Abstract
Objectives
Gram-positive infection is a common cause of sepsis worldwide. Our objective was to identify important pathogen recognition receptor (PRR) pathways regulating innate immune responses and outcome in Staphylococcus aureus sepsis.
Methods
We analyzed whether candidate PRR pathway genetic variants were associated with killed S. aureus-induced cytokine responses ex vivo and performed follow up in vitro studies. We tested the association of our top ranked variant with cytokine responses and clinical outcomes in a prospective multi-center cohort of patients with staphylococcal sepsis.
Results
An intronic TLR4 polymorphism and expression quantitative trait locus, rs1927907, was highly associated with cytokine release induced by stimulation of blood from healthy Thai subjects with S. aureus ex vivo. S. aureus did not induce TLR4-dependent NF-κB activation in transfected HEK293 cells. In monocytes TNF-α release induced by S. aureus was not blunted by a TLR4/MD-2 neutralizing antibody but in a monocyte cell line TNF-α was reduced by knockdown of TLR4. In Thai patients with staphylococcal sepsis, rs1927907 was associated with higher IL-6 and IL-8 levels, and with respiratory failure. S. aureus-induced responses in blood were most highly correlated with responses to Gram-negative stimulants.
Conclusions
A genetic variant in TLR4 is associated with cytokine responses to S. aureus ex vivo, and with cytokine levels and respiratory failure in staphylococcal sepsis. While S. aureus does not express lipopolysaccharide or activate TLR4 directly, the innate immune response to S. aureus does appear to be modulated by TLR4 and shares significant commonality with that induced by Gram-negative pathogens and lipopolysaccharide.
Keywords: immunity, innate, polymorphism, genetic, humans
Introduction
The incidence of Gram-positive sepsis has been continually increasing in recent years, and is the predominant type of sepsis in the United States [1]. Staphylococcus aureus is the most common Gram-positive organism implicated in severe sepsis, with a mortality rate of 30% [2]. Infection with S. aureus can be acquired in both community and healthcare settings, and methicillin resistance in both arenas is a major ongoing concern. A better understanding of the host-pathogen interaction that may lead to novel approaches to treating Gram-positive sepsis is therefore of paramount importance.
Pattern recognition receptors (PRRs) on host immune cells mediate much of the inflammatory response to infection upon activation by conserved pathogen associated molecular patterns (PAMPs) [3]. The PAMP-PRR interaction is a central driver of the host response in sepsis and a better comprehension of the PAMP-PRR axis in human sepsis may unveil new targets for therapeutic exploitation. As a Gram-positive organism, S. aureus cell wall components activate membrane-bound PRRs such as Toll-like-receptor (TLR) 2 (as a heterodimer with TLR1 or TLR6) and cytoplasmic NOD-like receptors [4]. Many of these PRRs are also activated by PAMPs of Gram-negative organisms [3]. Lipopolysaccharide (LPS) has long been considered to be an essential precipitant of the host inflammatory response to Gram-negative infection, however [5]. In contrast to Gram-negative organisms, S. aureus does not express LPS. Although various PRRs may be activated by different bacteria, downstream signaling pathways may converge or be inter-related [3]. It remains unclear whether and how the fundamental inflammatory pathways activated in Gram-positive and Gram-negative sepsis differ from one another and drive outcome [6].
Innate immune genetic variation modulates the host response to infection [7]. We hypothesized that key PRR genes regulating inflammatory responses to S. aureus sepsis could be identified by identifying human genetic variation associated with cytokine production and clinical outcome. To test this hypothesis, we assessed the relationship between ex vivo blood cytokine responses to S. aureus and PRR pathway genetic variation in a large cohort of healthy individuals, and then validated our findings in a prospective multi-center cohort of patients with staphylococcal sepsis.
Methods
Ex vivo blood stimulation assays
Three hundred healthy Thai subjects donating blood at the blood donation center at Sunpasitthiprasong Hospital, Ubon Ratchathani, Thailand were recruited for a blood sample as previously reported [8]. Those who met enrollment criteria gave written informed consent to participate and provided a post-donation blood sample in citrate tubes. A batch of ninety-six well immuno-assay plates were generated by adding 20 µl of stimuli in appropriate concentrations to each well. Plates were frozen at −80 °C until the day of use when they were thawed to 37 °C. 380 µl of fresh whole blood anticoagulated with citrate and mixed 1:1 with RPMI media was added to each well [8]. Final concentrations of stimuli were: whole heat-killed S. aureus strain Newman 2.5 × 107 CFU/ml and 5.0 × 107 CFU/ml, whole heat-killed Burkholderia pseudomallei K96243 2.5 × 107 CFU/ml, Salmonella typhimurium flagellin 500 ng/ml, Pam3CSK4 100 ng/ml (Invivogen), Pam2CSK4 100 ng/ml (Invivogen), MDP 10 µg/ml (Invivogen), TriDAP 10 µg/ml (Invivogen), Escherichia coli O111:B4 LPS 10 ng/ml (List Biologicals), and Salmonella minnesota Re595 LPS 10 ng/ml (List Biologicals). Plates were placed on a shaker at 37 °C under 5% CO2 for 6 hours before begun spun down and the plasma supernatant removed and frozen at −80 °C. Monocyte counts were performed in the hospital clinical laboratory.
Staphylococcal sepsis study
A prospective observational study of community onset S. aureus sepsis was conducted at four hospitals in northeast Thailand between March 2010 and December 2013 [9]. Study sites were Sunpasitthiprasong Hospital, Ubon Ratchathani; Udon Thani Hospital, Udon Thani; Srinagarind Hospital, Khon Kaen; and Khon Kaen Hospital, Khon Kaen. Potential study patients were identified by daily screening at each hospital diagnostic microbiology laboratory for clinical samples taken from sterile sites that grew a pure growth of S. aureus. Subsequent genetic analysis of isolates revealed that a minority subset (19%) were S. argenteus [9], a recently described closely related Staphylococcus species that is indistinguishable from S. aureus in the diagnostic laboratory. Written informed consent was obtained from all patients or their designee. Inclusion criteria were as follows: age at least 14 years (who are admitted to the adult wards), positive culture taken within two days of hospital admission, or after two days when sampling from a patient admitted with suspected infection was delayed, and at least two of four systemic inflammatory response (SIRS) criteria met within 48 hours of culture. SIRS criteria were: temperature <36 °C or >38 °C, heart rate >90 beats/min, respiratory rate >20 breaths/min or PaCO2 <32 mmHg or requiring mechanical ventilation, white blood cell count <4000 or >12000 cells/ml or >10% band forms. Baseline clinical information was obtained from the medical records. Blood was drawn on enrollment, and plasma was separated and frozen at −80 °C. Patients were followed until hospital discharge, and mortality was ascertained on the 28th day. Respiratory failure was defined as requirement for mechanical ventilation; shock was defined as requirement for vasopressors or inotropes.
Lipopolysaccharide and TLR4-stimulation assays
The presence of LPS in the heat-killed S. aureus strain Newman preparation was determined by Limulus Amebocyte Lysate gel-clot assay (Associates of Cape Cod) according to the manufacturer’s instructions (sensitivity 0.03 EU/ml). TLR4-dependent NF-κB activation by S. aureus was assessed by stimulation of hTLR4/hMD-2/hCD14-transfected HEK293 cells, using a modified version of a previous approach [10]. Cells were cultured in a 96-well flat-bottomed tissue culture plate at 35,000 cells/well in Dulbecco’s modified Eagle’s medium (DMEM plus 10% fetal bovine serum (FBS) and 1% L-glutamine). The following day, cells were transiently transfected with 100 µl of transfection mixture comprised of plasmids, transfection reagent (Polyfect, Qiagen), and DMEM. DNA was added in the following amounts per well: ELAM (firefly luciferase) (50 ng), pRL-TK (Renilla luciferase) (5 ng), hTLR4 or empty vector (10 ng), hCD14 (1 ng), and hMD-2 (1 ng). After 48 hours of incubation, transfected cells were stimulated with heat-killed S. aureus or E. coli O111:B4 LPS (List Biological Laboratories, Inc.) as a positive control. After 4 hours, cells were washed with PBS and lysed with passive lysis buffer (Promega, Madison, WI), and NF-κB activation was determined in 10 µl of lysate by the ratio of firefly to Renilla luciferase light emission by the use of the Dual-Luciferase reporter system (Promega, Madison, WI).
Primary monocyte/monocyte cell line stimulations and TLR4 knockdown
Peripheral blood mononuclear cells (PBMCs) were isolated from blood obtained from fasting subjects who avoided recent strenuous exercise drawn into 8 ml Vacutainer CPT tubes (BD, NJ). Subjects were between 18 and 65 years of age, weighed between 100 and 350 pounds, of southeast Asian ancestry, without any chronic medical conditions, non-pregnant and had not delivered in the last nine months, non-smokers, taking no medications other than oral contraceptives, and had not had recent illness or vaccinations. PBMCs were further processed to monocytes using the Monocyte Isolation Kit II (Miltenyi MACS, CA). Monocytes were plated in a 96 well plate at 50,000 cells/well and rested overnight in complete RPMI media (10% FBS, 1% L-glutamine). The next day, cells were treated with a monoclonal TLR4/MD-2 neutralizing antibody (mab-htlr4md2, Invivogen) or control isotype antibody (mabg1-ctrlm, Invivogen) at 20 µg/ml, 1 hour prior to addition of stimulus. Cells were stimulated for 6 hours with LPS from B. pseudomallei K92643 at a final concentration of 1 ng/ml or heat-killed S. aureus at a bacteria to cell ratio of 100 before supernatants were collected. THP-1-Dual Cells (Invivogen, CA), derived from the human THP-1 monocyte cell line, were differentiated over a period of 72 hours with vitamin D3 (final concentration 10 pM/ml), and plated at 50,000 cells per well in media containing D3. The next day, cells were transfected with Silencer Select siRNA (Ambion, MA) against TLR4 (#4392420) or Silencer Select Negative Control No. 1 siRNA (#4390843) at a final concentration of 5 nM using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies, CA). To evaluate efficiency of gene knockdown, total RNA was extracted from transfected cells after 48 hours using RNA lysis buffer (Promega, WI). Extracted RNA was converted into cDNA using iScript Reverse Transcription Supermix (BioRad, CA), according to the manufacturer’s protocol. Expression of TLR4 and UBC (housekeeping gene) was quantified by qPCR using the SsoAdvanced Universal SYBRGreen Supermix (BioRad, CA) run on a Chromo4 (BioRad, CA). Each reaction was run in triplicates - using 20 ng of template, in a total volume of 20 µl. Reactions were incubated at 95 °C for 3 minutes, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s. PrimePCR primers purchased from BioRad, CA for TLR4, Human (qHsaCED0037607) and UBC, Human (qHsaCED0023867) were used in the reactions. THP-1-Dual Cells were stimulated 48 hours after siRNA transfection with either B. pseudomallei LPS 100 ng/ml as a positive control or heat-killed S. aureus at a bacteria to cell ratio of 100 at 37 °C overnight. The following day, supernatants were collected.
Genotyping and SNP selection
DNA was extracted from whole blood of healthy subjects and patients using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany). Selection of ninety six non-synonymous coding single nucleotide polymorphisms in PRR pathway genes was performed by searching the HapMap project database (http://hapmap.ncbi.nlm.nih.gov) and based on functional prediction using a FastSNP analysis (http://fastsnp.ibms.sinica.edu.tw) [11]. Tag SNPs were selected from the Han Chinese in Beijing and Japanese in Tokyo populations in the HapMap database for variants with a minor allele frequency at least 2% using the HapMap tag-SNP picker option. Several inflammatory response SNPs previously associated with susceptibility or outcome from infection based on literature review were also included. Genotyping of healthy subjects was performed using Fluidigm SNPtype assays on a Biomark microfluidics real time PCR system (Fluidigm, South San Francisco, CA). rs1927907 was genotyped in patients using primers and probes designed by RealTimeDesign software from Biosearch Technologies (https://www.biosearchtech.com) and synthesized by Biosearch Technologies (Petaluma, CA, USA). The 147 bp region of TLR4 covering the variant position was amplified with forward primer 5' GGTAGACCACCTCTCCCTTTT 3' and reverse primer 5' GCTGGCCTCTCTGTAAGCA 3'. rs1927907 was genotyped using probes 5' FAM-AGAAGTAGTTTTTCACCAAACA-BHQ-1 3' for allele C and 5′ CAL Fluor Orange 560-CAAGAAGTAGTTTTTCATCAAAC-BHQ-1 for allele T. Real-time PCR was performed using CFX96 Touch Real-Time PCR Detection System (BioRad, Hercules, CA, USA) in a total volume of 20 µl.
Cytokine and chemokine measurements
Cytokine/chemokine assays were performed using Luminex multiplex bead system with R&D Systems reagents or ELISA (BD Biosciences and R&D Systems).
Statistical analyses
Continuous data following a normal distribution are reported as mean ± standard deviation; continuous data following a non-normal distribution are reported as median ± interquartile ranges. Plasma cytokine data were log10 transformed before analysis given their generally non-normal distribution. For the immuno-assay dataset, cytokine correlations were assessed using a Pearson correlation coefficient and correlation matrices were used to created heatmaps [12] and relatedness dendrograms. To maximize power for identifying genetic associations, a composite cytokine response was created. The monocyte-normalized responses for five cytokines (IL-6, IL-8, TNF-α, G-CSF, and IL-1β) were summed after subtracting the mean and dividing by the standard deviation of the 300 observations within each cytokine. These cytokines were chosen based on relatedness score in a cluster dendrogram. Linear regression of each SNP on composite cytokine response was performed assuming an additive genetic model, adjusting for log monocyte count. Eight of the 51 SNPs had observed minor allele frequencies <0.05 and were removed from the primary analysis due to low power in the face of multiple test adjustment, leaving 43 SNPs for testing with the composite cytokine outcome. Adjustment for multiple testing was done via permutation testing [13]. The distribution of the ordered p-values under the null hypothesis was produced by permuting the outcomes 2000 times and recomputing the 43 p-values for each permutation. The distribution of the minimum of the 43 p-values for each of the permutations was compared with the observed minimum p-value to obtain the probability of observing this minimum under the null hypothesis (i.e. a multiple-test corrected p-value).
Corrected p-values for the second- and third-most significant SNPs, etc, were obtained analogously. Associations were considered significant if the corrected p-value ≤0.05. Further tests of association of candidate SNPs with individual cytokines were performed on log10 transformed cytokine data using linear regression, adjusting for log monocyte count, again assuming an additive genetic model. For monocyte/monocyte cell line experiments, data were analyzed with the t test. For the clinical study, association of genotype with cytokine response or clinical outcome was performed with Fisher’s exact test (for clinical outcomes) or with regression, adjusting for age, sex, co-morbidity index, and site. The co-morbidity index is a 10 point score comprising one point each for lung disease, heart disease, kidney disease, liver disease, neurological disease, hematological disease, autoimmune disease, cancer, diabetes, and alcoholism. Secondary analyses were performed adjusting for bacterial species. Analyses were performed using Stata v11.2 (College Station, TX), or R (ww.r-project.org).
Ethics
Ethical approval was obtained from the Ethics Committees of the following institutions: Faculty of Tropical Medicine, Mahidol University, Bangkok; Sunpasitthiprasong Hospital, Ubon Ratchathani; Udon Thani Hospital, Udon Thani; Khon Kaen Hospital, Khon Kaen; and Faculty of Medicine (Srinagarind Hospital), Khon Kaen University, Khon Kaen, Thailand; and the University of Washington, Seattle.
Results
S. aureus induces robust inflammatory responses in blood
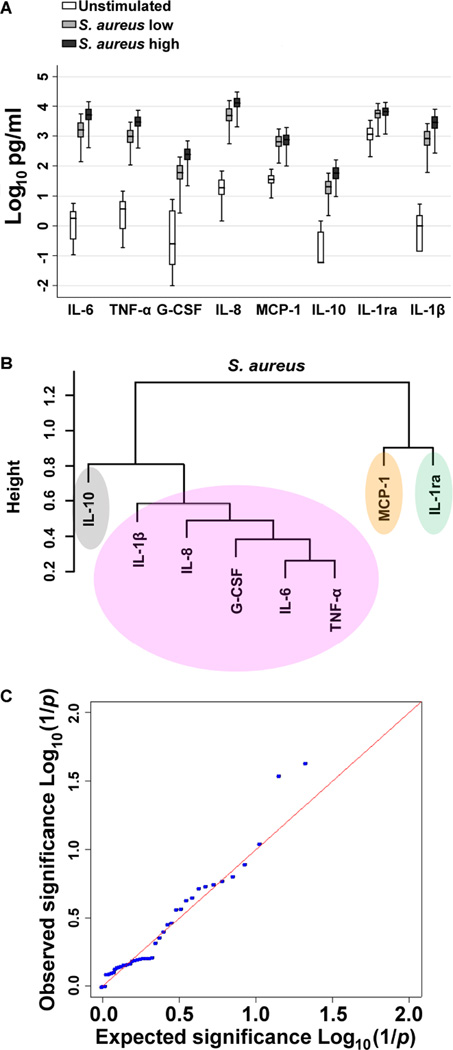
To evaluate the inflammatory response to S. aureus in human blood, we assayed pro- and anti-inflammatory cytokines and chemokines in plasma after stimulation of fresh whole blood from 300 healthy individuals with two different concentrations of heat killed S. aureus (Figure 1A). As expected, S. aureus induced significant increases above baseline values for all mediators (IL-1ra, IL-1β, IL-6, IL-8, IL-10, TNF-α, G-CSF, and MCP-1), with considerable inter-individual variation in responses.
Figure 1. A single nucleotide polymorphism in TLR4 is associated with S. aureus-induced cytokine release.
A: IL-1ra, IL-1β, IL-6, IL-8, IL-10, TNF-α, G-CSF, and MCP-1 were measured in plasma supernatants by multiplex bead assay after stimulation of fresh whole blood from 300 healthy individuals with heat killed S. aureus 2.5×107 CFU/ml (SA low) or 5 × 107 CFU/ml (SA high) for six hours at 37°C. Data are displayed on a log scale. Boxes show the median and interquartile range; whiskers show upper and lower adjacent values; outside values are not shown for clarity. All mediators induced by S. aureus are significantly greater than in unstimulated blood (P<0.001 for all). B: Cluster dendrogram created from relatedness scores calculated from pairwise comparisons between cytokines induced by stimulation of whole blood with S. aureus 2.5×107 CFU/ml used to derive a composite outcome measure of TNF-α, IL-6, IL-8, G-CSF and IL-1β responses. C: QQ plot (expected versus observed log10 (1/p values)) for the association of 43 single nucleotide polymorphisms with the composite outcome of S. aureus induced cytokine levels demonstrating several lower than expected p values (higher than expected log10 (1/p values)). The lowest p value is rs1927907, an intronic single nucleotide polymorphism in TLR4.
A human TLR4 variant is associated with cytokine responses to S. aureus
We next analyzed the association of candidate PRR pathway genetic variants with cytokine responses induced by S. aureus. We generated a composite cytokine response by performing pairwise comparisons between different cytokines produced from whole blood of each donor following activation with S. aureus and creating a cluster dendrogram based on inter-mediator relatedness scores (Figure 1B). The most closely related mediators were IL-1β, IL-8, G-CSF, IL-6, and TNF-α (r=0.6–0.9). We selected these mediators to use as the composite measure. We genotyped 96 single nucleotide polymorphisms in candidate PRR pathway genes, selected as described in the methods, in all 300 subjects. Five assays failed and 39 assays had no variation. One assay was excluded due to a call rate <97%. The remaining 51 variants are listed in Table S1. Eight variants had observed minor allele frequencies (MAF) <0.05 and were excluded due to low power. Forty three variants were tested for an association with cytokine response. We tested the association of each variant with the composite S. aureus-induced cytokine cluster (Figure 1C).The top ranked variant associated with cytokine response to S. aureus with P<0.05 (adjusted for multiple comparisons via the permutation test) and a false discovery rate <0.05 was rs1927907, an intronic TLR4 polymorphism that exchanges a C for a T. The minor allele frequency (MAF) of rs1927907 in our cohort was 0.10. To determine if a particular cytokine drove the observed composite effect, the association between rs1927907 and individual cytokine concentrations was tested. We found that four of the five cytokines induced by S. aureus – TNF-α, IL-1β, IL-6, and G-CSF – were significantly higher for carriers of the rs1927907 minor (T) allele (Table 1). By comparison, rs1927907 was also associated with cytokine responses to E. coli LPS (TNF-α, IL-1β, IL-6, and G-CSF) and Pam3CSK4 (TNF-α and IL-6) but not with cytokines induced by Pam2CSK4 or flagellin (data not shown).
Table 1.
Associations between TLR4 variant rs1927907 and inflammatory mediators induced by ex vivo stimulation of blood from 300 healthy subjects with S. aureus
| Median (IQR) in pg/ml | ||||||
|---|---|---|---|---|---|---|
| IL-6 | IL-8 | TNF-α | IL-1β | G-CSF | ||
| rs1927907 | ||||||
| Genotype | ||||||
| CC | 1480 (861–2656) | 4532 (3090–7610) | 927 (574–1433) | 763 (463–1328) | 55 (34–99) | |
| CT | 1852 (1304–4117) | 5687 (3990–10956) | 1128 (726–2684) | 1100 (699–1727) | 82 (49–148) | |
| TT | 3096 (701–5492) | 7658 (2772–12544) | 1833 (219–3447) | 2243 (599–3886) | 99 (27–170) | |
| P valuea | 0.012 | 0.065 | 0.017 | 0.002 | 0.001 | |
Linear regression model on log transformed cytokine concentrations assuming an additive genetic model and adjusting for monocyte count.
rs1927907 is a TLR4 expression quantitative trait locus (eQTL) in whole blood; the minor allele is associated with increased whole blood TLR4 expression [14]. Moreover, the minor allele of rs1927907 is associated with increased TLR4 protein expression on CD4+CD25high regulatory T cells of asthmatic subjects as determined by flow cytometry [15]. These data, which are concordant with our observations of increased blood cytokine responses in carriers of the rs1927907 minor allele, implicate TLR4 itself in the response to S. aureus. However, TLR4 is the canonical LPS receptor and therefore is not anticipated to be involved in sensing of a Gram-positive pathogen. This prompted us to consider contamination of our S. aureus preparations with LPS as a potential explanation for these findings. We assayed our S. aureus preparations using a sensitive Limulus Amebocyte Lysate assay but did not detect any LPS (data not shown).
S. aureus does not activate TLR4 in vitro
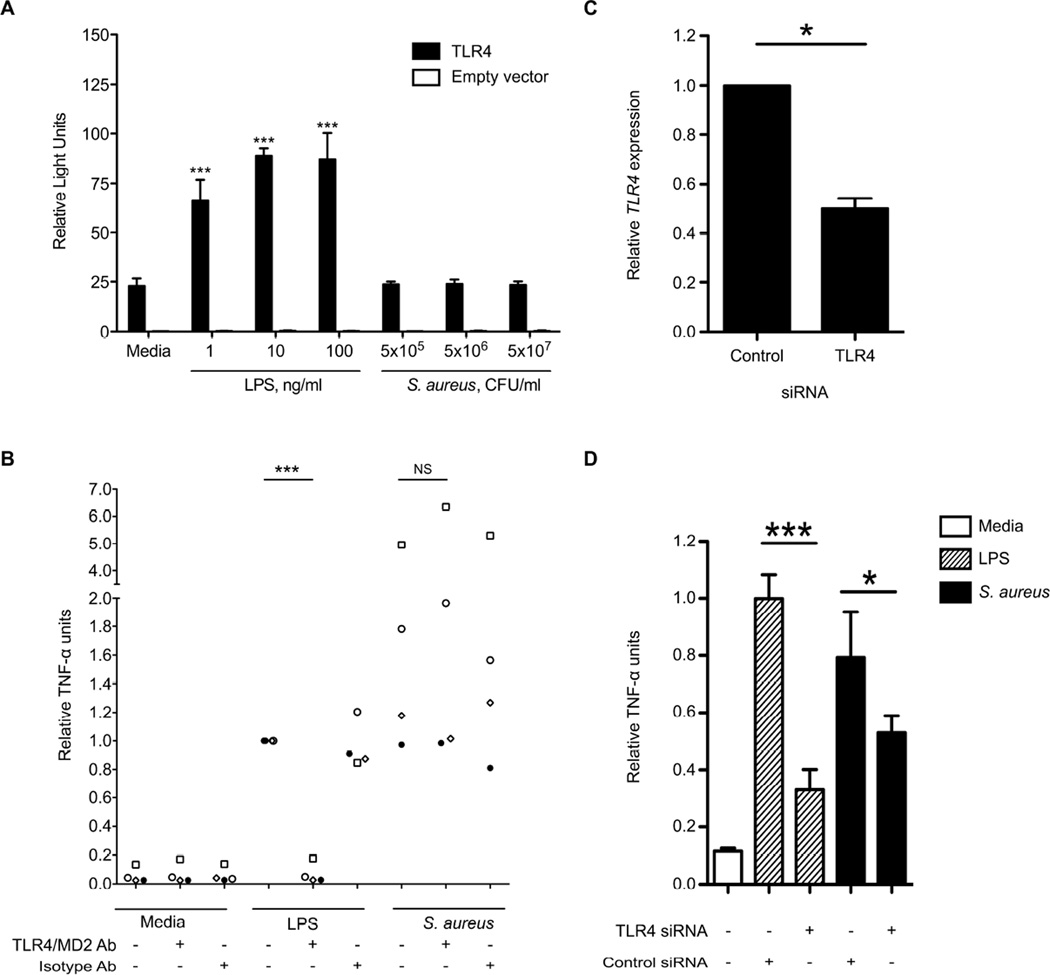
As we did not find any LPS contamination in our preparations, we postulated that S. aureus might express other factors that would serve as ligands for TLR4. To address this question, we stimulated hTLR4/MD-2/CD14-transfected HEK293 cells with our S. aureus preparations to assess for a TLR4-dependent signal. We measured NF-κB activation by luciferase reporter assay but found no S. aureus-induced TLR4-dependent light emission (Figure 2A). This observation suggested that S. aureus does not express a TLR4 ligand that signals through NF-κB. We next evaluated how blockade of TLR4 receptor function would impact cytokine responses to S. aureus. We stimulated primary blood monocytes from healthy subjects with S. aureus in the presence of a TLR4/MD-2 neutralizing antibody and assayed TNF-α concentrations in cell supernatants (Figure 2B). We observed no change in TNF-α release when TLR4 receptor function was impaired. Together, these experiments indicated that S. aureus does not activate TLR4 to induce inflammatory cytokine responses.
Figure 2. S. aureus does not activate NF-κB or induce TNF-α release via TLR4 ligation but TLR4 modulates S. aureus-induced TNF-α release.
A: HEK293 cells were transiently transfected with hTLR4 or equivalent amounts of empty vector, hMD-2, hCD14, ELAM (firefly luciferase), and pRL-TK (Renilla luciferase) before stimulation for four hours with E.coli LPS or heat killed S. aureus. NF-κB activation was determined in cell lysates by the ratio of firefly to Renilla luciferase light emission. Mean ± standard deviation values of quadruplicate samples are displayed. ***, P<0.001 compared to unstimulated TLR4-transfected cells. B: Primary blood monocytes from four healthy individuals were treated with a monoclonal TLR4/MD2 neutralizing antibody or control isotype antibody at 20 µg/ml, 1 hour prior to stimulation for six hours with LPS from B. pseudomallei K92643 1 ng/ml or heat-killed S. aureus at a bacteria to cell ratio of 100. TNF-α was assayed by ELISA in supernatants. Individual data (each with a unique symbol) representing means of duplicate or triplicate samples normalized to LPS-induced TNF-α release are displayed. ***, P<0.001. C: THP-1-Dual Cells were transfected with siRNA against TLR4 or negative control siRNA at 5 nM. After 48 hours TLR4 gene expression was determined by qPCR. TLR4 gene expression is presented relative to housekeeping gene UBC expression and normalized to control siRNA transfection results. Mean ± standard deviation values of quadruplicate samples from two independent experiments are displayed. *, P<0.05. D: THP-1-Dual cells transfected for 48 hours as in (C) were stimulated with B. pseudomallei LPS 100 ng/ml or heat-killed S. aureus at a bacteria to cell ratio of 100 overnight. TNF-α was assayed by ELISA in supernatants. Mean ± standard deviation values of quadruplicate samples normalized to LPS-induced TNF-α release are displayed, and the data comprise one representative example of three independent experiments. *, P<0.05; ***, P<0.001.
Knockdown of TLR4 inhibits the TNF-α response to S. aureus
To explain our observations of differential cytokine response associated with rs1927907 we next evaluated whether differential expression of TLR4 in the cell may modify the host response to S. aureus. We surmised that even if S. aureus did not directly activate TLR4, there may be downstream modulatory effects of TLR4 on other signaling pathways. We and others have observed such an effect of TLR5 on TLR4-dependent signaling [16, 17]. We developed an assay using human monocyte THP-1-Dual cells in which we transfected siRNA targeting TLR4 or control siRNA (Figure 2C). We achieved approximately 50% decrease in TLR4 gene expression in this assay. We next stimulated cells with S. aureus after transfection of siRNA against TLR4 (Figure 2D). We measured TNF-α concentration in cell supernatants and found that levels of this cytokine induced by S. aureus were significantly reduced after TLR4 knockdown. Collectively, these results supported the concept that S. aureus does not directly activate TLR4 but that TLR4 modulates the inflammatory response to S. aureus.
TLR4 variant rs1927907 is associated with increased cytokine responses during staphylococcal sepsis
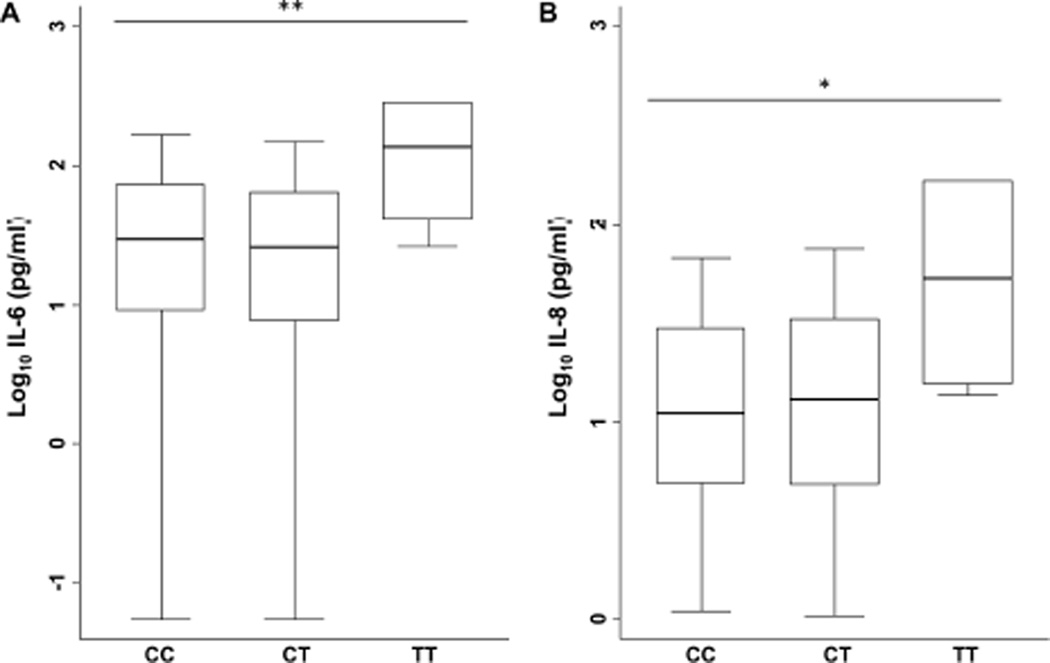
Regulators of the inflammatory response to heat killed S. aureus in experimental settings may be very different than host response determinants in clinical staphylococcal infection. To determine whether rs1927907 might predict cytokine/chemokine responses in a clinical setting, we genotyped the variant in a cohort of 327 individuals with community-onset staphylococcal sepsis recruited at four hospitals in northeast Thailand. The clinical characteristics of these patients are described in Table 2. After finding no evidence for deviation from Hardy Weinberg equilibrium in this cohort (P=0.61), we tested the association of rs1927907 with plasma cytokine concentration at enrolment. Minor allele homozygotes generated significantly higher levels of IL-6 and IL-8 in crude analyses and after adjusting for age, sex, co-morbidity index, and site (adjusted P values 0.01 and 0.045, respectively) (Figure 3).
Table 2.
Clinical characteristics and outcomes of 327 patients with staphylococcal sepsis
| Variable | Value |
|---|---|
| Median age in years (IQR) | 54 (42–65) |
| Female sex – no. (%) | 114 (34.9) |
| Pre-existing conditions – no. (%) | |
| Diabetes mellitus | 116 (35.5) |
| Heart disease | 17 (5.2) |
| Kidney disease | 53 (16.2) |
| Liver disease | 13 (4.0) |
| Lung disease | 18 (5.5) |
| Cancer | 7 (2.1) |
| Alcoholism | 5 (1.5) |
| Autoimmune disease | 5 (1.5) |
| Neurological disease | 8 (2.5) |
| Hematological disease | 9 (2.8) |
| Median co-morbidity index (IQR)a | 1 (0–1) |
| Outcomes – no. (%) | |
| Respiratory failure | 58 (17.7) |
| Shock | 38 (11.6) |
| Death at 28 days | 30 (9.2) |
Co-morbidity index is a 10 point score comprised of one point each for lung disease, heart disease, kidney disease, liver disease, neurological disease, hematological disease, autoimmune disease, cancer, diabetes, and alcoholism.
Figure 3. TLR4 variant rs1927907 is associated with plasma IL-6 and IL-8 in patients with staphylococcal sepsis.
IL-6 (A) and IL-8 (B) were assayed by ELISA in plasma samples from patients with staphylococcal sepsis and are displayed by TLR4 variant rs1927907 genotype. Boxes show the median and interquartile range; whiskers show upper and lower adjacent values; outside values are not shown for clarity. Values were log10 transformed and linear regression was performed using a recessive model (comparing CC and CT subjects to TT subjects), adjusting for age, sex, comorbidity index, and site. N=249 (CC), 69 (CT), 6 (TT). **, P ≤ 0.01; *, P < 0.05.
TLR4 variant rs1927907 is associated with increased respiratory failure during staphylococcal sepsis
We then tested whether rs1927907 genotype predicted clinical outcome from staphylococcal sepsis. In both crude and adjusted analyses, minor allele homozygotes developed significantly more respiratory failure (adjusted OR 9.65, 95% CI: 1.60–58.08, P=0.01) (Table 3). These individuals did not have significantly greater rates of shock or death, outcomes that were less frequent overall. Together these cytokine and clinical data in an independent cohort of patients offer confirmatory evidence in support of the role of TLR4 as a modulator of the host response in staphylococcal sepsis.
Table 3.
Association of TLR4 variant rs1927907 with clinical outcomes of patients with staphylococcal sepsis
| Genotype | Respiratory failure | Shock | Death | |||
|---|---|---|---|---|---|---|
| Yes | No | Yes | No | Yes | No | |
| CC | 42 (72%) |
210 (78%) |
29 (76%) |
223 (77%) |
22 (73%) |
230 (77%) |
| CT | 12 (21%) |
57 (21%) |
7 (18%) |
62 (21%) |
7 (23%) |
62 (21%) |
| TT | 4 (7%) |
2 (1%) |
2 (5%) |
4 (1%) |
1 (3%) |
5 (2%) |
| Unadjusted P valuea | 0.018 | 0.22 | 0.51 | |||
| Adjusted OR (95% CI) | 9.65 (1.60–58.08) | 4.31 (0.66–27.96) | 1.75 (0.18–17.11) | |||
| Adjusted P valueb | 0.01 | 0.13 | 0.63 | |||
Unadjusted analyses are performed using the exact test
Adjusted analyses are performed using logistic regression assuming a recessive genetic model adjusting for age, sex, co-morbidity index, and site.
Inflammatory responses to S. aureus correlate with responses to LPS and B. pseudomallei
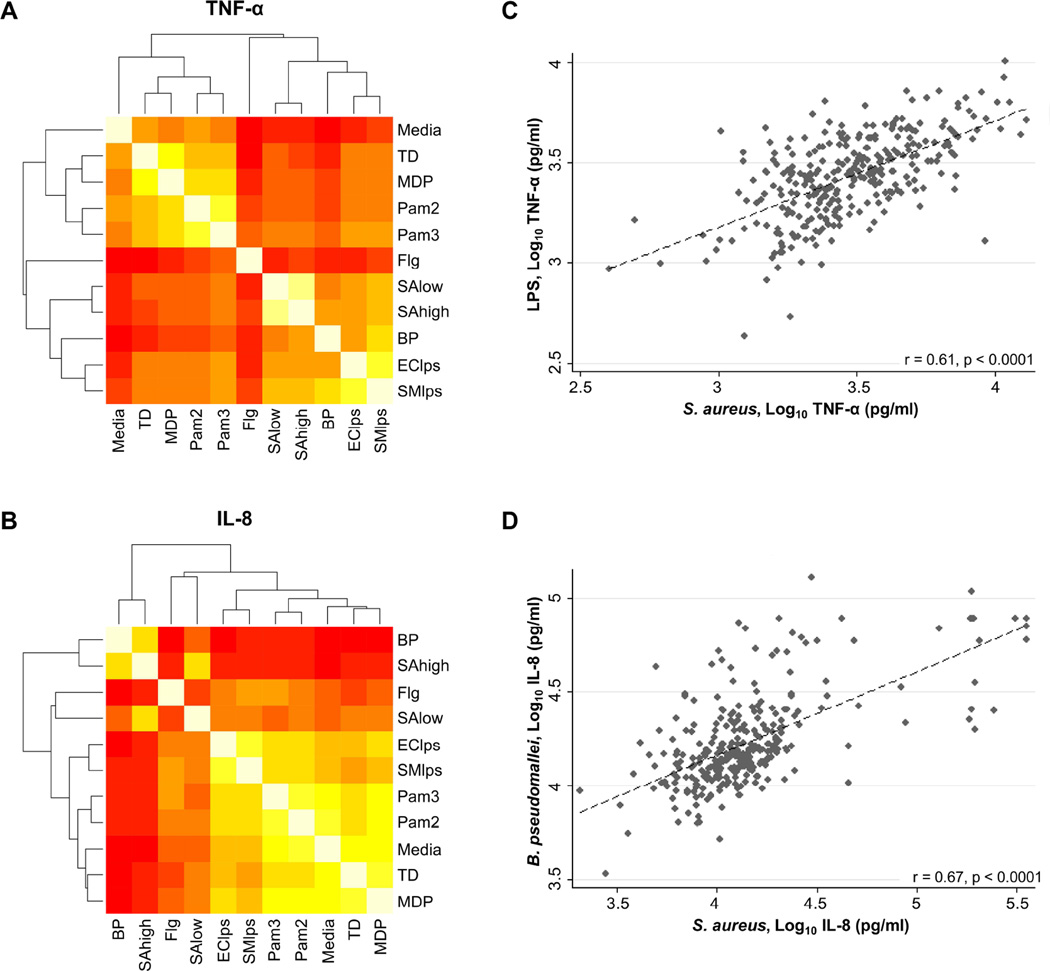
In light of our data implicating TLR4 in the immune response to S. aureus, we then sought to determine whether inflammatory responses induced by S. aureus in healthy individuals might be correlated with responses induced by LPS or by Gram-negative bacteria in our human blood immunoassay. We selected E. coli LPS, S. minnesota LPS and heat killed B. pseudomallei for this comparison. As control stimulants, we chose TLR2/6 agonist Pam2CSK4, TLR2/1 agonist Pam3CSK4, TLR5 agonist S. typhimurium flagellin, NOD1 agonist Tri-DAP, and NOD2 agonist MDP. We measured the correlations between each stimulus and created heat maps with derived relatedness dendrograms. Across all cytokines, the dendrograms indicated highest relatedness between responses to S. aureus and LPS or between S. aureus and B. pseudomallei. For example, the correlation between TNF-α induced by S. aureus and by S. minnesota LPS was 0.61, P<0.0001, and the correlation between IL-8 induced by S. aureus and by B. pseudomallei was 0.67, P<0.0001 (Figure 4).
Figure 4. High correlation between blood inflammatory responses induced by S. aureus and LPS or B. pseudomallei.
Heat maps of correlation between TNF-α (A) or IL-8 (B) responses induced by stimulation of whole blood from 300 healthy subjects with a panel of purified innate immune ligands and heat-killed bacteria for six hours at 37 °C. Stimulants were heat-killed S. aureus 2.5 × 107 CFU/ml (SAlow) and 5.0 × 107 CFU/ml (SAhigh), heat-killed B. pseudomallei 2.5 × 107 CFU/ml (BP), Salmonella typhimurium flagellin 500 ng/ml (Flg), Pam3CSK4 100 ng/ml (Pam3), Pam2CSK4 100 ng/ml (Pam2), Escherichia coli O111:B4 LPS 10 ng/ml (EClps), and Salmonella minnesota Re595 LPS 10 ng/ml (SMlps). A dendrogram of hierarchical clustering results for the correlation matrix for each mediator is shown. Lowest correlation is red (Pearson correlation coefficient r<0.1) and highest correlation is white (r=1.0). Scatterplots of individual TNF-α (C) or IL-8 (D) responses (log10 transformed) after stimulation of fresh whole blood from 300 healthy subjects with heat-killed S. aureus 5 × 107 CFU/ml, S. minnesota Re595 LPS (C), or heat-killed B. pseudomallei 2.5 × 107 CFU/ml (D) for six hours at 37°C. For each plot, the relationship between variables as determined by linear regression (dotted line) and Pearson correlation (correlation coefficient, r) are shown.
Discussion
The main findings of this study are that 1) blood inflammatory responses to S. aureus are associated with the TLR4 genetic polymorphism rs1927907, an eQTL, both ex vivo and in patients with staphylococcal sepsis; 2) TLR4 is not activated by S. aureus but TLR4 knockdown impairs TNF-α release induced by S. aureus, 3) rs1927907 predicts respiratory failure in patients with staphylococcal sepsis; and 4) blood inflammatory responses to S. aureus are tightly correlated with responses to LPS and a Gram-negative organism. These findings raise intriguing new questions about the role of TLR4 in human staphylococcal sepsis.
TLR4 is not usually considered a fundamental participant in the host response to Gram-positive infections. The S. aureus cell wall is composed of peptidoglycan in combination with other substances such as lipoteichoic acid (LTA) and lipoprotein. These components and secreted proteins can induce inflammation and may contribute to sepsis [4]. PRRs on host immune cells involved in recognition of S. aureus include TLR2 in combination with TLR1 or TLR6 and/or CD36 and CD14 [4]. While the cell wall of Gram-negative bacteria shares many of the same PAMPs as Gram-positive bacteria [3], a cell wall component unique to Gram-negative organisms is LPS [5]. LPS is known as a TLR4 agonist, and induces a robust pro-inflammatory response and septic shock [18], but is not expressed by S. aureus. Therefore, when we found associations between inflammatory responses and genetic variation in TLR4, and we observed tight correlation between responses to S. aureus and to LPS, we first excluded detectable LPS contamination in our experiments. We also considered that activation of TLR4 may occur by bacterial ligands other than LPS [19]. For example, the S. aureus exotoxin leukocidin has been reported to induce TLR4-dependent immune responses [20]. Yet, we did not find that TLR4 was activated by our preparations of S. aureus, at least via the transcription factor NF-κB.
Two other explanations may account for our findings. First, TLR4 may respond to endogenous ligands produced by the host [21]; during infection this effect may be sufficiently strong to result in altered TLR4-dependent inflammatory responses and clinical outcomes. This is supported by a report that hemoglobin (Hb), a host blood product, can significantly enhance macrophage secretion and expression of several cytokines and receptors in a TLR4-dependent pathway when activated with low levels of TLR-2 ligands such as LTA and Pam3CSK4. Hb can also lead to secretion of high mobility group box 1 protein (HMGB1), which synergizes with LTA to increase secretion of IL-6 [22]. Second, TLR4 may contribute to the inflammatory response without direct ligation of the receptor. This seems like the most likely explanation for our observations, as cytokine release in monocytes was not impaired by antibody-mediated blockade of TLR4, yet cytokine release in a monocyte cell line was blunted after transfection of siRNA reducing TLR4 gene expression. Cooperative behavior by TLRs may alter inflammatory responses even when the ligand is specific for only one of the interacting TLRs. For example, we and others have found that TLR5 may alter TLR4-dependent signaling [16, 17]. Furthermore, recent work suggests that TLR4 contributes to murine macrophage responses to S. aureus and that TLR2 and TLR4 may interact in this process [23]. Thus, although we present data that implicates TLR4 as a modulator of the host response to S. aureus, the mechanism by which this may occur requires further elucidation.
Genetic variation in innate immunity is well established as a modulator of inflammatory responses and outcome in sepsis [24–26]. Our focus on Thai subjects offers important new data as population differences in genetic association studies of sepsis are critical to consider. For example, functional TLR1 variants are associated with outcomes from sepsis in white North American subjects [24], but we have shown that TLR1 genetic architecture is profoundly different and variants are nonfunctional in southeast Asians [27]. rs1927907 is an intronic variant in TLR4 that is common throughout the world (MAF 0.11–0.25 [28]). Notably the clinical associations reported to date are restricted to Chinese populations: the variant is associated with late onset Alzheimer’s disease [29], asthma severity [15, 30], and tacrolimus pharmacokinetics after liver transplantation [31], neonatal early Crohn’s disease [32] and with chronic periodontitis in a TLR4 haplotype [33]. In the Southern Han Chinese population the variant is in high LD (r2=0.87) with a TLR4 promoter region variant, rs10983755 [28]. Ragnarsdottir et al have reported that rs10983755 disrupts a potential binding site for the transcription factor N-Myc and increases promoter activity in HEK293 cells upon infection with E. coli [34]. Moreover, rs1927907 is in high LD with rs960312, a TLR4 polymorphism that we have found to be associated with susceptibility to melioidosis, a common Gram-negative infection in northeast Thailand [35]. Thus, the haplotype tagged by rs1927907 is of considerable interest.
Several lines of evidence link rs1927907 to altered TLR4 expression, both at the gene and protein level. In a study of over 5,300 whole blood specimens, rs1927907 was identified as a TLR4 eQTL, with increasing TLR4 gene expression observed in carriers of the minor allele [14]. Moreover, the minor allele of rs1927907 is associated with increased TLR4 protein expression on CD4+CD25high regulatory T cells of asthmatic subjects as determined by flow cytometry [15]. While not excluding trans (more distant) effects of rs1927907 on expression of other genes, these findings complement our findings of increased cytokine responses in carriers of the rs1927907 minor allele and more concretely implicate TLR4 in this process. Further work is required to more completely understand the variation driving the observed associations and to replicate the clinical associations in other populations.
In light of the overlapping but not identical pathways activated by Gram-positive and Gram-negative bacteria, whether and how Gram-positive sepsis differs from Gram-negative sepsis is debated [36, 37]. In comparison to other PAMPs, we found that responses to LPS and to B. pseudomallei were most highly correlated with S. aureus-induced responses. This suggests a shared common pathway of inflammation precipitated by Gram-positive and Gram-negative bacteria. A study by Tang et al showed that the human sepsis transcriptome in Gram-positive sepsis is similar to the transcriptome in Gram-negative sepsis [38]. While this report also supports a common host response in the pathophysiology of sepsis regardless of infecting organism, a potentially confounding factor in clinical or in vivo studies is that LPS derived from gut bacteria may be released systemically during bowel hypoperfusion from sepsis of any etiology [39]. Our ex vivo data, obtained under controlled experimental conditions, avoid confounding by gut bacteria LPS release and provide corroborating protein-level evidence of shared pathway activation.
Several important limitations to our study deserve mention. First, the use of killed bacteria may not mimic the innate immune responses induced by live pathogens. Second, our analysis is restricted to the optimal concentrations of specific PAMPs chosen that induced detectable cytokine responses; conceivably there may be additional dose-dependent effects that have not been identified. Third, our genetic analyses may be confounded by population stratification, and may potentially differ in non-Thai subjects. Fourth, although we reported significant associations of rs1927907 with respiratory failure or plasma cytokines, the number of subjects with the recessive genotype in our clinical study was low and our results may be biased by unmeasured confounding. While the association of rs1927907 with cytokine responses in whole blood stimulation ex vivo was determined using an additive genetic model, the associations in the clinical study were apparent using a recessive genetic model. This may be due to inadequate power to distinguish between the relative contributions of these models in human infection or reflect different biological effects ex vivo versus in infected humans. Independent replication of these results in larger clinical cohorts is therefore important. Fifth, our study was likely underpowered to detect associations with shock or death but we note that IL-6 and IL-8 levels strongly predict these outcomes in our cohort. Finally, the pro-inflammatory cytokines investigated in our study are mainly driven by shared NF-κB/MAP-kinase signaling pathways downstream of the PRRs. We did not investigate interferon production, such as that mediated by interferon regulatory factors (IRFs). It has been recently reported that S. aureus RNA activates IRF5 via TLR8, and that this is antagonized by TLR2 activation [40]. How this sensing pathway relates to our findings necessitates further investigation.
Strengths of our immuno-assay study include the prospective nature of healthy subject recruitment from a blood donor pool, the large number of subjects assayed, highly standardized blood processing techniques, and batched generation of assays, measurement of cytokines, and genotyping. Furthermore, we replicated and extended our initial genetic associations in patients from the same region recruited in a prospective multi-site study of staphylococcal sepsis. To our knowledge, our study is one of the first to identify innate immune genetic variants that predict outcomes from Gram-positive sepsis in Asians.
In conclusion, in Thais a genetic variant in TLR4 that is an eQTL is associated with cytokine responses to S. aureus ex vivo, and with cytokine levels and respiratory failure in staphylococcal sepsis. While S. aureus does not express LPS – the canonical TLR4 ligand – or activate TLR4 directly, the innate immune response to S. aureus does appear to be modulated by TLR4 and shares significant commonality with that induced by Gram-negative pathogens and LPS. These data suggest a regulatory role for TLR4 beyond canonical LPS-receptor ligation in Gram-positive sepsis.
Supplementary Material
Acknowledgments
The authors acknowledge the support of the staff and patients at Sunpasitthiprasong Hospital, Udon Thani Hospital, Khon Kaen Hospital, and Srinagarind Hospital. We thank colleagues at Mahidol-Oxford Tropical Medicine Research Unit and the Department of Microbiology and Immunology, in the Faculty of Tropical Medicine, Mahidol University. We also thank Dr. Direk Limmathurotsakul and Dr. Tanin Hompluem for their administrative support. We are grateful for advice on immuno-assay development from Mark Wurfel and for technical assistance from Nicolle Myers, Deirdre Ducken, Lara Lovelace-Macon, and Shawna Okamoto.
Funding: This study was funded by the Wellcome Trust. NC is supported by a Wellcome Trust Career Development award in Public Health and Tropical Medicine (grant 087769/Z/08/Z). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was also supported by National Institute of Health award R01HL113382 (TEW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions: NC, SJP, and TEW designed the study. NC, ST, SS, CW, GW, PA, PS, NT, YJ, NS, PC, WM, PS, SP, MM, JA, SW, PC, and TEW generated the data. NC, MJE, and TEW analyzed the data. NC, SJP, and TEW wrote the first draft of the manuscript. All authors read and approved the final version. The authors declare no competing financial interests.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Opal SM, Garber GE, LaRosa SP, Maki DG, Freebairn RC, Kinasewitz GT, et al. Systemic host responses in severe sepsis analyzed by causative microorganism and treatment effects of drotrecogin alfa (activated) Clin Infect Dis. 2003;37:50–58. doi: 10.1086/375593. [DOI] [PubMed] [Google Scholar]
- 3.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 4.Fournier B, Philpott DJ. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev. 2005;18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7:167–202. [PubMed] [Google Scholar]
- 6.Opal SM, Cohen J. Clinical gram-positive sepsis: does it fundamentally differ from gram-negative bacterial sepsis? Crit Care Med. 1999;27:1608–1616. doi: 10.1097/00003246-199908000-00039. [DOI] [PubMed] [Google Scholar]
- 7.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 2008;114:347–360. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 8.Chantratita N, Tandhavanant S, Myers ND, Seal S, Arayawichanont A, Kliangsa-Ad A, et al. Survey of innate immune responses to Burkholderia pseudomallei in human blood identifies a central role for lipopolysaccharide. PLoS One. 2013;8:e81617. doi: 10.1371/journal.pone.0081617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chantratita N, Wikraiphat C, Tandhavanant S, Wongsuvan G, Ariyaprasert P, Suntornsut P, et al. Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin Microbiol Infect. 2016 doi: 10.1016/j.cmi.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West TE, Hawn TR, Skerrett SJ. Toll-like receptor signaling in airborne Burkholderia thailandensis infection. Infect Immun. 2009;77:5612–5622. doi: 10.1128/IAI.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armitage P, Berry G, Matthews JNS. Statistical Methods in Medical Research. 4th. Wiley-Blackwell; 2001. [Google Scholar]
- 14.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Xu AG, Zhao W, Xu QF, Zhao YM, Li DD, et al. A toll-like receptor 4 (TLR4) variant is associated with asthma severity. Int J Clin Exp Med. 2015;8:7849–7854. [PMC free article] [PubMed] [Google Scholar]
- 16.Amy KD, West TE. American Thoracic Society International Conference Abstracts: C59. New insights into the immunology of bacterial pneumonia: American Thoracic Society; 2013. Toll Like Receptor (TLR) 5 1174C>T Modulates The TLR 4-Dependent Innate Immune Response To Burkholderia pseudomallei ; p. A4556. [Google Scholar]
- 17.Collin GJ, Joseph S, Annette R, Jim A, Jaime C, Katarzyna B, et al. American Thoracic Society International Conference Abstracts: A101. Host defense, lung injury, and fibrosis: Innate mechanisms: American Thoracic Society; 2016. Toll-Like Receptor 5 Modulates MyD88-Dependent Toll-Like Receptor 4 Signaling; p. A2643. [Google Scholar]
- 18.Beutler B. TLR4 as the mammalian endotoxin sensor. Curr Top Microbiol Immunol. 2002;270:109–120. doi: 10.1007/978-3-642-59430-4_7. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 20.Inden K, Kaneko J, Miyazato A, Yamamoto N, Mouri S, Shibuya Y, et al. Toll-like receptor 4-dependent activation of myeloid dendritic cells by leukocidin of Staphylococcus aureus . Microbes Infect. 2009;11:245–253. doi: 10.1016/j.micinf.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. J Leukoc Biol. 2004;76:514–519. doi: 10.1189/jlb.0304127. [DOI] [PubMed] [Google Scholar]
- 22.Cox KH, Cox ME, Woo-Rasberry V, Hasty DL. Pathways involved in the synergistic activation of macrophages by lipoteichoic acid and hemoglobin. PLoS One. 2012;7:e47333. doi: 10.1371/journal.pone.0047333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Fu Y, Feng S, Zhang X, Liu Z, Cao Y, et al. Involvement of RP105 and Toll-Like Receptors in the Activation of Mouse Peritoneal Macrophages by Staphylococcus aureus . Scand J Immunol. 2013;78:8–16. doi: 10.1111/sji.12050. [DOI] [PubMed] [Google Scholar]
- 24.Wurfel MM. Genetic insights into sepsis: what have we learned and how will it help? Curr Pharm Des. 2008;14:1900–1911. doi: 10.2174/138161208784980554. [DOI] [PubMed] [Google Scholar]
- 25.Nakada TA, Russell JA, Boyd JH, Walley KR. IL17A genetic variation is associated with altered susceptibility to Gram-positive infection and mortality of severe sepsis. Crit Care. 2011;15:R254. doi: 10.1186/cc10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer NJ, Ferguson JF, Feng R, Wang F, Patel PN, Li M, et al. A functional synonymous coding variant in the IL1RN gene is associated with survival in septic shock. Am J Respir Crit Care Med. 2014;190:656–664. doi: 10.1164/rccm.201403-0586OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chantratita N, Tandhavanant S, Myers ND, Chierakul W, Wuthiekanun V, Mahavanakul W, et al. Common TLR1 genetic variation is not associated with death from melioidosis, a common cause of sepsis in rural Thailand. PLoS One. 2014;9:e83285. doi: 10.1371/journal.pone.0083285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YC, Yip PK, Huang YL, Sun Y, Wen LL, Chu YM, et al. Sequence variants of toll like receptor 4 and late-onset Alzheimer’s disease. PLoS One. 2012;7:e50771. doi: 10.1371/journal.pone.0050771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Qian FH, Zhou LF, Wei GZ, Jin GF, Bai JL, et al. Polymorphisms in toll-like receptor 4 gene are associated with asthma severity but not susceptibility in a Chinese Han population. J Investig Allergol Clin Immunol. 2011;21:370–377. [PubMed] [Google Scholar]
- 31.Wang Z, Wu S, Chen D, Guo F, Zhong L, Fan J, et al. Influence of TLR4 rs1927907 locus polymorphisms on tacrolimus pharmacokinetics in the early stage after liver transplantation. Eur J Clin Pharmacol. 2014;70:925–931. doi: 10.1007/s00228-014-1673-2. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L, Liu Y, Lu Y, Zhang W. Correlation between TLR4 genetic polymorphisms and susceptibility to neonatal early Crohn disease. Int J Clin Exp Pathol. 2016;9:2345–2349. [Google Scholar]
- 33.Ding YS, Zhao Y, Xiao YY, Zhao G. Toll-like receptor 4 gene polymorphism is associated with chronic periodontitis. Int J Clin Exp Med. 2015;8:6186–6192. [PMC free article] [PubMed] [Google Scholar]
- 34.Ragnarsdottir B, Jonsson K, Urbano A, Gronberg-Hernandez J, Lutay N, Tammi M, et al. Toll-like receptor 4 promoter polymorphisms: common TLR4 variants may protect against severe urinary tract infection. PLoS One. 2010;5:e10734. doi: 10.1371/journal.pone.0010734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V, Emond MJ, et al. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun. 2012;13:38–46. doi: 10.1038/gene.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opal SM, Patrozou E. Translational research in the development of novel sepsis therapeutics: logical deductive reasoning or mission impossible? Crit Care Med. 2009;37:S10–S15. doi: 10.1097/CCM.0b013e3181921497. [DOI] [PubMed] [Google Scholar]
- 37.Wang JE, Dahle MK, McDonald M, Foster SJ, Aasen AO, Thiemermann C. Peptidoglycan and lipoteichoic acid in gram-positive bacterial sepsis: receptors, signal transduction, biological effects, and synergism. Shock. 2003;20:402–414. doi: 10.1097/01.shk.0000092268.01859.0d. [DOI] [PubMed] [Google Scholar]
- 38.Tang BM, McLean AS, Dawes IW, Huang SJ, Cowley MJ, Lin RC. Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med. 2008;36:1125–1128. doi: 10.1097/CCM.0b013e3181692c0b. [DOI] [PubMed] [Google Scholar]
- 39.Marshall JC, Foster D, Vincent JL, Cook DJ, Cohen J, Dellinger RP, et al. Diagnostic and prognostic implications of endotoxemia in critical illness: results of the MEDIC study. J Infect Dis. 2004;190:527–534. doi: 10.1086/422254. [DOI] [PubMed] [Google Scholar]
- 40.Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, et al. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-beta Production via a TAK1-IKKbeta-IRF5 Signaling Pathway. J Immunol. 2015;195:1100–1111. doi: 10.4049/jimmunol.1403176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.