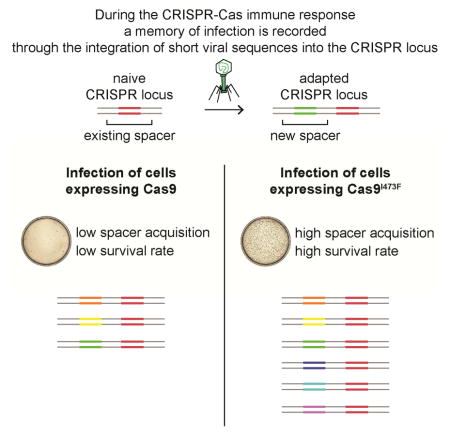
SUMMARY
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and their associated (Cas) proteins encode a prokaryotic immune system that protects against viruses and plasmids. Upon infection, a low fraction of cells acquire short DNA sequences from the invader. These sequences (spacers) are integrated in between the repeats of the CRISPR locus and immunize the host against the matching invader. Spacers specify the targets of the CRISPR immune response through transcription into short RNA guides that direct Cas nucleases to the inavding DNA molecules. Here we performed random mutagenesis of the RNA-guided Cas9 nuclease to look for variants that provide enhanced immunity against viral infection. We identified a mutation, I473F, which increases the rate of spacer acquisition by more than two orders of magnitude. Our results highlight the role of Cas9 during CRISPR immunization and provide a useful tool to study this rare process and develop it as a biotechnological application.
eTOC BLURB
Heler et al. screened a library of cas9 mutants for mutations that enhanced the CRISPR immune response. The I473F substitution increases CRISPR immunity by more than two orders of magnitude. This is due to an increase in the rate of the acquisition of viral sequences as new CRISPR spacers.
INTRODUCTION
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and their associated (Cas) proteins protect bacteria and archaea against their viruses (Barrangou et al., 2007) and plasmids (Marraffini and Sontheimer, 2008). In the first step of the CRISPR immune response, a very low proportion of the infected cells acquire a short sequence, known as a spacer sequence, of the invading genome in between the repeats of the CRISPR array (Barrangou et al., 2007). Spacer acquisition is catalyzed by the Cas1/Cas2 integration complex (Nunez et al., 2014; Nunez et al., 2015; Yosef et al., 2012) and results in the immunization of the host (Barrangou et al., 2007). In the second step of the CRISPR immune response, spacer sequences are transcribed and processed into a small RNA known as the CRISPR RNA (crRNA) (Brouns et al., 2008; Carte et al., 2008; Deltcheva et al., 2011). The crRNA is used as a guide by Cas nucleases to find its complementary sequence, known as the protospacer, in the invading viral or plasmid genome (Gasiunas et al., 2012; Jinek et al., 2012; Jore et al., 2011; Samai et al., 2015). Target recognition through base-pairing between the crRNA and the target DNA results in the destruction of the invader and host immunity (Garneau et al., 2010).
Based on their cas genetic repertoire, CRISPR-Cas systems have been classified into six types, I through VI (Makarova et al., 2015; Shmakov et al., 2015). Cas9 is the crRNA-guided nuclease of the type II-A CRISPR-Cas system of Streptococcus pyogenes (Jinek et al., 2012). In addition to protospacer recognition by the crRNA, Cas9 target cleavage requires a 5′-NGG-3′ protospacer adjacent motif (PAM) immediately downstream of the target (Anders et al., 2014; Deveau et al., 2008; Jiang et al., 2013; Jinek et al., 2012). Cas9 is also required for the immunization step of the CRISPR response (Heler et al., 2015; Wei et al., 2015), using its PAM binding domain to specify functional spacer sequences that are flanked by the required NGG motif (Heler et al., 2015). In support of its role in spacer acquisition, Cas9 can associate in vivo with the other proteins encoded by the type II-A CRISPR-Cas system: Cas1, Cas2 and Csn2 (Heler et al., 2015).
To further study the role of Cas9 in spacer acquisition, we decided to change its PAM specificity. Earlier work from our lab revealed a weak but detectable activity of Cas9 for targets flanked by NAG PAMs (Jiang et al., 2013). We decided to increase this activity to understand how Cas9 affects the acquisition of spacers flanked by non-NGG motifs. After structural analysis that identified the PAM interacting domain of Cas9 (Anders et al., 2014; Jinek et al., 2014), other researchers specifically mutated this domain to obtain an NAG-recognizing Cas9 (Kleinstiver et al., 2015). We took a different approach and searched for mutations in any region of the nuclease that would increase its specificity for NAG-flanked targets. We found one such mutation, I473F, which provided partial immunity against NAG-flnked viral targets. Importantly, we found that this mutation also enhanced the CRISPR-Cas adaptive immune response by more than two orders of magnitude through a mechanism that increases the rate of spacer acquisition but not of Cas9 target cleavage. Our results highlight the role of Cas9 during CRISPR immunization and provide a useful tool to study this otherwise rare process.
RESULTS
Directed evolution of Cas9 yields a mutant with altered PAM specificity and enhanced CRISPR-Cas immunity
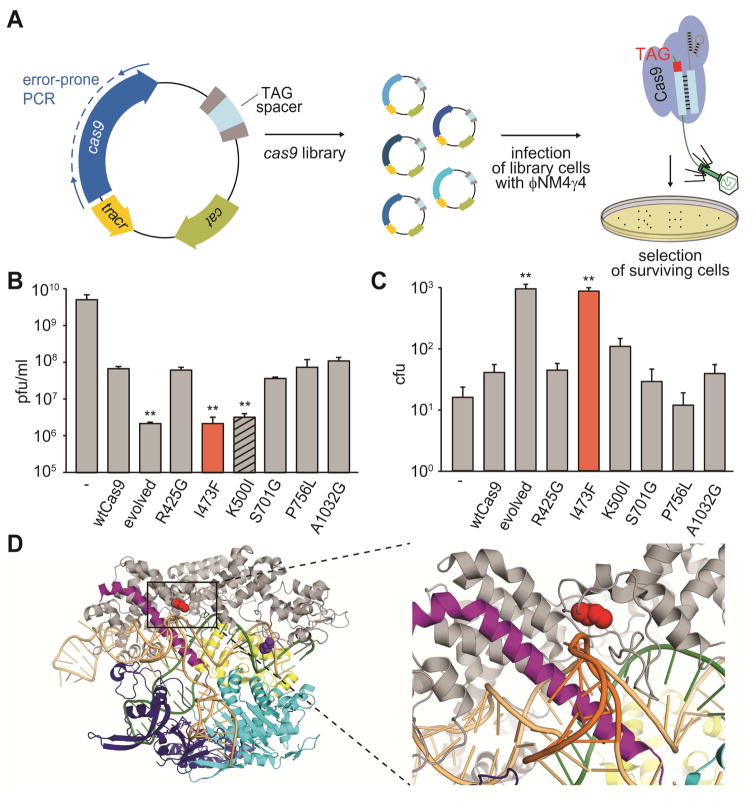
S. pyogenes Cas9 has an innate ability to cleave NAG-adjacent targets, but with much lower efficiency than it cleaves canonical (NGG) targets (Jiang et al., 2013). To improve its specificity for NAG PAMs, we constructed a library of plasmids carrying cas9 variants generated by error-prone PCR (Fig. 1A). The library plasmids also harbor the trans-activating crRNA (tracrRNA) gene (Deltcheva et al., 2011) and a single-spacer CRISPR array targeting a TAG-adjacent protospacer on the genome of the lytic staphylococcal bacteriophage ϕNM4γ4 (Goldberg et al., 2014). The library was transformed into Staphylococcus aureus RN4220 cells that were subjected to two rounds of phage infection on soft-agar plates to select for phage-resistant bacterial colonies. Several colonies were obtained and we proceeded with a more extensive analysis of one of the “evolved” mutants that gained phage resistance. Sequencing of the plasmid revealed the presence of six single-nucleotide substitutions in the cas9 gene (see Extended Experimental Procedures) producing the following missense mutations: R425G, I473F, K500I, S701G, P756L and A1032G. To evaluate the importance of each of these mutations in the gain-of-function phenotype we introduced them individually into the cas9 gene and tested the ability of the resulting plasmid to prevent ϕNM4γ4 propagation by measuring the number of plaque forming units (pfu) that result after infection of the host cells (Fig. 1B). Cas9 harboring the R425G, S701G, P756L and A1032G mutations allow wild-type levels of phage propagation and therefore do not contribute to the gain-of-function-phenotype of the evolved cas9 allele we isolated. In contrast, cells containing Cas9 with the I473F or K500I mutations decrease phage propagation by about four orders of magnitude. This is close to the levels of immunity provided by wild-type Cas9 when programmed against NGG-flanked targets (a reduction of ~ 5 orders of magnitude, see Fig. 3B). Similar results were obtained when other NAG PAMs were tested (AAG, CAG, GAG, Figs. S1A–B). Therefore the I473F and K500I mutations enhance the ability of Cas9 to recognize targets with NAG PAMs. The pfu count was similar in all mutant and control strains when infected with ϕ85, a lytic phage that lacks the target sequence (Fig. S1C), corroborating that the decrease in phage propagation observed for the I473F and K500I mutations is a direct consequence of Cas9 targeting and not due to cell toxicity induced by the various mutants.
Figure 1. Directed evolution of cas9 generates mutants with increased specificity for NAG targets.
See also Figure S1.
(A) Schematic diagram of the directed evolution assay. S. pyogenes cas9 was mutagenized by error-prone PCR and library amplicons were cloned into a plasmid carrying a spacer matching a TAG-adjacent target sequence on the ϕNM4γ4 phage. Library cells were infected with lytic phage to screen for mutants displaying improved NAG cleaving efficiency.
(B) Phage propagation was measured as the number of plaque forming units (pfu) per ml of stock, on cells targeting the NAG-adjacent protospacer and harboring plasmids with different mutations on cas9: one of the “evolved” alleles or each of the six mutations present in this allele. Mutations with pfu values significantly different than wild type are highlighted (**, p-value < 0.05 compared to wtCas9).
(C) Colony forming units (cfu) obtained after phage infection of naïve cells (not programmed to target any viral sequence) harboring plasmids with different mutations in cas9. Mutations with cfu values significantly different than wild type are highlighted.
(D) Location of residues I473 and K500 on the Cas9:single-guide RNA ribonucleoprotein (PDB 4UN3). Red, I473; purple, K500; orange, sgRNA; green, target DNA (the GG PAM highlighted in red); grey, alpha-helical (REC) lobe; yellow, HNH domain; light blue, RuvC domain; blue, PAM-interacting CTD.
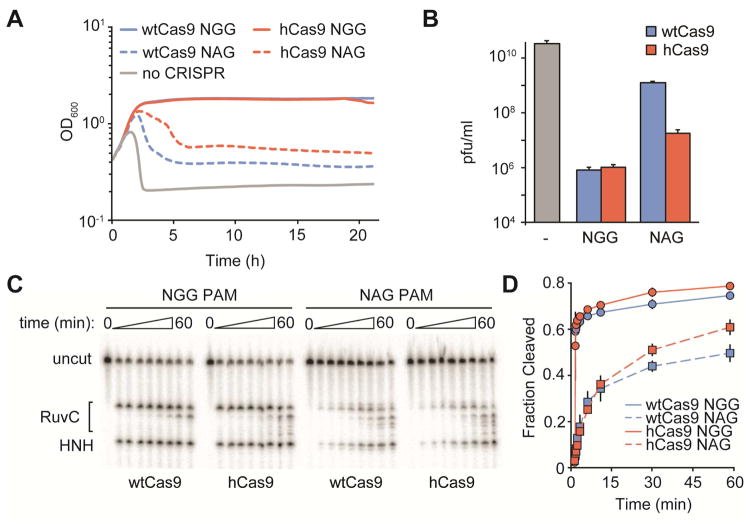
Figure 3. hCas9 has increased interference efficiency against NAG- but not NGG-adjacent targets.
See also Figure S3.
(A) Growth curves of cultures infected with ϕNM4γ4 harboring the wtCas9 or hCas9 (but not Cas1, Cas2 and Csn2) programmed to target either NAG- or NGG-flanked viral sequences.
(B) Phage propagation, measured in pfu/ml, of the bacteria presented in A.
(C) Cleavage of radiolabeled dsDNA targets flanked by either NGG or NAG PAMs, by wtCas9 or hCas9.
(D) Quantification of the cleavage results shown in C. Data are represented as mean ± SD of 3 representative biological replicates.
Given the requirement of Cas9 for the immunization phase of the CRISPR-Cas immune response, i.e. the acquisition of virus-derived spacer sequences (Heler et al., 2015; Wei et al., 2015), we wondered whether the evolved Cas9 as well as the individual mutants affected this process. To test this, we introduced the different alleles of cas9 into a plasmid harboring the tracrRNA gene, the S. pyogenes SF370 CRISPR array (containing six spacers, none of them matching the genome of ϕNM4γ4) and the type II-A genes exclusively involved in the acquisition of new spacers, cas1, cas2 and csn2 (Heler et al., 2015; Wei et al., 2015). S. aureus cells containing the different plasmids were infected with ϕNM4γ4 and the number of survivors were enumerated as colony forming units (cfu) (Fig. 1C). Only a small fraction of cells containing wild-type Cas9 are able to acquire new spacers, about 2-fold over a CRISPR-less control. In contrast, the evolved cas9 allele containing all six mutations increased the number of CRISPR-surviving cells by about 60-fold. Analysis of single mutants revealed that this highly significant increase was provided almost exclusively by the I473F mutation (Fig. 1C). Due to the sharp enhancement of the CRISPR-Cas immune response conferred by the I473F mutation we decided to name the Cas9I437F mutant “hyper-Cas9”, or hCas9. I473 is located close to the surface of Cas9, outside of the PAM-interacting domain, and it is part of a projection from the Helical III domain that interacts with the nexus of the guide RNA (Jiang et al., 2016) (Fig. 1D). This position does not suggest an evident effect of the I473F mutation on Cas9 activity and therefore we decided to investigate the basis for its phenotype by performing a detailed comparison with the CRISPR-Cas immune response mediated by wild-type Cas9.
hCas9 enhances the CRISPR-Cas adaptive immune response by two orders of magnitude
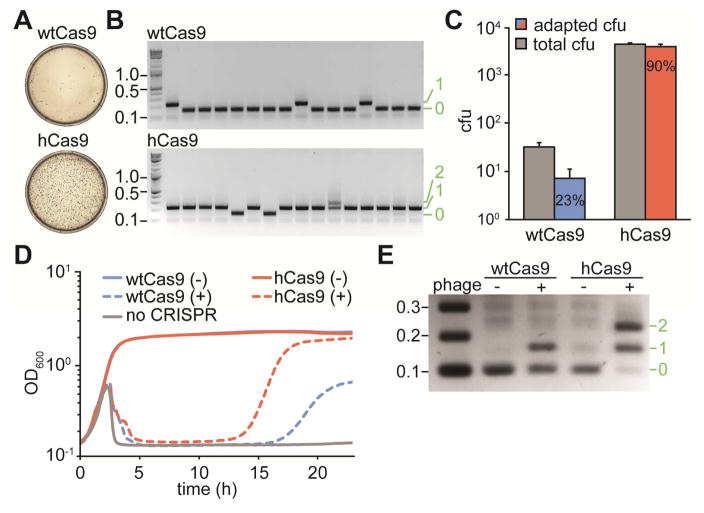
To perform a more accurate comparison between wild-type (wtCas9) and hCas9, we counted the number of CRISPR-mediated, phage resistant cells that arise after phage infection. Figure 2A shows representative plates of infected cells containing plasmids with the wtCas9 or hCas9 S. pyogenes CRISPR-Cas locus, showing a striking difference in the number of surviving colonies. Most of these colonies arise from single cells that were able to acquire a new spacer matching the ϕNM4γ4 genome. However, a fraction of the surviving cells repel phage attack by non-CRISPR related mechanisms, such as envelope resistance (Heler et al., 2015). To make a more accurate quantification of the CRISPR-Cas response, we analyzed individual colonies by PCR of the CRISPR array (Heler et al., 2015; Yosef et al., 2012) to detect those in which new spacers were acquired, i.e. “adapted” cells (Fig. 2B). Not only did many more resistant colonies originated from cells harboring hCas9 (an average of 31 cfu for wtCas9 vs 4,312 cfu for hCas9, Fig. 2C), but also most of them showed CRISPR-mediated phage resistance (23% for wtCas9 vs 90% for hCas9, Fig. 2C). We wondered whether this was a consequence of the specific substitution of I473 by phenylalanine. To test this we introduced an I473A mutation into Cas9 (Fig. S2). We found that cells harboring the I473A mutant produced a number of CRISPR-mediated immune cfu comparable to cells carrying wtCas9, but 10 times lower than the cfu obtained from infection of cells expressing hCas9. Therefore we conclude that the I473F mutation increases the CRISPR-adaptive immune response through a specific effect of the phenylalanine residue in position 473 and by more than two orders of magnitude: on average, approximately 7 cfu (31×0.23) per experiment for infected wtCas9-containing cells, and approximately 3,863 cfu (4,312×0.90) for infected hCas9-expressing bacteria. We sequenced PCR products to determine the PAM of the spacers acquired by 40 colonies expressing wtCas9 (Table S1) or hCas9 (Table S2). Interestingly, all 40 spacers acquired by cells expressing hCas9 matched targets with an NGG PAM, suggesting that this nuclease can still target sequences followed by the canonical PAM in addition to targets with NAG PAMs.
Figure 2. Cas9I473F, or hyper-Cas9 (hCas9) mounts an enhanced CRISPR adaptive immune response.
See also Figure S2.
(A) Representative plates obtained after lytic infection of cells harboring the full CRISPR system of S. pyogenes with wtCas9 or hCas9, showing the number of surviving colonies.
(B) Agarose gel electrophoresis of PCR products of the amplification of the CRISPR of arrays of surviving cells to detect newly acquired spacers (asterisks). Molecular markers (in kb) are indicated in black, number of new spacers added in green.
(C) Quantification of total surviving colonies (gray bars) and surviving colonies with newly incorporated spacers, as detected by PCR (blue and red bars). Data are represented as mean ± SEM of 3 representative biological replicates.
(D) Growth curves of cultures of cells harboring the full CRISPR system of S. pyogenes with wtCas9 or hCas9, with (+) or without (−) phage infection.
(E) PCR-based analysis of the liquid cultures shown in C (at 24 hours post-infection) to check for the acquisition of new spacer sequences in the presence (+) or the absence (−) of phage ϕNM4γ4 infection, by cells expressing wtCas9 or hCas9. Molecular markers (in kb) are indicated in black, number of new spacers added in green. Image is representative of three technical replicates.
Similar results were observed when cells in culture carrying naïve wtCas9 or hCas9 CRISPR-Cas systems were infected with phage. Upon addition of ϕNM4γ4, the cultures lyse, as the vast majority of cells do not undergo spacer acquisition (Fig. 2D). Nonetheless, hCas9 cultures were able to regrow much earlier (~14 hours post-infection) than wtCas9 cultures (~17 hours post-infection). PCR analysis using DNA extracted from the whole culture at 24 hours post-infection corroborated the earlier observation that hCas9 cells mount a more robust CRISPR immune response (Fig. 2E). Whereas the PCR products derived from wtCas9 staphylococci showed the presence of both adapted and non-adapted CRISPR arrays in the surviving population, the PCR results from cultures carrying hCas9 showed very little non-adapted CRISPR arrays, with the great majority of the cells acquiring one or two new spacers. Altogether these data show that the I473F mutation in Cas9 allows for a more robust CRISPR-Cas immune response due to a specific effect of the phenylalanine residue.
hCas9 displays a modest increase in the cleavage efficiency of targets with NAG PAMs
Next, we investigated if the enhanced immunity phenotype of hCas9 documented in Figure 2 was due to an increase in the frequency of spacer acquisition, a more robust cleavage by hCas9 of its targets, or both. First we considered the possibility that hCas9 could provide better cleavage of the infecting viral DNA. In this scenario both wtCas9 and hCas9 populations can acquire a similar number of new spacers but a more robust cleavage of the target DNA by hCas9 would lead to a faster recovery of the bacteria that acquired the spacers. To test this hypothesis, we infected cells carrying plasmids with either wtCas9 or hCas9 programmed to target the ϕNM4γ4 virus and the tracrRNA gene, but without the spacer acquisition machinery (cas1, cas2 and csn2). This genetic background supports CRISPR-Cas anti-viral defense but does not allow the acquisition of new spacer sequences (Heler et al., 2015). Because our data suggested that hCas9 can still target protospacers followed by NGG PAMs, we tested the immunity of cells programmed to attack targets with either an NAG or an NGG PAM located in the same region of the ϕNM4γ4 genome (Fig. S3A). Bacteria containing different plasmids were infected with phage during exponential growth and the optical density of the culture was followed over time to measure the immunity provided by Cas9 cleavage of the viral genome (Fig 3A). As expected, cells harboring a vector control were rapidly lysed by the addition of phage. On the other hand, cells expressing wtCas9 or hCas9 programmed against an NGG target cleared the infection efficiently and continued the exponential growth, indicating that the I473F mutation does not affect the recognition and targeting of NGG-flanked sequences. In contrast, both cultures display poor survival when NAG-flanked protospacers were targeted by either Cas9 version, with cells expressing wtCas9 suffering a more substantial lysis than cells expressing hCas9. Similar results were obtained when we tested the same cultures for their ability to limit phage propagation (pfu/ml) (Fig. 3B).
Both in vivo experiments measuring bacterial survival (Fig. 3A) and phage propagation (Fig. 3B) suggest that hCas9 has not improved efficiency of cleavage of NGG-flanked targets, and displays only a small increase in the cleavage of NAG-flanked sequences. To unequivocally demonstrate this, we performed in vitro cleavage assays with purified wtCas9 and hCas9 (Fig. 3C). In this case, we were able to compare cleavage of radiolabeled oligonucleotides containing the same protospacer sequence followed by either a TGG or TAG PAM (Fig. S3B). Consistent with in vivo data, experiments showed similar cutting rates of the NGG target for wtCas9 and hCas9. Quantification of the cleavage products showed that hCas9 cleaved more of the NAG target than wtCas9 over longer timescales (Fig. 3D). Altogether, the data presented in Figure 3 indicate that while there is a modest increase in the NAG-targeting properties of hCas9, this cannot explain the rise in the number of CRISPR-resistant colonies mediated by the I473F mutation (Fig. 2C).
hCas9 promotes higher rates of spacer acquisition
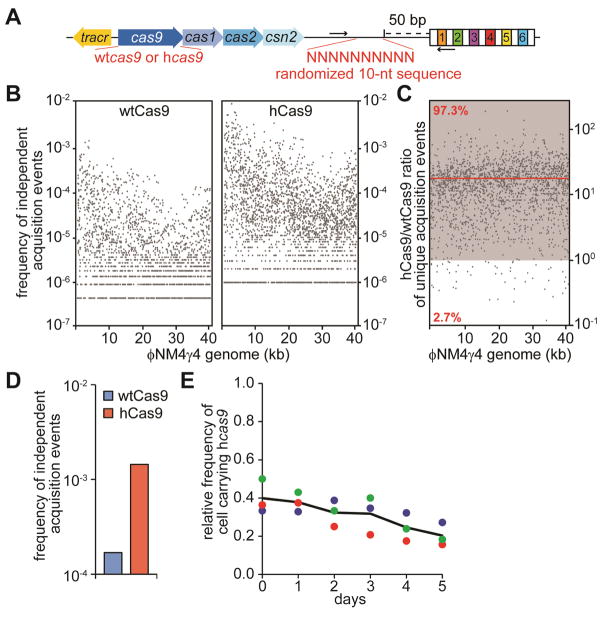
A second hypothesis that could explain the increase in CRISPR-Cas immunity conferred by hCas9 is an increase in the frequency of spacer acquisition by the cells expressing this mutant. To test this we performed a comparison of the spacer repertoires acquired by cells harboring wtCas9 or hCas9. We made two plasmid libraries, carrying the spacer acquisition genes cas1, cas2 and csn2 and wtcas9 or hcas9, the tracrRNA gene and the S. pyogenes CRISPR array preceded by a “barcode” sequence of 10 nucleotides 50 bp immediately upstream of the CRISPR array (Fig. 4A). Cells harboring each library were infected with phage ϕNM4γ4 and DNA from the survinving cells was used to amplify the CRISPR array via PCR and collect sequence information of all the new acquired spacers using next generation sequencing. The primers used also amplify the barcode sequence (Fig. 4A) and therefore each new spacer sequence can be associated with a unique barcode, allowing us to count how many times a given spacer was independently acquired in each bacterial population. Over three million reads belonging to either library were analyzed. The frequency of reads corresponding to each acquired spacer sequence was plotted according to its position in the ϕNM4γ4 genome (Fig. S4A). Analysis of the PAMs of the acquired spacers showed that over 99.5% of the spacers matched NGG targets in both libraries (Fig. S4B), corroborating our in vivo data showing that hCas9 retained NGG PAM specificity. In addition, we looked at the repertoire of unique different spacers independently of the number of reads per sequence (Fig. S4C). Consistent with our previous finding that the PAM specificity of Cas9 is responsible for the PAM sequence of the new protospacers, the hCas9 library showed a 5-fold increase in the acquisition of spacers matching NAG-flanked targets. Even with this increase these spacers represent less than 0.05% of the total acquisition events, most likely due to the fitness cost associated with the low efficiency of NAG-target cleavage observed for hCas9 when compared with its cleavage of NGG targets. We also observed an increase in the total number of different spacer sequences, from 1980 for wtCas9 cells to 2500 for the hCas9 sample. All together, these findings show that hCas9 provides the host bacterium with highly efficient spacer acquisition, thus enhancing CRISPR-Cas immunity.
Figure 4. hCas9 promotes higher rates of spacer acquisition.
See also Figure S4.
(A) Schematic diagram of the S. pyogenes CRISPR locus showing the barcode and primers (arrows) used to measure the number of independent spacer acquisition events.
(B) Cultures expressing wtCas9 or hCas9 were infected with ϕNM4γ4 phage, surviving cells were collected after 24 hours, DNA extracted and used as template for PCR of the CRISPR arrays. Amplification products were separated by agarose gel electrophoresis (not shown) and the DNA of the expanded CRISPR array was subject to MiSeq next-generation sequencing. The number of barcodes for each spacer sequence across the phage genome, normalized by the total number of spacer reads obtained, was plotted.
(C) The hCas9/wtCas9 frequency of independent acquisition events ratio for 1938 common spacer sequences was plotted across the phage genome. The zone where the ratio is greater than one is shown in grey. The red line shows the average ratio.
(D) Same as (B) but without phage infection; i.e. a measure of acquisition of spacers derived form the host chromosome and resident plasmids.
(E) Pair-wise competition between staphylococci expressing wtCas9 or hCas9. The change in the relative frequency of cells carrying the hcas9 allele (y-axis) is plotted against the number of culture transfers (one transfer per day, x-axis).
To calculate the frequency of acquisition of every spacer we divided the number of different barcodes for a given spacer sequence by the total number of reads. This value was plotted according to its position in the ϕNM4γ4 genome (Fig. 4B). The data show a drastic increase in the frequency of acquisition in hCas9 cells. For all 1938 newly acquired spacer sequences shared between the two libraries, we calculated the ratio of unique adaptation events (i.e. number of different barcodes) for hCas9 reads compared to wtCas9 (Fig. 4C). We found that more than 97% of the spacers were acquired more frequently in the hCas9 library (ratio > 1), with an average ratio of ~18. This experiment indicates that hCas9 enhances the rate of spacer acquisition during the CRISPR adaptation phase. To rule out any effect that the phage selection imposed on adapted cells could have on our experiments we looked at the rates of spacer acquisition in the absence of phage infection. Using our barcoded system, we passaged cells expressing wtCas9 or hCas9 for 10 days, and subjected a PCR product containing the CRISPR locus of each culture to next generation sequencing. The frequency of acquisition was one order of magnitude higher for hCas9-expressing cells (Fig. 4D). Altogether, these findings show that hCas9 provides the host bacterium with more efficient spacer acquisition, and suggest that this is a major contributor to the enhanced CRISPR-Cas immunity granted by hCas9.
Higher levels of immunization during the CRISPR-Cas response to phage infection provides better host defense. However, this could also lead to detrimental effects in the absence of infection, leading to high levels of CRISPR “autoimmunity”. Consistent with this scenario, the I473F mutation was not found in type II-A cas9 gene variants (Fig. S4D). To explore the possible detrimental effects of hCas9 we looked at the rates of plasmid loss in the absence of phage infection since in our barcoded experiment without viral infection, as well in other similar experiments (Heler et al., 2015; Levy et al., 2015; Yosef et al., 2012), most of the acquired self-spacers match plasmid sequences. To test this we plated cells after 10 days of growth with and without chloramphenicol to calculate the frequency of plasmid loss as the number of chloramphenicol-resistant cfu relative to the total cfu count (Fig. S4E). Whereas most staphylococci expressing wtCas9 maintained the plasmid, about 30% of the cells producing hCas9 lost it. This decrease was dependent on the presence of an active Cas1-Cas2 spacer integrase, demonstrating that plasmid loss was caused by CRISPR autoimmunity. Higher autoimmunity in hCas9-expressing cells resulted in a fitness cost, as shown by pairwise competition assays in which wtCas9- and hCas9-expressing cells were grown together and the relative proportion of each strain was measured over time (staphylococci harboring the hcas9, but not the wtcas9, plasmid also carried an erythromycin-resistance gene in their chromosome). We detected a decrease in the proportion of erythromycin-resistant cfu over time (Fig. 4E), demonstrating that a “hyper-acquiring” type II CRISPR-Cas system confers a fitness cost to the cells that carry it.
DISCUSSION
Here we performed random mutagenesis on the entire cas9 gene and found a mutant with an expanded CRISPR-Cas response. This “hyper” Cas9 version (hCas9) harbors the mutation I473F. Compared to wild-type staphylococci, cells harboring hcas9 displays a modest increase in NAG-target recognition but a substantial increase (more than two orders of magnitude) in the frequency of spacer acquisition. The molecular mechanism by which the I473F mutation enables this increase in spacer acquisition is not clear. We previously reported the existence of a complex between the four Cas proteins encoded by the type II-A CRISPR locus, namely Cas9, Cas1, Cas2 and Csn2 (Heler et al., 2015). We hypothesized that this complex functions in spacer acquisition, with Cas9 selecting sequences flanked by NGG PAMs (Heler et al., 2015) and Cas1 and Cas2 (Nunez et al., 2014; Nunez et al., 2015) being involved in the integration of these sequences into the CRISPR array. Given its location on the surface of hCas9, F473 could interact with other Cas proteins and increase the abundance or the stability of the complex, thus enhancing the rate of spacer acquisition. To test this we incubated the four proteins along with a single-guide RNA (Jinek et al., 2012) and subjected them to gel filtration to detect the formation of the complex. However, we did not observe significant amounts of stable complexes neither in the presence of wtCas9 nor hCas9. In wtCas9, the isoleucine residue is in direct contact with bases of the tracrRNA (Fig. 1D) that are equivalent to the nexus in the single-guide RNA (Briner et al., 2014). Specifically, nucleotide U59 of the tracrRNA inserts into a hydrophobic pocket lined by I473 and its adjacent residues (Jiang et al., 2016). It is possible that the bulkier phenylalanine residue could interfere with the tracrRNA:Cas9 association, affecting the involvement of Cas9 in the immunization step of the CRISPR-Cas response. This hypothesis is supported by the wild-type phenotype of the I473A mutation (Fig. S2), since the smaller alanine residue most likely will not interfere with the tracrRNA interaction. Another mutation in a residue close to I473, K500I, also seems to affect Cas9 target specificity, but not the rate of spacer acquisition. K500 is located in the minor groove of the PAM-distal crRNA-target DNA duplex (Fig. S1D), near the backbone of nucleotide 12 of the DNA protospacer and nucleotide 3 of the crRNA (Anders et al., 2014). The loss of a basic residue in this region might alter target binding and recognition, analogous to the increase in specificity resulting from mutations of other residues making nonspecific DNA contacts (Kleinstiver et al., 2016). Future work will explore the importance of this region in Cas9 activity during the different phases of CRISPR-Cas immunity.
In spite of the enhanced immune response provided by the I473F substitution, we could not find cas9 genes harboring this mutation in the genome of bacteria sequenced so far. Two studies have shown that Cas9 is required for the acquisition of self-targeting spacers (Heler et al., 2015; Wei et al., 2015), a situation that leads to “auto-immunity” and to the death of the host (Bikard et al., 2014; Jiang et al., 2013). Here we show that the enhanced rate of spacer acquisition of hCas9 results in an increase in the autoimmunity events and therefore leads to a fitness cost for the host cell. We believe that this prevents the evolution of the I473F mutation into Cas9.
The phenotype of the I473F mutation in Cas9 further demonstrates the involvement of this nuclease in the acquisition of new spacers in type II CRISPR-Cas systems and provides a new tool that could facilitate the study of CRISPR immunization, making this process more frequent and easier to detect. In addition, hCas9 provides a useful tool for the development of technologies that use the incorporation of spacers to develop synthetic biology devices that can record different cellular events (Shipman et al., 2016). Currently, the low adaptation frequency limits the number of stimuli that can be captured as new spacers in the CRISPR array. Using an enhanced CRISPR adaptation machinery such as hCas9 could boost the spacer acquisition frequency and thus facilitate the development of this and other related synthetic biology technologies.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Cultivation of S. aureus RN4220 (Kreiswirth et al., 1983) was carried out in heart infusion broth (BHI) at 37 °C. Whenever applicable, media were supplemented with chloramphenicol at 10 μg ml−1 to ensure pC194-derived plasmid maintenance or 5mM CaCl2 for phage adsorption.
Directed evolution of cas9
The cas9 gene was mutagenized at a low rate of 0–4.5 mutations/kb by error prone PCR using GeneMorph II Random Mutagenesis Kit. The mutant cas9 amplicons were cloned into a backbone plasmid containing a spacer matching a TAG-adjacent target on ϕNM4γ4. The library was subjected to soft-agar lytic phage infection and surviving colonies were re-streaked on fresh plates. The TAG-cleaving efficiency of surviving colonies was individually assessed by phage propagation assays.
High-throughput sequencing
Plasmid DNA was extracted from adapted cultures using the in-liquid spacer acquisition assay described in Experimental Procedures. 200 ng of plasmid DNA was used as template for Phusion PCR to amplify the CRISPR locus with primer pairs H372-H373 and H376-H377 (Supplementary table S3) for the wtcas9 and hcas9 libraries, respectively. Following gel extraction and purification of the adapted bands, samples were subject to Illumina MiSeq sequencing. Data analysis was performed in Python: first, all newly acquired spacer sequences were extracted from raw MiSeq FASTA data files. Next, the frequency (number of different barcode sequences), the phage target location and the flanking PAM were determined for each unique spacer sequence (see Supplemental Data Set 1).
Supplementary Material
HIGHLIGHTS.
A library of cas9 alleles was screened for clones with enhanced CRISPR-Cas immunity.
The I473F substitution in Cas9 enhances the CRISPR immune response by a 100-fold.
This phenotype is due to an increase of the rate of spacer acquisition.
Cas9 is involved in spacer acquisition and the I473F mutation is a tool to study it.
Acknowledgments
We thank members of the lab for critical discussion of the experiments and their results. R.H. is the recipient of a Howard Hughes International Student Research Fellowship. L.A.M is supported by the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01). L.A.M. is a founder of Intellia Therapeutics and a member of its scientific advisory board. A.V.W. is supported by a graduate student fellowship from the National Science Foundation; J.A.D. acknowledges support from the National Science Foundation, the Paul Allen Foundation and the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
R.H., D.B. and L.A.M. conceived the study. R.H. and A.V.W. executed the experimental work. M.V., D.B. and R.H. analyzed MiSeq data. R.H. and L.A.M. wrote the paper with the help of the rest of the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman SL, Nivala J, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016 doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.