Abstract
Salmonella genomic island 1 (SGI1) harbors an antibiotic resistance gene cluster and was previously identified in the multidrug-resistant Salmonella enterica serovars Typhimurium DT104, Agona, Paratyphi B, and Albany. This antibiotic resistance gene cluster is a complex class 1 integron and most often confers resistance to ampicillin (Ap), chloramphenicol (Cm)/florfenicol (Ff), streptomycin (Sm)/spectinomycin (Sp), sulfonamides (Su), and tetracycline (Tc) (ApCmFfSmSpSuTc profile). Recently, variant SGI1 antibiotic resistance gene clusters conferring different antibiotic resistance profiles have been identified in several S. enterica serovars and were classified as SGI1-A to -G. We identified a new variant SGI1 antibiotic resistance gene cluster in two multidrug-resistant S. enterica serovar Newport strains isolated from humans in France. In these strains, the Sm/Sp resistance gene cassette aadA2 inserted at the first attI1 site was replaced by two other aminoglycoside resistance gene cassettes. The first one contains a new resistance gene encoding an AAC(3)-I aminoglycoside 3-N-acetyltransferase that confers resistance to gentamicin (Gm) and sisomicin (Sc). This gene has been named aac(3)-Id. The second one harbors the Sm/Sp resistance gene aadA7. This gene cassette replacement in the SGI1 complex integron of serovar Newport strains constitutes a new variant SGI1 antibiotic resistance gene cluster named SGI1-H. The occurrence of SGI1 in different S. enterica serovars, now including serovar Newport, strengthens the hypothesis of horizontal transfer of SGI1.
A chromosomal genomic island called Salmonella genomic island 1 (SGI1) has been initially described in epidemic multidrug-resistant Salmonella enterica serovar Typhimurium phage type DT104 strains (serovar Typhimurium DT104) (4). SGI1 contains an antibiotic resistance gene cluster conferring the common multidrug resistance profile ApCmFfSmSpSuTc (i.e., ampicillin [Ap], chloramphenicol [Cm]/florfenicol [Ff], streptomycin [Sm]/spectinomycin [Sp], sulfonamides [Su], and tetracycline [Tc]) of epidemic multidrug-resistant serovar Typhimurium DT104 (1, 2, 4-7, 12, 13, 23, 24, 30, 31). Multidrug-resistant serovar Typhimurium DT104 emerged during the 1980s as a global health problem because of its involvement in diseases in animals and humans (7, 12, 13, 23, 24, 30, 31).
The 43-kb SGI1 is located between the thdF and int2 genes of the chromosome of serovar Typhimurium. The int2 gene is part of a retron sequence, which has been reported to date only in serovar Typhimurium strains (4, 5). In other S. enterica serovars, SGI1 is located between the thdF and yidY genes (3, 4, 9-11, 17, 18). The antibiotic resistance gene cluster is located near the 3′ end of SGI1 and constitutes a complex class 1 integron that belongs to the In4 group (3). The In4 group has a 3′ conserved segment (3′-CS) that includes a copy of IS6100 but no transposition genes, and most members are bound by 25-bp inverted repeats IRi and IRt (20, 21). The antibiotic resistance gene cluster of SGI1 is indeed bound by IRi and IRt and thus can be considered a complex In4-type integron (3). Further, the multidrug resistance region is surrounded by 5-bp direct repeats, strongly suggesting it was integrated by a transposition event (3, 20, 21). Interestingly, in SGI1 there is a duplication of a part of the 5′ conserved segment (5′-CS) of the integron that leads to a second attI1 site, followed by a gene cassette. At the first attI1 site, the cassette carries the aadA2 gene, which confers resistance to Sm and Sp, and a 3′-CS with a truncated sul1 (sul1Δ) gene. At the second attI1 site, the cassette contains the β-lactamase gene pse-1 conferring resistance to Ap and a 3′-CS with a complete sul1 gene conferring resistance to Su. Flanked by the two cassettes are the floR gene, which confers cross-resistance to Cm and Ff, and the Tc resistance genes tetR and tet(G) (1, 2, 4, 6).
Variant SGI1 antibiotic resistance gene clusters have recently been described in the serovars Typhimurium DT104, Agona, and Albany. SGI1 variants were accordingly classified in SGI1-A to SGI1-G (3, 9, 11). These clusters of antibiotic resistance genes were likely generated, according to sequence analysis, after chromosomal recombinational events or by antibiotic resistance gene cassette replacement at one of the attI1 sites. In variants SGI1-A, -D, and -G, the 3′-CS, designated 3′-CS1, is followed by the orf513/dfrA10 region originally described in In6 and In7 (19, 29). Following the orf513/dfrA10 region is a second copy of the 3′-CS, designated 3′-CS2. The variant SGI1-F from serovar Albany represented the first example in which gene cassette replacement took place at one of its attI1 sites (11).
SGI1 has been identified in another serovar Typhimurium phage type, DT120, and in the S. enterica serovars Agona, Paratyphi B, and Albany, indicating the horizontal transfer potential of SGI1 (3, 4, 8, 11, 17). SGI1-carrying serovar Agona, Paratyphi B, and Albany strains were isolated from different animal species, such as poultry in Belgium, tropical fish from Singapore, and food fish imported from Thailand in France, respectively (3, 4, 8, 11, 17). Recently, we have reported the first human case infected by a serovar Agona strain harboring SGI1-A (10).
We examined here 292 strains of serovar Newport isolated from humans in France between 2000 and 2002 by antibiotic susceptibility testing. Two serovar Newport strains isolated in 2001 displayed the multidrug resistance profile ApCmFfSmSpSuTc typical of the SGI1 antibiotic resistance gene cluster but with additional resistances to gentamicin (Gm) and nalidixic acid (Nal). The identification of SGI1 and characterization of its antibiotic resistance gene cluster were thus undertaken for these two serovar Newport strains.
MATERIALS AND METHODS
Clinical and control strains.
A total of 292 S. enterica serovar Newport strains isolated from humans in France between 2000 and 2002 (101 strains in 2000, 125 strains in 2001, and 66 strains in 2002) were studied at the French National Reference Center for Salmonella. Serovar Newport isolates 01-2174 and 01-5348 further characterized in the present study were isolated in 2001 from stools from two French patients with gastroenteritis. They had been infected by these serovar Newport strains during two different travels to Egypt in April and July, respectively. The strains were isolated at their return to France. These strains, control strains harboring SGI1, i.e., serovars Typhimurium DT104 BN9181, Agona 959SA97, Paratyphi B 44, and Albany 7205.00 and other serovar Newport control strains, and Escherichia coli cloning strains TOP10 (Invitrogen, Cergy-Pontoise, France) and XL1-Blue were grown overnight at 37°C in brain heart infusion broth or agar plates.
Serotyping.
Isolates were serotyped on the basis of somatic O and phase 1 and phase 2 flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and WHO Collaborative Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France) according to the White-Kauffman-Le Minor scheme (22).
Antibiotic susceptibility testing.
Isolates were screened for resistance to 35 antibiotics by the disk diffusion method on Mueller-Hinton agar according to the guidelines of the Antibiogram Committee of the French Society of Microbiology (communiqué 2003 [www.sfm.asso.fr/nouv/general.php?pa=2]) (27). The following antibiotics were tested: Ap, amoxicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cephalothin, cefamandole, cefoperazone, cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, moxalactam, imipenem, Sm, Sp, kanamycin, neomycin, tobramycin, netilmicin, Gm, amikacin, dibekacin (Dk), isepamicin, Nal, pefloxacin, ciprofloxacin, Su, Tm, Cm, Ff, and Tc. All antibiotic disks except for Ff were purchased from Bio-Rad. Ff disks were obtained from Schering-Plough Animal Health (Segré, France). The MICs of Gm and Sc were determined by the standard agar doubling dilution method. The MIC breakpoints for Gm and Sc were defined by the Antibiogram Committee of the French Society of Microbiology, i.e., susceptible (MIC ≤ 4 μg/ml) or resistant (MIC > 8 μg/ml) (27).
PCR mapping, cloning, and sequencing.
Detection of SGI1 and its location were performed with primers corresponding to the left and right (with or without retron) junctions in the chromosome as described previously (Fig. 1) (3, 9, 11, 17). PCR mapping of the typical antibiotic resistance genes associated with SGI1 was performed by using conditions and primers described previously (Fig. 1) (3, 9, 11, 17). The antibiotic resistance gene organization was also assessed by Southern blot of genomic DNA cut by HindIII (Qbiogene, Illkrich, France) by using as a probe the XbaI fragment of recombinant plasmid pSTF3, comprising nearly the entire serovar Typhimurium DT104 SGI1 antibiotic resistance gene cluster (Fig. 1). The presence of entire SGI1 was also assessed by Southern blotting of genomic DNA cut by XbaI (Qbiogene) by using probe p1-9 as previously described (4). This probe corresponds to a 2-kb EcoRI fragment comprising parts of the S023 and S024 open reading frames (ORFs), which encode putative helicase and exonuclease proteins (4).
FIG. 1.
Genetic organization of the antibiotic resistance gene cluster of SGI1 and the new SGI1-H variant of serovar Newport strains 01-2174 and 01-5348. The direction of transcription of the genes is indicated by arrows. Black and gray arrows correspond to SGI1 antibiotic resistance genes and chromosomal genes flanking SGI1, respectively. DR-L and DR-R are the left and right direct repeats, respectively, bracketing SGI1. IRi and IRt are 25-bp imperfect inverted repeats defining the left and right ends of complex class 1 integrons. PCRs used to assess the genetic organization of the antibiotic resistance genes (i.e., PCRs floR, A, B, C, D, E, and F) and the SGI1 junctions to the chromosome (i.e., PCRs LJ and RJ for left and right junctions, respectively) are indicated. Restriction sites: X, XbaI; H, HindIII.
To assess the genetic exchange of resistance gene cassettes at the first attI1 site of the SGI1 complex integron in serovar Newport strains 01-2174 and 01-5348, PCR amplification was also performed with a forward primer of PCR A and a reverse primer of PCR B (Fig. 1). Cloning of this PCR product in plasmid pCR2.1-TOPO was performed by using the TOPO TA cloning kit (Invitrogen). The recombinant plasmid obtained was named pBD09. Nucleotide sequencing of the insert of pBD09 was achieved by Genome Express (Meylan, France). Sequence analysis was done by using BLAST (http://www3.ncbi.nlm.nih.gov/BLAST/). PCR amplification of the novel aminoglycoside acetyltransferase gene cassette identified at the first attI1 site was also performed by using the forward primer of PCR A and the reverse primer aacR (5′-GCGGGCACTTTTTCACTCAT-3′). Cloning of this PCR product in plasmid pCR2.1-TOPO was performed by using the TOPO TA cloning kit. The EcoRI fragment containing this PCR product from pBD11 recombinant plasmid was subcloned in plasmid pGEM-7Zf. This recombinant plasmid, named pBD12, was electroporated in E. coli XL1-Blue to further determine the antibiotic resistance profile and the Gm and Sc resistance levels conferred by this novel aminoglycoside acetyltransferase resistance gene.
PFGE.
Strains of serovar Newport were grown overnight on trypto-casein soy agar (Bio-Rad) at 37°C. Preparation of agarose plugs was carried out as described previously (33), except that cells were resuspended in HEPES buffer (16 mM HEPES-NaOH, 16 mM sodium acetate [pH 7.5]). Genomic DNA was digested with XbaI (Roche Diagnostics, Mannheim, Germany) overnight at 37°C. Fragments of DNA were separated by pulsed-field gel electrophoresis (PFGE) in a 1% agarose gel (Seakem Gold; BMA) in HEPES buffer (15) containing 50 μM thiourea by using a CHEF-DR III (Bio-Rad, Hemel Hempstead, United Kingdom). The running conditions were 4 V/cm at 12°C for 27 h, with pulse times ramped from 2.2 to 63.8 s. Lambda Ladder PFG Marker (New England Biolabs, Beverly, Mass.) was used as the molecular size marker.
Nucleotide sequence accession number.
The nucleotide sequence of the serovar Newport strain 01-2174 integron fragment harboring the two aminoglycoside resistance gene cassettes aac(3)-Id and aadA7 has been deposited in the GenBank database under accession no. AY458224.
RESULTS
Multidrug resistance profile.
A total of 292 serovar Newport human strains isolated in France between 2000 and 2002 were tested for their antibiotic susceptibilities. Only two strains showed a multidrug resistance profile similar to that of serovar Typhimurium, serovar Agona, or serovar Paratyphi B harboring SGI1, i.e., ApCmFfSmSpSuTc (4, 8, 17, 18). These strains were also resistant to Gm and Nal and showed a decreased susceptibility to Dk. This multidrug resistance profile suggested the possible occurrence of SGI1 in these serovar Newport isolates.
Identification of SGI1.
To assess the presence of SGI1 and its location in the chromosome of serovar Newport strains 01-2174 and 01-5348, PCRs were realized by using primers corresponding to the left and right junctions of SGI1 in the Salmonella chromosome (Fig. 1). PCR was also performed to detect the presence or absence of the retron sequence, which is located downstream of SGI1 only in serovar Typhimurium DT104 (4). PCR results were positive for the left junction with the thdF gene of the chromosome with primer U7-L12 specific to the thdF gene and primer LJ-R1 specific to the left-end int gene of SGI1 (4). For the right junction, if a sequence of the int2 gene of the retron was used as a reverse primer, the PCR results were negative for the right junction of SGI1 but positive if the sequence of yidY gene was used (3, 9, 11, 17). The two serovar Newport strains thus lacked the retron sequence found only to date in serovar Typhimurium DT104. Thus, these results indicated that serovar Newport strains 01-2174 and 01-5348 harbor SGI1 at the same chromosomal location, i.e., between the thdF and yidY genes, as serovars Typhimurium DT104, Agona, Paratyphi B, and Albany (4, 11, 17, 18).
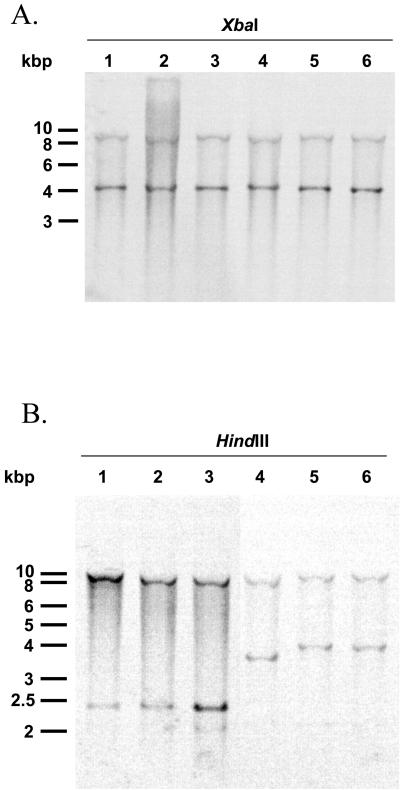
In addition to the antibiotic resistance gene cluster presented below, the presence of the entire SGI1 in serovar Newport strains was confirmed by Southern blot hybridization of XbaI-digested genomic DNA with the p1-9 probe described previously (4). This probe corresponds to a 2-kb EcoRI fragment located outside of the antibiotic resistance gene cluster comprising parts of the S023 and S024 ORFs in the central region of SGI1 (4). The p1-9 probe showed two XbaI fragments of the expected 4- and 9-kb sizes as in SGI1-carrying serovar Typhimurium DT104, Agona, Paratyphi B, and Albany strains (Fig. 2).
FIG. 2.
(A) Southern blot hybridization with the p1-9 probe of XbaI-digested genomic DNAs of serovar Typhimurium DT104 strain BN9181 (lane 1), serovar Agona strain 959SA97 (lane 2), serovar Paratyphi B strain 44 (lane 3), serovar Albany strain 7205.00 (lane 4), serovar Newport strain 01-2174 (lane 5), and serovar Newport strain 01-5348 (lane 6). (B) Southern blot hybridization of HindIII-digested genomic DNAs of serovar Typhimurium DT104 strain BN9181 (lane 1), serovar Agona strain 959SA97 (lane 2), serovar Paratyphi B strain 44 (lane 3), serovar Albany strain 7205.00 (lane 4), serovar Newport strain 01-2174 (lane 5), and serovar Newport strain 01-5348 (lane 6) with the pSTF3 probe containing all SGI1 antibiotic resistance genes (Fig. 1).
New variant antibiotic resistance gene cluster.
PCR mapping of the typical antibiotic resistance gene cluster associated with SGI1 is schematized in Fig. 1. PCRs A to I and floR were performed on genomic DNA extracted from serovar Newport strains 01-2174 and 01-5348. The two strains yielded fragments B, C, D, E, F, and floR of the sizes expected from the serovar Typhimurium DT104 control strain BN9181 harboring SGI1. However, PCR A specific for the aadA2 resistance gene cassette was not obtained. Thus, these results indicated the presence of the floR, tetR, and tet(G) genes and the second gene cassette carrying pse-1. The positive result for PCR B nevertheless indicated the presence of the 3′-CS with the truncated sul1 gene. To assess the gene cassette array at the first attI1 site of the SGI1 complex integron, a PCR with the forward primer of PCR A, targeting the intI1 gene, and the reverse primer of PCR B, targeting the floR gene, was performed (Fig. 1). The two serovar Newport strains yielded a fragment 600 bp larger than that of the serovar Typhimurium DT104 control strain BN9181 (data not shown). This PCR product was cloned and sequenced (GenBank accession no. AY458224). The recombinant plasmid obtained, named pBD09, conferred resistance to Gm, Sm, and Sp in a susceptible E. coli host strain. Sequence analysis identified two resistance gene cassettes showing 100% nucleotide identity to those found in class 1 integrons from a Vibrio fluvialis strain and from an S. enterica serovar Kentucky strain recently deposited under GenBank accession numbers AB114632 and AY463797, respectively. The first resistance gene cassette was named aac(3)-Id (GenBank accession no. AB114632) or aacC-A5 (GenBank accession no. AY463797). The resistance gene was annotated to confer Gm, Sc, and fortimicin resistance (GenBank accession no. AB114632). The second resistance gene cassette, named aadA7 (GenBank accession numbers AB114632 and AY463797), was previously reported to confer Sm and Sp resistance in an E. coli strain of the ECOR collection (16). Thus, instead of the aadA2 resistance gene cassette classically found at the first attI1 site of the SGI1 complex integron, two resistance gene cassettes, i.e., a novel 3′-N-aminoglycoside acetyltransferase gene cassette, named aac(3)-Id or aacC-A5, and the aadA7 resistance gene cassette were found in both multidrug-resistant serovar Newport strains of the present study (Fig. 1). The 3′-CS was 100% identical to that of the SGI1 complex integron of serovar Typhimurium DT104 with a truncated sul1 gene (Fig. 1). Thus, the antibiotic resistance gene cluster from serovar Newport strains 01-2174 and 01-5348 constitutes a new SGI1 variant antibiotic resistance gene cluster; we propose the name of SGI1-H according to the previously proposed nomenclature of SGI1 variants (3, 9, 11).
The presence of this new SGI1 variant in both serovar Newport strains was confirmed by Southern blotting of HindIII-digested genomic DNA with the pSTF3 probe containing nearly the entire SGI1 antibiotic resistance gene cluster of serovar Typhimurium DT104 control strain BN9181 (corresponding to the XbaI fragment shown in Fig. 1). Indeed, identical HindIII Southern blot profiles were obtained for both serovar Newport strains 01-2174 and 01-5348 (Fig. 2). This profile was clearly distinct from that obtained for previously characterized SGI1 variant control strains and was in accordance with the nucleotide sequence of the insert of recombinant plasmid pBD09 described above (GenBank accession no. AY458224) (Fig. 2).
Cloning and expression in E. coli of the aac(3)-Id/aacC-A5 gene cassette.
The deduced amino acid sequence from the novel 3′-N-aminoglycoside acetyltransferase gene, named aac(3)-Id or aacC-A5, exhibits 47 to 54% identity to that of known AAC(3)-I proteins, i.e., AAC(3)-Ia, AAC(3)-Ib, and AAC(3)-Ic (Fig. 3) (25, 26, 34). The aac(3)-Id or aacC-A5 gene product likely constitutes a fourth enzyme of this group. According to the aminoglycoside acetyltransferase nomenclature, and more particularly to that of the AAC(3)-I group of enzymes, the new gene and gene product should be named aac(3)-Id rather than aacC-A5 and AAC(3)-Id rather than AAC(3)-Ie, respectively (GenBank accession numbers AB114632, AY463797, and AY458224) (28, 32).
FIG. 3.
Amino acid sequence alignment of the AAC(3)-Id protein encoded by the aac(3)-Id resistance gene cassette of SGI1-H with that of other AAC(3)-I proteins, including AAC(3)-Ia (GenBank accession no. CAA33850), AAC(3)-Ib (GenBank accession no. AAA88422), and AAC(3)-Ic (GenBank accession no. CAD53575). The alignment was generated by CLUSTAL W (http://www.ebi.ac.uk/clustalw/) and modified manually. Identical amino acids among AAC(3)-Id and another AAC(3)-I protein are highlighted in white on a black background.
A PCR fragment corresponding to the resistance gene cassette aac(3)-Id with the integron promoter was amplified and cloned into the pGEM-7Zf vector. This recombinant plasmid, named pBD12, was introduced into an aminoglycoside-susceptible E. coli XL1-Blue host strain to determine the aminoglycoside susceptibility patterns. Serovar Newport strains 01-2174 and 01-5348 tested by the disk diffusion method showed the classical SGI1 multidrug resistance profile ApCmFfSmSpSuTc but were also resistant to Gm and Sc and showed a decreased susceptibility to Dk (Table 1). Both serovar Newport strains showed the same level of resistance with Gm MICs of 16 μg/ml and Sc MICs of 32 μg/ml. E. coli XL1-Blue strain carrying the recombinant plasmid pBD12 was resistant to Gm and Sc and showed a decreased susceptibility to Dk as the serovar Newport strains 01-2174 and 01-5348 (Table 1). This strain showed resistance to Gm and Sc with MICs of 32 and 128 μg/ml, respectively. Susceptibilities to amikacin, kanamycin, neomycin, netilmicin, Sm, Sp, and tobramycin were not affected. These results thus indicated that the aac(3)-Id gene contributes to aminoglycoside resistance with a typical resistance profile of AAC(3)-I enzymes, i.e., Gm and Sc (28).
TABLE 1.
Antibiotic resistance profiles and MICs of aminoglycosides for S. enterica serovar Newport and E. coli strains
| Strain | Antibiotic resistance profile | MIC (μg/ml)
|
|
|---|---|---|---|
| Gm | Sc | ||
| S. enterica serovar Newport 01-2174 | ApCm(Dk)FfGmScSmSpSuTc | 16 | 32 |
| S. enterica serovar Newport 01-5348 | ApCm(Dk)FfGmScSmSpSuTc | 16 | 32 |
| E. coli XL1-Blue (pBD12) | ApDkGmScTc | 32 | 128 |
| E. coli XL1-Blue (pGEM-7Zf) | ApTc | <0.5 | <0.5 |
Macrorestriction analysis of SGI1-carrying serovar Newport strains.
Macrorestriction analysis by PFGE of serovar Newport isolate DNAs with XbaI showed that both SGI1-carrying strains 01-2174 and 01-5348 were indistinguishable (Fig. 4), whereas other serovar Newport strains that did not carry SGI1 exhibited different but closely related XbaI macrorestriction patterns. Thus, these results indicated that serovar Newport strains 01-2174 and 01-5348 are clonally related but are genetically distinct from the other SGI1-carrying S. enterica serovars Typhimurium DT104, Agona, Paratyphi B, and Albany according to their PFGE XbaI patterns described elsewhere (data not shown) (8, 11, 17).
FIG. 4.
Macrorestriction analysis by PFGE of genomic DNAs cut by XbaI of serovar Newport strains: SGI1-H-carrying strain 01-2174 (lane 1), SGI1-H-carrying strain 01-5348 (lane 2), non-SGI1 strain 01-8181 isolated in France in 2001 (lane 3), and serovar Newport control strains from the French National Reference Center for Salmonella collection (lanes 4 and 5).
DISCUSSION
SGI1 is the first genomic island containing an antibiotic resistance gene cluster identified in S. enterica. Its acquisition may have been an important trait in the worldwide epidemic of the multidrug-resistant serovar Typhimurium DT104 clone causing disease in animals and humans (7, 12, 13, 23, 24, 30, 31). SGI1 has been further identified in other S. enterica serovars, such as Agona, Paratyphi B, and Albany from different animal species (cattle, poultry, and fish) and from humans around the world (9-11, 17, 18). The identification of SGI1 in several S. enterica serovars indicates a large diffusion of this genomic island and also leads to the hypothesis of the SGI1 horizontal transfer potential. Moreover, the same chromosomal location of SGI1 in different S. enterica serovars at the 3′ end of the thdF gene strengthens the hypothesis of the SGI1 horizontal transfer via a site-specific integrational recombination mechanism into the chromosome, as has been reported for integrating conjugative elements such as the SXT element in Vibrio cholerae (14). The two serovar Newport strains isolated from humans in France in the present study thus represent the fifth S. enterica serovar in which SGI1 has been identified. The two serovar Newport strains were clonally related, as shown by XbaI macrorestriction analysis, and were probably linked from an epidemiological point of view. Gastroenteritis occurred in the two French patients during travels to Egypt in April and July 2001, and they were probably infected by the same serovar Newport strain during their travel.
The serovar Newport isolates studied here also represent a new SGI1 variant antibiotic resistance gene cluster, named SGI1-H according to the previously proposed nomenclature (3, 9, 11). The aac(3)-Id and aadA7 gene cassettes found instead of aadA2 are the second example of gene cassette replacement at one of the SGI1 attI1 sites. The first case of gene cassette replacement reported for the SGI1 complex class 1 integron was that of the serovar Albany SGI1-F variant, in which the aadA2 gene cassette was replaced by the dfrA1 and ORF gene cassettes (11). The aac(3)-Id and aadA7 gene cassettes of serovar Newport may have been introduced in the SGI1 complex class 1 integron by homologous recombination with another class 1 integron containing the same array of resistance gene cassettes from another bacterium. Another explanation is the exchange of resistance gene cassettes mediated by the integron-encoded integrase, implying excision of the aadA2 gene cassette and insertion of the aac(3)-Id and aadA7 gene cassettes. The partial intI1 sequence from serovar Newport showed four point mutations compared to the classical SGI1 intI1 gene (99% nucleotide identity) (data not shown). Moreover, the array of resistance gene cassettes found in the integron of the serovar Newport strain were the same as those recently reported in an integron of a Vibrio fluvialis strain (GenBank accession no. AB114632). When we consider these data, the first hypothesis of a homologous recombination between the SGI1 complex integron of serovar Newport and the class 1 integron of another bacterium seems to be more likely. In the present study, the contamination origin by the serovar Newport strains harboring SGI1 has not been elucidated. According to the natural aquatic environment of pathogenic V. fluvialis strains, antibiotic resistance gene exchange between different bacterial species such as Vibrio and Salmonella probably takes place in such aquatic environments.
The new variant SGI1-H of the present study was shown to contain a novel 3-N-aminoglycoside acetyltransferase gene, aac(3)-Id, conferring resistance to Gm and Sc. The aminoglycoside resistance profile of the AAC(3)-I enzymes group is characterized by resistance to Gm, Sc, and astromicin (also called fortimicin [not tested in the present study]). The nucleotide identity of the aac(3)-Id gene is ca. 40 to 60% compared to that of the other acetyltransferase genes, aac(3)-Ia, aac(3)-Ib, and aac(3)-Ic, that encode the same AAC(3)-I aminoglycoside resistance profile (25, 26, 28, 34). Like the other aac(3)-I resistance genes, the aac(3)-Id gene is located within a class 1 integron. The AAC(3)-Id protein represents a fourth evolutionary lineage of the AAC(3)-I group and shares 54% amino acid sequence identity with that of the AAC(3)-Ic protein, which has recently been described (25). As shown in the present study, the newly described aac(3)-Id resistance gene thus contributes to aminoglycoside resistance with a typical profile of AAC(3)-I enzymes, i.e., resistance to Gm and Sc. To our knowledge, the combination of this gene cassette with the aadA7 gene cassette in SGI-H confers for the first time the extended aminoglycoside resistance profile, i.e., resistance to Gm, Sc, Sm, and Sp in serovar Newport.
In conclusion, as appears from this and previous studies, the SGI1 complex class 1 integron may contribute to the capture in the Salmonella chromosome of a wide diversity of resistance gene cassettes and thus generate diverse antibiotic resistance gene clusters (3, 9, 11). Moreover, the growing number of S. enterica serovars for which SGI1 has been identified and the identical chromosomal location of SGI1 in these serovars suggest that SGI1 is a new emerging mobile element that integrates site specifically into the chromosome. Studies are under way to examine experimentally the capacity of horizontal transfer of SGI1.
Acknowledgments
We thank C. Mouline for expert technical assistance. We also thank D. Boyd and M. R. Mulvey for their collaboration.
This study was supported by a grant from the French Institut National de la Recherche Agronomique (Action Transversalité 2001-2003).
REFERENCES
- 1.Arcangioli, M. A., S. Leroy-Sétrin, J. L. Martel, and E. Chaslus-Dancla. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174:327-332. [DOI] [PubMed] [Google Scholar]
- 2.Bolton, L. F., L. C. Kelley, M. D. Lee, P. J. Fedorka-Cray, and J. J. Maurer. 1999. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. Sidi Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D. A., G. A. Peters, L. K. Ng, and M. R. Mulvey. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 189:285-291. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., and S. Schwarz. 2001. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica serotype Typhimurium DT104. Vet. Res. 32:301-310. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., K. Sidi Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:A2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublet, B., P. Butaye, H. Imberechts, J. M. Collard, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella Agona harboring genomic island 1-A. Emerg. Infect. Dis. 10:756-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, D., T. Besser, J. Gay, D. Rice, M. Davis, and C. Gay. 2000. The global epidemiology of multiresistant Salmonella enterica serovar Typhimurium DT104, p. 217-243. In C. Brown and C. Bolin (ed.), Emerging diseases of animals. American Society for Microbiology, Washington, D.C.
- 14.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 15.Koort, J. M., S. Lukinmaa, M. Rantala, E. Unkila, and A. Siitonen. 2002. Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:3497-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulvey, M. R., D. Boyd, A. Cloeckaert, R. Ahmed, L. K. Ng, et al. 2004. Emergence of multidrug-resistant Salmonella enterica serovar Paratyphi B dT+, Canada. Emerg. Infect. Dis. 10:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid DGO100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge, S. R., H. J. Brown, H. W. Stockes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborative Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 23.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescott. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 24.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riccio, M. L., J. D. Docquier, E. Dell'Amico, F. Luzzaro, G. Amicosante, and G. M. Rossolini. 2003. Novel 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ic, from a Pseudomonas aeruginosa integron. Antimicrob. Agents Chemother. 47:1746-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SFM Antibiogram Committee. 2003. Comité de l'antibiogramme de la société francaise de microbiologie report 2003. Int. J. Antimicrob. Agents 21:364-391. [DOI] [PubMed] [Google Scholar]
- 28.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 30.Tauxe, R. V. 1999. Salmonella Enteritidis and Salmonella Typhimurium DT104: successful subtypes in the modern world, p. 37-53. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infectious 3. American Society for Microbiology, Washington, D.C.
- 31.Threlfall, E. J. 2000. Epidemic Salmonella Typhimurium DT104: a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7-10. [DOI] [PubMed] [Google Scholar]
- 32.Vakulenko, S. B., and S. Mobashery. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. SHV-12-like extended-spectrum-β-lactamase-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohlleben, W., W. Arnold, L. Bissonnette, A. Pelletier, A. Tanguay, P. H. Roy, G. C. Gamboa, G. F. Barry, E. Aubert, and J. Davies. 1989. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC(3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217:202-208. [DOI] [PubMed] [Google Scholar]