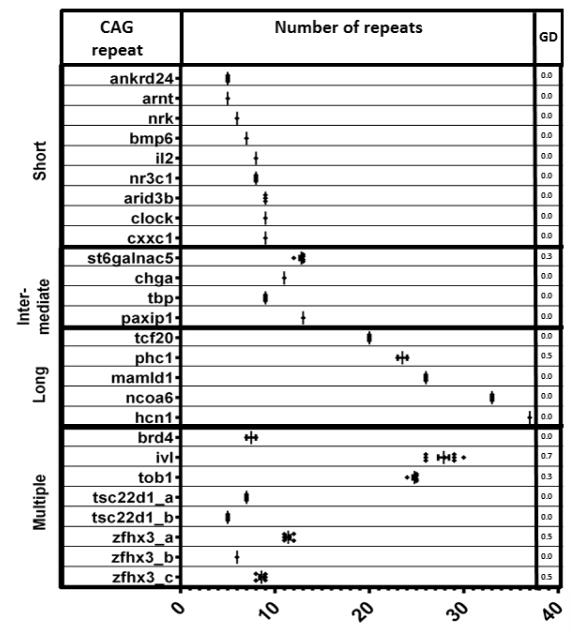
To quantitatively estimate CAG repeat variability in the striatum, we calculated the gene diversity score (GD) for each repeat (Figure 2). This estimation was only performed in old mice since there were not observable size differences of the CAG containing amplicons in young mice by regular electrophoresis. As described previously, GD estimates repeat length variability based on the frequency of each length size [19]. For this GD calculation only the band with the expected PCR amplicon, or the band with the closest to expected band size, was included per mouse, which in the case of multiple amplicons, will exclude bands with other sizes. Some multiple short and intermediate CAG repeats such as ivl _a (19 n) and ivl_b (6 n), tob1_a (7 n), tob1_b (6 n) and tob1_c (10 n), and zfhx3_a (11 n) and zfhx3_c (9 n) were found to have increased length variability (0.7 for ivl, 0.3 for tob1 and 0.5 for zfhx3). The calculated GD for ivl and tob1 corresponds to more than one repeat located in the same PCR amplicon and therefore it was not possible to determine which specific repeat was responsible for increased variability. However, the variability observed for zfhx3_a and zfhx3_c corresponds to two different PCR amplicons and therefore the GD scores reflect repeat variability for each repeat. Only two unique CAG repeats were found to be variable in the striatum of old mice, st6galnac5 (10 n, GD =0.3) and phc1 (23 n, GD=0.5).
Figure 2. Length variability of polymorphic CAG repeats in the striatum of old mice.
The range of variability is represented graphically for each repeat and a measure of the gene diversity (GD) is shown.
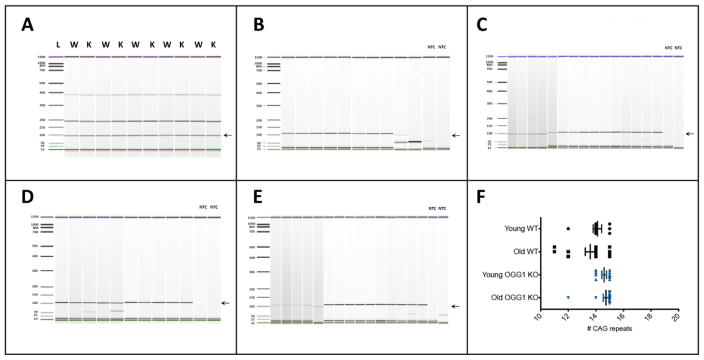
Given the amount of starting genomic DNA used for conventional PCR, this reaction tends to favor the amplification of dominant alleles against less common alleles. Repeat variability at the somatic level may occur at a low level and, therefore, less common unstable alleles (repeat expansions or contractions) may be missed by conventional PCR. To test for repeat variability as the result of somatic instability, we performed SP-PCR which uses one genome equivalent per reaction in multiple reactions for the same sample. We selected four samples for SP-PCR: young WT, old WT, young ogg1 KO and old ogg1 KO to be able to compare repeat length differences associated with age and ogg1 activity in the striatum. Since tbp has the only polymorphic CAG repeat that has an ortholog associated with a human trinucleotide disorder, we decided to perform SP-PCR for this repeat. In contrast to what was observed by conventional PCR (Figure 3A), the CAG repeat of tbp was found to be variable by SP-PCR (Figure 3B to 3F). Variation was higher in the striatum of old WT (13.6±1.4 repeats and 10.3% CV) compared to old ogg1 KO (14.7±0.8 CAG repeats and 5.4% CV) (P=0.0088, F=7.503. Figure 3F). Since the variable bands are displaced to the left, we assume that such variations correspond to small repeat deletions occurring with very low frequency in the striatum of old mice.
Figure 3. Age-related variability of the length of the CAG repeats of tbp gene.
A. Comparative microfluidic electropherograms of the CAG repeat region of tbp in the striatum of old mice in wild-type (W) and ogg1 KO (K) mice as determined by conventional PCR. B to E. Comparative microfluidic eletropherograms of the CAG repeat in young WT (B), old WT (C), young ogg1 KO (D) and old ogg1 KO (E) mice obtained by SP-PCR. The arrow indicates the expected 97bp PCR fragment. NTC: non-template control. F. Quantitative analysis of the number of CAG repeats obtained by SP-PCR in B to E.
4. DISCUSSION
Somatic instability of CAG repeats is a potential mechanism underlying the loss of vulnerable cell populations in the brain in trinucleotide repeat disorders. Many factors have been associated with increased somatic instability in mice including brain region, age, and/or DNA repair activity. The striatum has been found to be highly susceptible to CAG repeat instability compared with other brain regions [4, 24, 25], and somatic CAG expansion has also been observed to increase with age and ogg1 activity; however, these studies have evaluated extremely long transgenic CAG repeats and little is known about naturally polymorphic repeats [4, 13–15]. To study how aging and ogg1 activity play a role in the stability of naturally polymorphic CAG repeats in susceptible brain regions, we evaluated the length of known polymorphic CAG repeats in the striatum of aged WT and ogg1 KO mice. We detected some degree of variability in multiple CAG repeats (ivl, tob1 and zfhx3), one intermediate (st6galnac5) and one long CAG repeat (phc1); but, this variability was not associated with age or ogg1 activity.
We further explored whether repeat variability was occurring at a low level and therefore undetectable by conventional PCR, as it may be the case of age-related somatic expansion, by performing SP-PCR of the CAG repeat in tbp. We detected increased repeat length variation in the striatum of old mice that was not observed by conventional PCR suggesting that age may influence the somatic stability of the tbp CAG repeat. This variation was higher in WT compared to Ogg1 KO mice suggesting that, at least for the tbp CAG repeat, ogg1 activity increases somatic instability but this observation will require to be confirmed with a larger experimental cohort. Nonetheless, the observed variation is consistent with small repeat deletions occurring somatically in old mice. Given that such small deletions were observed in both types of tissue and genotype, they are likely to be a normal aging process. It is known that normal aging is accompanied by DNA damage, most commonly 8-oxoG accumulation [26] and also that the attempt of the BER system to repair the oxidized CAG repeat may result in the formation of a hairpin on the template strand, pol β slippage, pol β hairpin bypass, FEN1 alternate flap cleavage and repeat deletion [27–29].
Interestingly, tbp CAG repeat is the only repeat that is polymorphic in mice and has also been associated with a CAG repeat-related neurodegenerative disorder in humans. While in mice tbp has 9 to 16 synonymously interrupted CAG repeats depending on the mouse strain, in humans TBP has an uninterrupted CAG repeat that is 25 to 42 repeats long. Most of expansions over 35 to 40 repeats manifest phenotypically as Spinocerebellar ataxia 17 (SCA17), a rare form of Huntington Disease-like and late onset PD disease [30].
As previously noticed, the presence of repeat interruptions has an important impact in repeat instability [19, 31]. It is probable that, by reducing the length of pure CAG tracts, synonymous and non-synonymous interruptions are protective against expansion. Interestingly, human pathogenic CAG repeats are predominantly long uninterrupted CAG repeats [32] whereas long polymorphic CAG repeats with interruptions have been reported to be stable in mice [19]. With some important exceptions, our results support this idea since 11n repeats or shorter were found to be uninterrupted or terminally interrupted whereas CAG repeats longer than 13n were all found to be interrupted. An important exception is tbp which has 13 repeats and was found unstable with age despite having many synonymous interruptions. Additionally, a few short and uninterrupted repeats were observed to be variable in the striatum but having a common characteristic: they were multiple (ivl, tob1 and zfhx3) and two of them were in close proximity to each other (ivl and tob1) which raises the possibility that repeat instability may be not only being affected by the length and presence of interruptions but also by their proximity to other repeat regions. Indeed, we identified that these multiple CAG repeats form more stable secondary structures than unique CAG repeats (Supplementary table) which may favor the formation of slipped-strand DNA and repeat expansion [33] and this may explain the high GD score of these repeats in the striatum. Previous studies have suggested that there is a threshold length over which repeats have an increased tendency to expand [4, 6, 34, 35]. The observed higher variability of naturally polymorphic CAG repeats longer than 11n contrasts with the stability observed of short CAG repeats such as htt [13] and other transgenic CAG repeats [36] and seem to support that there is a repeat threshold over which repeats are more susceptible to age-related instability. However, the above-mentioned exceptions to this threshold rule also suggest that other intrinsic characteristics of repeat sequences play a role on length variability and stability. A better understanding of these characteristics and their effect on age-related somatic instability may help elucidating the early phenomena that leads to repeat expansion and neurodegeneration.
5. CONCLUSIONS
The study of factors inducing trinucleotide repeat instability may help understand the progression of repeat neurodegenerative disorders. Age and DNA repair activity have been proposed to influence somatic instability of in transgenic models; however, it is uncertain whether these factors can also induce somatic instability of naturally polymorphic repeats. Our study suggests that naturally polymorphic CAG repeats present somatic repeat variability that is associated with repeat number, proximity and length. Additionally, low levels of variation of the tbp repeat were detected in the striatum of aged mice suggesting that somatic instability takes place in this naturally polymorphic repeat in an age-related manner. Also, the lack of Ogg1 influenced the variability in repeats suggesting a role for DNA repair processes. Future studies are required to understand how age induces repeat instability of tbp and its implications for neurodegenerative repeat disorders.
Supplementary Material
The sequences of polymorphic CAG repeats and 100bp flanking sequence was blatted on UCSC to determine the presence of other repeats nearby. STR=simple tandem repeat. SR=simple repeat. The polymorphic CAG repeats and 10bp flanking sequence was analyzed by mFOLD and the free energy (G) of the most stable structure including the CAG hairpin is shown. When repeat sequences were identified nearby the mFOLD analysis included the repeat sequence.
Highlights.
Naturally polymorphic CAG repeats present somatic variability in mice
The length of the CAG repeat located in tbp, the TBP homologue, in striatum was age-dependent, but not influenced by the activity of the DNA repair enzyme Ogg1
Somatic stability of naturally polymorphic CAG repeats may be influenced by repeat length, the specific repeat sequence and its surrounding sequence context
Acknowledgments
Supported by NIA grant RO1AG031184-01 to F.C.P. and NIH NCRR P20 RR17670
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Ashley CT, Jr, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Human Molecular Genetics. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 4.Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RL, Bates GP. DNA instability in postmitotic neurons. Proc Natl Acad Sci U S A. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shelbourne PF, Keller-McGandy C, Bi WL, Yoon SR, Dubeau L, Veitch NJ, Vonsattel JP, Wexler NS, Arnheim N, Augood SJ. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Human Molecular Genetics. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 6.Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 7.Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, Wheeler VC. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 9.Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 10.Ashizawa T, Wong LJ, Richards CS, Caskey CT, Jankovic J. CAG repeat size and clinical presentation in Huntington’s disease. Neurology. 1994;44:1137–1143. doi: 10.1212/wnl.44.6.1137. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci. 2012;37:162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovtun IV, Johnson KO, McMurray CT. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging (Albany NY) 2011;3:509–514. doi: 10.18632/aging.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 15.Hubert L, Jr, Lin Y, Dion V, Wilson JH. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Human Molecular Genetics. 2011;20:4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mollersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveteras L, Bjolgerud A, Wilson DM, 3rd, Bjoras M, Klungland A. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Human Molecular Genetics. 2012;21:4939–4947. doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoulson I, Young AB. Milestones in huntington disease. Mov Disord. 2011;26:1127–1133. doi: 10.1002/mds.23685. [DOI] [PubMed] [Google Scholar]
- 18.Barnes GT, Duyao MP, Ambrose CM, McNeil S, Persichetti F, Srinidhi J, Gusella JF, MacDonald ME. Mouse Huntington’s disease gene homolog (Hdh) Somat Cell Mol Genet. 1994;20:87–97. doi: 10.1007/BF02290678. [DOI] [PubMed] [Google Scholar]
- 19.Ogasawara M, Imanishi T, Moriwaki K, Gaudieri S, Tsuda H, Hashimoto H, Shiroishi T, Gojobori T, Koide T. Length variation of CAG/CAA triplet repeats in 50 genes among 16 inbred mouse strains. Gene. 2005;349:107–119. doi: 10.1016/j.gene.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell CD, Atha DH, Jakupciak JP, Amos JA, Richie K. Standardization of PCR amplification for fragile X trinucleotide repeat measurements. Clin Genet. 2002;61:13–20. doi: 10.1034/j.1399-0004.2002.610103.x. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macdonald AJ, Sarre SD, Fitzsimmons NN, Aitken N. Determining microsatellite genotyping reliability and mutation detection ability: an approach using small-pool PCR from sperm DNA. Mol Genet Genomics. 2011;285:1–18. doi: 10.1007/s00438-010-0577-9. [DOI] [PubMed] [Google Scholar]
- 24.Watase K, Venken KJ, Sun Y, Orr HT, Zoghbi HY. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Human Molecular Genetics. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, Joyner AL, MacDonald ME. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Human Molecular Genetics. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 26.Cardozo-Pelaez F, Song S, Parthasarathy A, Hazzi C, Naidu K, Sanchez-Ramos J. Oxidative DNA damage in the aging mouse brain. Mov Disord. 1999;14:972–980. doi: 10.1002/1531-8257(199911)14:6<972::aid-mds1010>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroutil LC, Kunkel TA. Deletion errors generated during replication of CAG repeats. Nucleic Acids Res. 1999;27:3481–3486. doi: 10.1093/nar/27.17.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M, Gabison J, Liu Y. Trinucleotide repeat deletion via a unique hairpin bypass by DNA polymerase beta and alternate flap cleavage by flap endonuclease 1. Nucleic Acids Res. 2013;41:1684–1697. doi: 10.1093/nar/gks1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevanin G, Brice A. Spinocerebellar ataxia 17 (SCA17) and Huntington’s disease-like 4 (HDL4) Cerebellum. 2008;7:170–178. doi: 10.1007/s12311-008-0016-1. [DOI] [PubMed] [Google Scholar]
- 31.Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 32.Menon RP, Nethisinghe S, Faggiano S, Vannocci T, Rezaei H, Pemble S, Sweeney MG, Wood NW, Davis MB, Pastore A, Giunti P. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9:e1003648. doi: 10.1371/journal.pgen.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Human Molecular Genetics. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 35.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 36.Reddy PH, Charles V, Williams M, Miller G, Whetsell WO, Jr, Tagle DA. Transgenic mice expressing mutated full-length HD cDNA: a paradigm for locomotor changes and selective neuronal loss in Huntington’s disease. Philos Trans R Soc Lond B Biol Sci. 1999;354:1035–1045. doi: 10.1098/rstb.1999.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of polymorphic CAG repeats and 100bp flanking sequence was blatted on UCSC to determine the presence of other repeats nearby. STR=simple tandem repeat. SR=simple repeat. The polymorphic CAG repeats and 10bp flanking sequence was analyzed by mFOLD and the free energy (G) of the most stable structure including the CAG hairpin is shown. When repeat sequences were identified nearby the mFOLD analysis included the repeat sequence.