Abstract
Improved treatments for chronic hepatitis C virus (HCV) infection are needed due to the suboptimal response rates and deleterious side effects associated with current treatment options. The triphosphates of 2′-C-methyl-adenosine and 2′-C-methyl-guanosine were previously shown to be potent inhibitors of the HCV RNA-dependent RNA polymerase (RdRp) that is responsible for the replication of viral RNA in cells. Here we demonstrate that the inclusion of a 7-deaza modification in a series of purine nucleoside triphosphates results in an increase in inhibitory potency against the HCV RdRp and improved pharmacokinetic properties. Notably, incorporation of the 7-deaza modification into 2′-C-methyl-adenosine results in an inhibitor with a 20-fold-increased potency as the 5′-triphosphate in HCV RdRp assays while maintaining the inhibitory potency of the nucleoside in the bicistronic HCV replicon and with reduced cellular toxicity. In contrast, while 7-deaza-2′-C-methyl-GTP also displays enhanced inhibitory potency in enzyme assays, due to poor cellular penetration and/or metabolism, the nucleoside does not inhibit replication of a bicistronic HCV replicon in cell culture. 7-Deaza-2′-C-methyl-adenosine displays promising in vivo pharmacokinetics in three animal species, as well as an acute oral lethal dose in excess of 2,000 mg/kg of body weight in mice. Taken together, these data demonstrate that 7-deaza-2′-C-methyl-adenosine is an attractive candidate for further investigation as a potential treatment for HCV infection.
Hepatitis C virus (HCV) infects an estimated 170 million people worldwide. In the United States alone, approximately 3.9 million people are infected, roughly five times the number of people infected with human immunodeficiency virus (HIV) (21). HCV is transmitted primarily through direct percutaneous exposure to blood and is the most common chronic blood-borne infection in the United States. The Centers for Disease Control and Prevention estimate that the introduction of routine blood screening reduced the rate of HCV infection in the United States from an average of 240,000 new cases per year in the 1980s to approximately 25,000 in 2001 (15). Blood transfusions account for a substantial portion of those currently infected, but injection drug use is a continuous source of infection and accounts for ∼60% of new HCV infections in the United States (12). Other sources of exposure are linked to occupational accidents, hemodialysis, high-risk sexual activity, and perinatal sources of infection. Unfortunately, no vaccine is currently available for HCV infection.
The majority of HCV infections progress to chronic infection, which can lead to cirrhosis, liver failure, and hepatocellular carcinoma, and HCV is currently the leading cause of liver transplantation in Europe and the United States (25, 26, 30). The current standard therapy, a combination of pegylated interferon alpha (pIFN-α) and the nucleoside analog ribavirin, produces viral response rates of less than 50% in the majority of patients who are infected with genotype 1 virus (25), the most prevalent strain in the United States and Europe. In addition, treatment with pIFN-α-ribavirin results in serious and unfavorable side effects, which include flu-like symptoms, hemolytic anemia, fatigue, depression, and even suicide (25). Ribavirin is also teratogenic and therefore must be used with significant caution in women of child-bearing age.
The HCV NS5B protein, the RNA-dependent RNA polymerase, is responsible for the synthesis of the viral RNA genome and is therefore an attractive target for the development of antiviral agents (4, 14). Viral polymerases are validated targets for antiviral drug development. Nucleoside analogs are known inhibitors of HIV, hepatitis B virus, and herpes simplex virus polymerases and are approved therapeutics for treatment of these viral infections. Chain-terminating nucleoside analogs are converted in cells to the corresponding triphosphates. These are incorporated by the viral polymerase into nascent nucleic acid chains, where they prevent the incorporation of the next nucleotide and result in the formation of incomplete, nonfunctional RNA or DNA products.
Previously, we reported that the triphosphates of 2′-C-methyl-adenosine and 2′-C-methyl-guanosine are effective in vitro inhibitors of HCV NS5B polymerase (23). However, these compounds would not be effective therapeutics because the plasma half-life of the adenosine analog is short, and although the guanosine analog has excellent oral bioavailability, it lacks robust cell penetration and is inefficiently converted intracellularly to the active triphosphates (23). Here we describe a 7-deaza modification of several purine nucleoside triphosphates which results in increased inhibitory potency in assays of the purified HCV RNA-dependent RNA polymerase (RdRp). 7-Deaza-2′-C-methyl-adenosine, in particular, is a potent inhibitor of HCV replication with low cellular toxicity and excellent pharmacokinetic parameters.
MATERIALS AND METHODS
Nucleoside analogs.
3′-Deoxy-guanosine (TriLink BioTechnologies, San Diego, Calif.), 2′-araguanosine (R.I. Chemical Inc., San Diego, Calif.), 2′-O-methyl-guanosine (TriLink BioTechnologies), 2′-O-methyl-adenosine (TriLink BioTechnologies), and 7-deaza-adenosine (Sigma-Aldrich, St. Louis, Mo.) are commercially available. All other nucleoside analogs were synthesized as previously described (9, 16). In general, triphosphates were synthesized from the corresponding nucleosides and purified according to the general procedure for triphosphate synthesis and purification previously described (16). 3′-Deoxy-ATP was purchased from Sigma-Aldrich, and arabino-ATP was purchased from both TriLink BioTechnologies, Inc. and IBA GmbH (Göttingen, Germany). Ultrapure ATP, UTP, GTP, and CTP were purchased from Amersham Biosciences. Additionally, a nonnucleotide inhibitor of NS5B, compound A (32), was purchased from Interbioscreen Ltd. (Moscow, Russia).
Radiolabeled material.
Tritiated 7-deaza-[7-3H]2′-C-methyl-adenosine was prepared as previously described for 2′-C-methyl-adenosine (23). [2′,3′-3H]dideoxyinosine (ddI) and [8-3H]acyclovir were supplied by Moravek (Brea, Calif.), and [2-3H]adenosine and [8-3H]tubercidin were purchased from Amersham Biosciences.
NS5B enzyme assays.
HCV (genotype 1b-BK strain) wild-type (WT) NS5BΔ21, WT NS5BΔ55, and S282T NS5BΔ55 (23) were expressed in Escherichia coli and purified as previously described (10, 11).
RNA polymerase activity and inhibition displayed by nucleoside triphosphate (NTP) analogs were determined in reactions catalyzed by NS5BΔ21, NS5BΔ55, or S282T NS5BΔ55 by measuring the incorporation of radiolabeled NTPs onto a heteromeric RNA template, t500, as previously described (11). No significant differences in the potency of inhibition were determined for inhibition of NS5BΔ21 compared to that of NS5BΔ55. The incorporation and extension of nucleoside triphosphates were also analyzed in reactions utilizing a 5′-32P-labeled oligoribonucleotide template (5′-AGAUGGCCCGGUUUUCCGGGCC-3′, synthesized by Dharmacon (Lafayette, Colo.) that was designed to fold into a hairpin structure, with the first available template base being a U (underlined) followed by an A, as previously described (11).
BVDV RNA-dependent RNA polymerase assays.
Bovine viral diarrhea virus (BVDV) NS5BΔ21 was purified as previously described (31). The RNA polymerase activity of BVDV NS5BΔ21 was measured in reaction mixtures containing 20 mM Tris-HCl, pH 7.5, 50 μM EDTA, 2 mM MgCl2, 5 mM dithiothreitol (DTT), 80 mM KCl, 0.4 U of RNasin (Promega)/μl, 0.2% polyethylene glycol 8000, 0.03 μg of t500/μl, and 1 μM enzyme. Reactions were carried out and products were analyzed by using the same conditions as described above for HCV NS5BΔ21.
Human DNA polymerases.
Human DNA polymerases α, β, and γ were assayed for inhibition by 7-deaza-2′-C-methyl-ATP as previously described (11).
Human RNA polymerase II.
Human RNA polymerase II was purified as previously described (18). In vitro polymerase activity was measured in reaction mixtures containing 70 mM Tris, pH 8, 70 mM ammonium sulfate, 6 mM MgCl2, 2 mM DTT, 5 mM spermidine, 0.4 U of RNasin (Promega)/μl, 0.04 μg of M13 single-stranded DNA (Amersham Biosciences)/μl, and 0.09 μg of enzyme/μl. Reactions were initiated by the addition of an NTP mixture containing 20 μM ATP, 500 μM GTP, UTP, and CTP, and ∼2 μCi of [α-33P]ATP (NEN Life Sciences) per reaction, with or without an NTP analog. Reactions proceeded for 1 h at 37°C and were quenched by the addition of ∼140 mM EDTA (final concentration). Product formation was determined by DE-81 filter binding (Whatman) as previously described (11).
Poly(A) polymerase.
The activity of yeast poly(A) polymerase (U.S. Biochemicals) in vitro was measured in reaction mixtures containing 25 mM Tris-HCl, pH 7, 40 mM KCl, 0.5 mM MnCl2 or MgCl2, 0.05 mM EDTA, 0.5 mM DTT, 0.2 mg of bovine serum albumin/ml, 10% glycerol, 1 U of enzyme/μl, and 200 nM of the heteromeric RNA template, t500. Reactions were initiated by the addition of 50 μM ATP (Amersham Biosciences) and ∼2 μCi [α-33P]ATP (NEN Life Sciences) per reaction, with or without an ATP analog. Reactions proceeded for 30 min at 30°C and were quenched by the addition of 20 μl of 0.5 M EDTA, for a final concentration of ∼220 mM EDTA. Product formation was determined by DE-81 filter binding (Whatman) as previously described (23, 34).
Tissue culture, replication analysis, selection, and sequencing of resistant replicons.
Huh-7 and HBI10A cells were cultured and transfected by electroporation with in vitro-transcribed RNAs as described previously (23, 34). Transient-transfection assays were performed using cells highly competent for HCV replication, obtained by curing HBI10A cells of the endogenous replicons with human alpha 2b interferon as described previously (23, 34). The effect of compounds on viral replication was monitored by in situ RNase protection assay or cell enzyme-linked immunosorbent assay as described previously (23, 34). Clones resistant to 7-deaza-2′-C-methyl-adenosine were selected as described previously (23, 34). Replicon RNAs extracted from resistant clones were retrotranscribed and amplified by PCR, and their sequence was determined by direct automated sequencing of the PCR products (23, 34).
Plasmids.
Plasmid pHCVNeo17.B encodes an HCV replicon identical to I377neo/NS3-3′/wt (EMBL-GenBank no. AJ242652) but contains two tissue-culture-adaptive mutations as described previously (34). Plasmid pHCVNeo17.S282T is identical to plasmid pHCVNeo17.B but contains replacement of the triplet AGC (nucleotides 8441 to 88443) coding for serine 282 in NS5B, with triplet ACC, coding for threonine.
Antiviral activity with additional viruses.
In vivo, non-HCV antiviral assays, with the exception of BVDV assay, were performed at the Utah State University Institute for Antiviral Research under direction of Robert Sidwell. Fifty percent effective and cytotoxic concentrations (EC50s and CC50s) of 7-deaza-2′-C-methyl-adenosine were determined in cell culture antiviral assays against the following 13 viruses (strains): West Nile virus (New York isolate), dengue type 2 virus (New Guinea), yellow fever virus (17D), rhinovirus type 2 (HGP), rhinovirus type 14 (Tow), poliovirus type 3 (Wyo3), Western equine encephalitis virus (California), Venezuelan equine encephalitis virus (Trinidad), respiratory syncytial virus (A2), measles virus (Chicago), human cytomegalovirus (AD-169), herpesvirus type 1 (McCrae), and herpesvirus type 2 (E194). The preceding assays, with the exception of that for human cytomegalovirus, are described by Sidwell and Huffman (27) and were used in the reports of in vitro antiviral activity of ribavirin (19, 28). Once cellular monolayers were established, compound dilutions were added, corresponding to four-point or seven-point titrations, followed by the addition of virus after approximately 5 min. Toxicity measurements were determined with cells treated with compound without the addition of virus. Cells were incubated with compound and virus until viral controls showed adequate cytopathic effect (CPE) (≥3 days for all viruses) for measurement. The cells were then microscopically examined for both virus-induced CPE and compound-related toxicity. The EC50s and CC50s were calculated by regression analysis of the viral CPE data and toxicity control data. The CPE results were confirmed using a Neutral Red assay (3) (Table), except for the herpesvirus data, which are visual CPE results, and human cytomegalovirus data, which are based on a plaque reduction assay (8, 9). In vitro BVDV potency determinations were performed as previously described (31).
Intracellular metabolism studies.
The intracellular metabolism of nucleoside analogs was examined as previously described (11).
Pharmacokinetics.
All animal studies described in this report were approved by Merck Research Laboratories Institutional Animal Care and Use Committee. Pharmacokinetic studies were performed with male beagle dogs, Sprague-Dawley rats, and rhesus macaques dosed orally via gavage at 2 mg/kg of body weight and intravenously by bolus injection at 1 mg/kg (n = 2/route). Compound was formulated at 4 mg/ml in ethanol-PEG 400-water (20:30:50 [vol/vol/vol]). Intravenous dosing was performed with rats, beagles, and rhesus macaques via implanted catheters in the femoral, cephalic, and saphenous veins, respectively. Blood samples were obtained at 5, 15, and 30 min and 1, 2, 4, 6, and 8 h, placed in heparin-coated tubes, and centrifuged, and the plasma was decanted and stored at −70°C until analysis. An additional sample from dogs and monkeys was obtained at 24 h. Liver concentration studies were performed with male Sprague-Dawley rats dosed orally at 2, 20, and 200 mg/kg, using 0.5% methylcellulose in water as the vehicle. The doses were formulated so that subjects were dosed at a volume of 5 ml/kg. Plasma and liver samples were obtained at 8 and 24 h and stored at −70°C until analysis.
Plasma samples (50 μl) were precipitated with acetonitrile, centrifuged, decanted, dried under nitrogen, and reconstituted with mobile-phase material. Liver samples were prepared by homogenization with four equivalents of distilled water. A 250-μl aliquot was then processed in the same manner as the plasma samples. Separate calibration curves were constructed for each matrix using plasma and liver samples from control animals. Sample analysis was performed using a 4.6-by-150-mm Zorbax SB-CN column (Mac-Mod Analytical, Chadds Ford, Pa.) connected to a Perkin-Elmer Series 200 liquid chromatography system with an isocratic mobile phase of 5 mM ammonium formate with 0.1% formic acid in 80/20 acetonitrile-water at a flow rate of 1 ml/min. A Sciex API 3000 (Toronto, Canada) triple quadripole instrument operated in the positive-ion APCI (atmospheric pressure chemical ionization) mode using multiple-reaction monitoring was used to detect the analyte and internal standard. Data reduction was performed by using Sciex Analyst software (version 1.1). Pharmacokinetic calculations were performed using Watson 6.02.0.02 for Windows (Pharmaceutical Software Systems, Inc., Wayne, Pa.).
Jurkat cytotoxicity assays.
Cytotoxicity was determined by using Jurkat T cells which were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 1% penicillin-streptomycin (Pen/Strep). Clear-bottom 96-well microtiter plates were preformatted with 50 μl of medium containing compounds of interest solubilized in Me2SO at a concentration twofold higher than the final testing concentration and a Me2SO content of 2%. Individual plates were prepared for each time point to be assayed. Jurkat cells were diluted to a concentration of 2 × 105 cells/ml, and 50 μl of cells were added to each test well for a final incubation volume of 100 μl and a final Me2SO content of 1%. At the indicated time points, viable cells were assayed by using the CellTiter 96 AQueous One Solution cell proliferation assay (Promega Corporation, Madison, Wis.) according to the manufacturer's directions. Briefly, 20 μl of MTS [3-(4,5-dimethyl-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] reagent was added directly to each well, and cells were incubated for 2.5 h. Absorbance at 490 nm, which is directly proportional to the number of live cells in culture, was read in a Molecular Devices ThermoMax microplate reader.
[14C]thymidine uptake.
[14C]thymidine uptake assays were performed by using the thymidine uptake 14C cell proliferation assay kit from Amersham (Piscataway, N.J.) according to the manufacturer's directions. Huh-7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 2 mM l-glutamine, 1% nonessential amino acids, and 1% Pen/Strep. Ninety-six-well Cytostar plates were preformatted with 100 μl of medium containing compounds of interest solubilized in Me2SO at a concentration twofold higher than the final testing concentration and a Me2SO content of 2%. Huh-7 cells were diluted to 40,000 cells/ml in medium containing 100 nM [14C]thymidine, of which 100 μl was added to each test well, bringing the total volume to 200 μl. Cells were incubated at 37°C in 5% CO2. Detection of [14C]thymidine incorporation into cells was determined at indicated times by reading plates on a Packard TopCount NXT instrument.
RNA incorporation.
The incorporation of tritiated nucleosides into cellular RNA was determined using the following analogs: [3H]adenosine, [3H]tubercidin, [3H]ddI, [3H]acyclovir, and 7-deaza-7-[3H]2′-C-methyl-adenosine. The tritium radiolabel was on the ribose for ddI while on the base for all other nucleosides. Huh-7 cells were cultured in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1% nonessential amino acids, and 1% Pen/Strep. One T-75 flask was seeded with 105 Huh-7 cells (20% confluency) for each nucleoside tested. Labeled nucleosides were added to a 0.5 μM concentration at time of seeding except for the negative control. Cells were incubated at 37°C in 5% CO2 for 3 days. Media were removed, and cell monolayers were washed with phosphate-buffered saline. RNA was isolated by extraction of the cell monolayers with 4 ml of a trizol reagent (Gibco BRL). As a negative control, [3H]adenosine was added to the trizol lysate of Huh-7 cells that had been grown in the absence of labeled nucleoside. Phase separation was achieved by the addition of 0.8 ml of chloroform to each lysate, followed by centrifugation for 15 min at 12,000 × g at 4°C. RNA was precipitated from the aqueous phase by the addition of 2 ml of isopropanol and centrifugation for 15 min at 12,000 × g at 4°C. Pellets were washed in 75% ethanol and air dried for 5 min. RNA pellets were dissolved in 150 μl of H2O. DNase digestion buffer and RNase-free DNase were added, and samples were incubated at 37°C for 30 min. RNAs were further purified by spin column purification using the RNeasy RNA purification kit (QIAGEN) as per the manufacturer's protocol and were recovered in a 50-μl final volume. RNA concentrations were determined by absorbance. Purified RNA (5 μl for adenosine and tubercidin and 20 μl for dideoxyinosine, acyclovir, and 7-deaza-2′-C-methyl-A) was diluted into 5 ml of Ready-Safe scintillation fluid (Beckman-Coulter, Fullerton, Calif.), and radioactivity was quantified using a Packard LS 6000SC scintillation counter.
Acute oral toxicity study with mice.
Female mice [Crl:CD-1(ICR)BR; Charles River Laboratories, Raleigh, N.C.] ranging from 6 to 9 weeks old and from 22 to 35 g in weight were used to determine the acute oral toxicity of both 7-deaza-adenosine (tubercidin) and 7-deaza-2′-C-methyl-adenosine following a single oral dose. Mice were housed three per box containing hardwood chip bedding in a climate-controlled room with a 12-h light cycle. Rodent diet and fresh water were available at all times. A single oral dose of either 7-deaza-2′-C-methyl-adenosine at 2,000 mg/kg or tubercidin at either 20 or 50 mg/kg was administered to three mice in a 60-mg/ml suspension in 0.5% aqueous methylcellulose. Oral doses were given by gastric intubation, using a metal catheter attached to a syringe. All animals were examined daily for mortality and physical signs of drug effect for 14 days. Body weights were recorded pretest and on days 7 and 14 postadministration. All animals were euthanatized with carbon dioxide and discarded without necropsy after the 14-day observation period.
RESULTS
Inhibition of NS5B polymerase and HCV replicon activity by 7-deazapurine analogs.
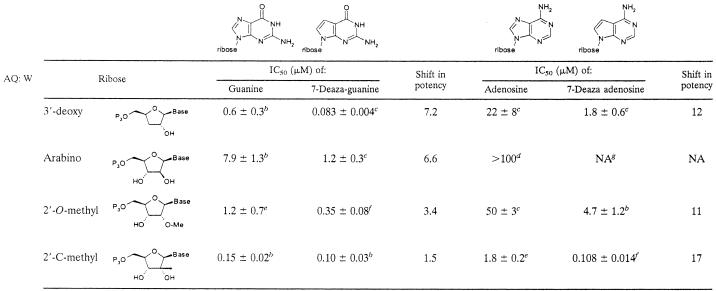
The inhibitory potencies of 15 purine nucleoside triphosphate analogs were determined for HCV RdRp (Table 1). Of the four modified ribose motifs studied, the most potent inhibitors of the polymerase were the 2′-C-methyl-modified sugars. This structure activity relationship (SAR) held whether the base was a purine or a 7-deazapurine. The data also demonstrate that the 7-deaza modification of purines caused a significant improvement in inhibitory potency with each of the ribose modifications examined (Table 1).
TABLE 1.
In vitro IC50 determinations of NTP analogs of interest with HCV NS5BΔ21 in the t500 polymerization assaya
Reported values are a means from two or more independent experiments, and standard deviations were calculated by Microsoft excel XP using the “nonbiased” or “n − 1” method. The shift in potency is calculated as the ratio of the IC50 for the aza-nucleotide to that for the deaza-nucleotide.
n = 3.
n = 2.
Four determinations were conducted. In each experiment the IC50 exceeded 100 μM.
n = 5.
n = 4.
NA, compound not available.
The abilities of both 7-deaza-2′-C-methyl-adenosine and 7-deaza-2′-C-methyl-guanine to inhibit HCV replication was examined with a cell line, HBI10A, that harbors an HCV subgenomic bicistronic replicon (24). Although the corresponding guanosine nucleoside analogs were, in general, more potent in the in vitro enzyme assay than the corresponding adenosine analogs, 7-deaza-2′-C-methyl-guanine was not inhibitory in the HCV replicon assay (Table 2). This was presumably due to a lower level of cell penetration and/or subsequently less-efficient phosphorylation of the guanine analogs (23). We therefore focused our efforts on the characterization of the replicon active inhibitor, 7-deaza-2′-C-methyl-adenosine.
TABLE 2.
Effect of nucleoside analogs on wild-type and S282T replicons and NS5B polymerasesa
| Nucleoside analog | EC50 (μM) for HBI10A | EC50 (μM) for HBI10A-S282T | CC50 (μM) for MTS | IC50 (μM) for WT NS5BΔ55 | IC50 (μM) for S282T NS5BΔ55 |
|---|---|---|---|---|---|
| 7-Deaza-A (Tubercidin) | ≤0.4* | ND | 3 | ND | ND |
| 2′-C-Me-A | 0.25 ± 0.12 | 2.7 ± 0.6 | >100 | 1.9 ± 0.3 | >100 |
| 7-Deaza-2′-C-Me-A | 0.3 ± 0.16 | 10.1 ± 0.6 | >100 | 0.07 ± 0.1 | 25 ± 0.4 |
| 7-Deaza-2′-C-Me-G | >50 | ND | >100 | ND | ND |
The cytotoxic (MTS) and inhibitory effect on replicons was estimated in a 24-h cell culture assay. The effect on NS5BΔ55 was determined in the t500 polymerization assay. All data are averages for at least two experiments. *, toxicity observed at 0.4 μM concentrations and above. ND, not determined. A, adenosine; Me, methyl; G, guanosine.
The potency of inhibition of HCV replicon replication by 7-deaza-2′-C-methyl-adenosine was similar to that of the previously reported 2′-C-methyl-adenosine (Table 2). No cellular toxicity was associated with 7-deaza-2′-C-methyl-adenosine when tested at 24 or 72 h at a 100 μM concentration. In contrast, the des-methyl analog, tubercidin (7-deaza-adenosine), displayed striking cytotoxicity, with CC50s of 3 μM at 24 h and 0.15 μM at 72 h.
Selection and characterization of mutants resistant to 7-deaza-2′-C-methyl-adenosine.
The strategy employed for resistance selection was similar to that described earlier (23). Cell clones resistant to 7-deaza-2′-C-methyl-adenosine were obtained by culturing HBI10A cells in the presence of G418 and compound at concentrations increasing from 4- to 20-fold higher than its EC50. Selection experiments yielded several replicons, all of which expressed HCV RNA and proteins at levels either lower than or comparable to those of parental cells (data not shown). More importantly, replication of the cognate replicons was resistant to inhibition by 7-deaza-2′-C-methyl-adenosine and to other 2′-C-methyl-modified nucleosides, displaying an approximate 10-fold loss of potency compared to parental cell lines. Conversely, clones were still sensitive to inhibition by alpha interferon and to a benzimidazole-based nonnucleoside inhibitor, compound A (data not shown) (33), demonstrating that resistance was specific for 2′-C-methyl-modified nucleosides.
To identify the mutations responsible for resistance, the nucleotide sequence of the NS5B coding region of replicons extracted from four resistant clones was determined and compared to that of the RNA sequence from parental cells. All four resistant clones contained a common mutation, resulting in the change of serine 282 of NS5B to threonine (S282T). This substitution had been previously identified (23) and shown to be responsible for resistance to other 2′-C-methyl-modified nucleosides. To investigate whether the S282T substitution was sufficient for resistance to 7-deaza-2′-C-methyl-adenosine, nucleoside potency, on a replicon carrying this single amino acid change, was determined in a transient replication assay (Table 2). Despite its low replication competency, the modified replicon exhibited significant resistance to 7-deaza-2′-C-methyl-adenosine. The EC50s measured for these replicons were similar to those determined for the clones obtained under selective pressure by 7-deaza-2′-C-methyl-adenosine, suggesting that substitution of serine 282 with threonine was sufficient to recapitulate the resistance phenotype displayed by selected clones.
In vitro resistance to 7-deaza-2′-C-methyl-ATP.
To confirm that the S282T substitution also resulted in resistance to 7-deaza-2′-C-methyl-ATP in vitro, inhibition with S282T NS5BΔ55 was examined (Table 2). Resistance to inhibition by 7-deaza-2′-C-methyl-ATP was engendered by the single amino-acid change, as evidenced by a 350-fold loss in potency compared to the WT enzyme. Although resistance was displayed, incorporation of the 7-deaza motif into 2′-C-methyl-ATP also resulted in an improved potency against the mutant enzyme compared to the parental analog, 2′-C-methyl-ATP. This suggests that the increase in potency observed for 7-deaza-modified compounds is independent of the S282T change.
Mode of inhibition.
Inhibition of polymerase activity by 7-deaza-2′-C-methyl-ATP was competitive with respect to substrate ATP. The Ki value, as determined from a replot of the slopes of the double reciprocal plot, was 0.024 μM (data not shown), >30-fold lower than the Ki value obtained for 2′-C-methyl-ATP (11).
Gel-based incorporation assay.
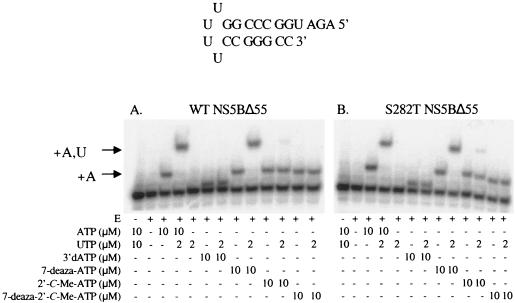
The mechanism of inhibition of 7-deaza-2′-C-methyl-ATP was further investigated using a gel-based incorporation-and-extension assay (Fig. 1). In this assay, NS5BΔ55-catalyzed polymerization can be visualized using a 5′-32P RNA template capable of incorporating AMP, followed by incorporation of UMP in a subsequent extension step. Tubercidin monophosphate, 2′-C-methyl-AMP, and 7-deaza-2′-C-methyl-AMP are incorporated into the growing RNA strand opposite uridine in reactions catalyzed by NS5BΔ55. Tubercidin monophosphate, once incorporated, can support further extension by the incorporation of UMP. In contrast, both 2′-C-methyl-AMP and 7-deaza-2′-C-methyl-AMP are incorporated into the growing RNA strand, but the HCV RdRp is inefficient at catalyzing the addition of the next nucleotide. Therefore, 2′-C-methyl-ATP and 7-deaza-2′-C-methyl-ATP act as functional chain terminators. These results indicate that the ability of these nucleoside analogs to function as chain terminators is due to the 2′-C-methyl substituent and not the 7-deaza modification. As reported previously (23), a trace of product resulting from the extension of incorporated 2′-C-methyl-AMP by the WT enzyme is evident (Fig. 1A, lane 11) and is increased in reactions catalyzed by S282T (Fig. 1B, lane 11). In contrast, no extension of the incorporated 7-deaza-2′-C-methyl-AMP is detected in reactions catalyzed by either WT or S282T (Fig. 1A and B, respectively, lane 13).
FIG. 1.
PhosphorImager analysis of the incorporation and extension of nucleotide analogs by HCV WT NS5BΔ55 and S282T NS5BΔ55. Reactions included 600 nM 5′-32P-end-labeled RNA template (sequence shown as a hairpin configuration) and 1 μM WT or S282T NS5BΔ55 in reaction buffer as described in Methods. The first lane of both A and B is a control with no enzyme added. Reactions included nucleoside triphosphates, as indicated in the figure. The arrows indicate the positions of products corresponding to the addition of one AMP (or an adenosine analog) or the addition of one AMP and one UMP to the RNA template.
Antiviral activity and specificity of polymerase inhibition by 7-deaza-nucleosides.
7-Deaza-2′-C-methyl-adenosine displayed significant antiviral activity against other positive-strand-RNA viruses belonging to the Flaviviridae and Picornaviridae families, including BVDV, West Nile virus, dengue virus type 2, yellow fever virus, rhinovirus type 2, rhinovirus type 14, and poliovirus type 3, but none against the more distantly related positive-stranded-RNA viruses western equine encephalitis virus and Venezuelan equine encephalitis virus (Table 3). In addition, 7-deaza-2′-C-methyl-adenosine did not have antiviral activity against several minus-stranded-RNA and double-stranded-DNA viruses, including respiratory syncytial virus, measles virus, herpesvirus, and human cytomegalovirus. 7-Deaza-2′-C-methyl-ATP exhibited an IC50 of 3.7 μM against BVDV polymerase, a value 10-fold lower than that for 2′-C-methyl-ATP and consistent with the shift observed for HCV NS5B.
TABLE 3.
Antiviral activity of 7-deaza-2′-C-methyl-adenosine against viruses encoding phylogenetically related polymerases
| Virusa | Result for compound
|
Antiviralb activity | |
|---|---|---|---|
| EC50 (μM) | CC50 (μM) | ||
| (+) Strand RNA viruses | |||
| Flaviviridae | |||
| BVDV1 | 0.3 | >50 | Y |
| West Nile virus2a | 4.5 | 250 | Y |
| Dengue virus type 22a | 15 | >320 | Y |
| Yellow fever virus2a | 15 | >320 | Y |
| Picornaviridae | |||
| Rhinovirus type 23a | 0.5 | >100 | Y |
| Rhinovirus type 144a | 6 | >200 | Y |
| Poliovirus type 32a | 5.9 | >200 | Y |
| Togaviridae | |||
| Western equine encephalitis virus2a | 200 | >320 | N |
| Venezuelan equine encephalitis virus2a | >320 | >320 | N |
| (−) Strand RNA viruses | |||
| Paramyxoviridae | |||
| Respiratory syncytial virus5a | >320 | >320 | N |
| Measles virus6a | >320 | 180 | N |
| dsDNA Viruses | |||
| Herpesviridae | |||
| Herpes virus type 12a^ | >320 | >320 | N |
| Herpes virus type 22a^ | >320 | >320 | N |
| Human CMV7a′ | 50 | >320 | N |
Antiviral assays were performed in triplicate in the following cell lines: 1, Madin-Darby bovine kidney; 2, African green monkey kidney (Vero); 3, human oral epidermoid carcinoma (KB); 4, human cervical epitheloid carcinoma (HeLa Ohio-1); 5, embryonic African green monkey kidney (MA-104); 6, African green monkey kidney (CV-1); 7, Human embryonic lung (MRC-5). Superscript “a,” antiviral assays were performed at Utah State University as described in “Methods,” and reported values are neutral red results, visual CPE (^), or plaque reduction (′).
Antiviral activity obtained from CC50/EC50. Y, yes; N, no.
Interestingly, 7-deaza-2′-C-methyl-ATP had no effect on the activity of human DNA polymerases α, β, and γ, human RNA polymerase II, and yeast poly(A) polymerase in vitro when tested up to a 50 μM concentration.
Intracellular metabolism.
The intracellular metabolism of 7-deaza-2′-C-methyl-adenosine was determined in cell culture experiments by incubating compound radiolabeled with tritium at the 5-position of the pyrrolopyrimidine ring with both Huh-7 and HBI10A cell lines for 3 and 23 h. The metabolism of tritiated adenosine was monitored in parallel for comparison. The efficiency of uptake of 7-deaza-2′-C-methyl-adenosine is significantly less than that of adenosine, yet the ratio of conversion from the nucleoside to the triphosphate remains similar in the two cell lines, suggesting that the intracellular phosphorylation efficiency is not adversely affected by the 7-deaza and 2′-C-methyl modifications of adenosine (Table 4). In addition, the ratio of conversion of 7-deaza-2′-C-methyl-adenosine to the triphosphate remained constant at the 3- and 23-h time points, resulting in an overall increase in the total concentration of triphosphate with time, while the total concentration of radiolabeled intracellular ATP decreased from 3 to 23 h.
TABLE 4.
Intracellular metabolism of 7-deaza-2′-C-methyl-adenosine and adenosine in cell culturea
| Analog administered | Intracellular metabolite(s) detected | Amt of metabolite
|
|||
|---|---|---|---|---|---|
| Huh-7
|
HB-1
|
||||
| 3 h | 23 h | 3 h | 23 h | ||
| 7-Deaza-2′-C-methyl-adenosine | Total drug | 2.8 | 5.6 | 3.7 | 10 |
| Triphosphate | 1.7 | 3.4 | 2.2 | 6.3 | |
| Triphosphate/total drugb | 0.61 | 0.61 | 0.59 | 0.63 | |
| Adenosine | Total drug | 290 | 130 | 260 | 290 |
| Triphosphate | 190 | 63 | 160 | 150 | |
| Triphosphate/total drugb | 0.66 | 0.48 | 0.62 | 0.52 | |
Two micromolar nucleoside was incubated for 3 or 23 h on Huh-7 or HB-1, and the intracellular components were analyzed by high-performance liquid chromatography. Reported values represent picomoles/million cells and are averages for at least two separate experiments.
Ratio of triphosphate concentration to total drug concentration.
Pharmacokinetics.
7-Deaza-2′-C-methyl-adenosine exhibited good oral bioavailability in male beagle dogs, Sprague-Dawley rats, and rhesus macaques (Table 5). The rate of compound clearance was moderate in all three animal species. The elimination half-life was 1.6 h in the rat but was significantly longer in monkeys and dogs, consistent with allometric scaling predictions. The concentration of 7-deaza-2′-C-methyl-adenosine and related material in liver was determined for male Sprague-Dawley rats dosed orally at 2, 20, and 200 mg/kg (Table 6). Plasma and liver concentrations were dose proportional at 2 and 20 mg/kg. However, both the plasma and liver concentrations for the rodents dosed at 200 mg/kg were approximately one-half of the values that would be obtained if a linear relationship were maintained at all dosing levels.
TABLE 5.
Pharmacokinetic properties of 7-deaza-2′-C-methyl-adenosine in male beagle dogs, Sprague-Dawley rats, and rhesus macaques dosed orally at 2 mg/kg and intravenously at 1 mg/kg
| Parametera | Value for animal
|
||
|---|---|---|---|
| Rhesus | Rat | Dog | |
| Clp (ml/min/kg) | 15 | 25 | 6.2 |
| t1/2 (h) | 9.0 | 1.6 | 14 |
| CmaxOral (μM) | 0.3 | 1.2 | 5.0 |
| TmaxOral (h) | 2.0 | 0.5 | 0.5 |
| F % | 51 | 51 | 98 |
Clp, plasma clearance; t1/2, elimination half-life; CmaxOral, maximum concentration of orally administered drug in plasma; TmaxOral, time to maximum concentration of orally administered drug in plasma; F %, percent oral bioavailability.
TABLE 6.
Liver concentrations of 7-deaza-2′-C-methyl-ATP and plasma concentrations of 7-deaza-2′-C-methyl-adenosine in male Sprague-Dawley rats dosed orally at 2, 20, and 200 mg/kg
| Dose (mg/kg) | Time (h) | Concn in liver (μmol/kg) | Concn in plasma (μM) | Liver/plasma ratioa |
|---|---|---|---|---|
| 2 | 8 | 11.93 | 0.100 | 119.3 |
| 24 | 1.26 | 0.015 | 82.0 | |
| 20 | 8 | 92.29 | 1.018 | 90.7 |
| 24 | 14.81 | 0.176 | 83.9 | |
| 200 | 8 | 506.62 | 4.781 | 106.0 |
| 24 | 75.64 | 0.772 | 98.0 |
Ratio of concentration of 7-deaza-2′-C-methyl-ATP in liver to concentration of 7-deaza-2′-C-methyl adenosine in plasma.
Cellular toxicity.
The effect of high concentrations of nucleoside analogs on cell viability and proliferation was determined by using two different measurements of cytotoxicity. An MTS assay in a Jurkat T-cell line and an assay which determines the rate of incorporation of [14C]thymidine into the nucleic acid of compound-treated Huh-7 cells revealed a cytotoxic profile for 7-deaza-2′-C-methyl-adenosine wherein high concentrations of compound (0.8 to 1 mM) and a 72-h incubation were required before significant cytotoxicity was detected (Table 7). Of the analogs tested, 7-deaza-2′-C-methyl-adenosine was the least toxic, while the closely related compound tubercidin (7-deaza-adenosine) displayed striking toxicity at submicromolar concentrations at all time points in both assay formats. Another closely related compound, 2′-C-methyl-adenosine, displayed a 30-fold-lower CC50 in the [14C]thymidine uptake assay after a 72-h incubation than 7-deaza-2′-C-methyl-adenosine. These results suggest that the CC50 of 7-deaza-2′-C-methyl-adenosine is well above the EC50 of the compound (>2,000-fold higher than replicon EC50 at 24 h).
TABLE 7.
Cytotoxicity of nucleoside analogs in cell culture as measured using MTS or [14C]thymidine uptake
| Nucleoside analoga | Jurkat MTS CC50 (μM)
|
Huh-7 [14C]thymidine uptake CC50 (μM)
|
||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| 7-Deaza-2′-C-methyl-A | >1,000 | >1,000 | ∼1,000 | >1,000 | >1,000 | ∼800 |
| 7-Deaza-A | 0.125 | 0.029 | 0.014 | 0.086 | 0.046 | |
| 2′-C-methyl-A | >100 | >100 | >100 | >100 | >100 | 26 |
A, adenosine.
RNA incorporation.
The extent of nucleoside incorporation into total RNA isolated from Huh-7 cells following treatment for 3 days with 0.5 μM [3H]adenosine, [3H]tubercidin (7-deaza-adenosine), [3H]acyclovir, [3H]ddI, or [3H]7-deaza-2′-C-methyl-adenosine was determined (Table 8). Adenosine and tubercidin were efficiently incorporated into RNA. Incorporation of ddI was approximately 15-fold lower than that of adenosine, while acyclovir and 7-deaza-2′-C-methyl-adenosine showed minimal incorporation. The lack of incorporation displayed by 7-deaza-2′-C-methyl-adenosine provides further evidence that the incorporation and subsequent chain termination of the corresponding monophosphate is specifically catalyzed by viral polymerases and in particular by HCV RdRp.
TABLE 8.
Incorporation of tritiated analogs into cellular RNAa
| Nucleoside analog | Incorporation (pmol of cmpd/mg of RNA) | Mol of endogenous nt per mol of incorporated cmpd |
|---|---|---|
| [7-3H]-7-deaza-2′-C-methyl-A | 3.8 | 720,000 |
| [2-3H]adenosine | 5,800 | 470 |
| [8-3H]acyclovir | 14 | 200,000 |
| [2′,3′-3H]2′,3′-dideoxyinosine | 390 | 7,000 |
| [8-3H]7-deaza-A (Tubercidin) | 2,480 | 1,100 |
| (−) Control | 0.016 | 1.7 × 108 |
cmpd, compound; nt, nucleotides; A, adenosine; (−) control, negative control.
Acute oral toxicity study with mice.
The approximate oral 50% lethal dose (LD50), following a 14-day observation period, of 7-deaza-2′-C-methyl-adenosine when administered as a single dose to female mice was greater than 2,000 mg/kg. No deaths or treatment-related physical signs were seen throughout the duration of the 14-day study. The three mice dosed with 50 mg of 7-deaza-adenosine (tubercidin)/kg died on days 3, 5, and 6. Prior to death, animals showed urine staining, decreased activity, ptosis, and/or an appearance of being hunched and unkempt. No deaths occurred in mice dosed with 20 mg of tubercidin/kg, and they appeared normal throughout the 14-day study.
DISCUSSION
The current therapeutic regimen for HCV consists of a combination of pIFN-α and ribavirin, which achieves a sustained viral response of less than 50% in most patients (25). Although the exact mechanism of action of ribavirin is still under investigation, neither it nor pIFN-α is believed to function as a direct antiviral agent against viral proteins. One approach to the discovery of superior therapies for treatment of HCV infection includes developing direct inhibitors of virally encoded enzymes, such as HCV RdRp. Previous work had identified a class of 2′-C-methyl-modified nucleosides as potent inhibitors of HCV replication that act as chain terminators. The lead compound in the series, 2′-C-methyl-adenosine, suffered from poor pharmacokinetic properties that prevented its further development.
The initial SAR that was developed by examining nucleoside triphosphates in the polymerase assay pointed to improvements in inhibitory potency with a 7-deaza substitution of the purine ring. This increase in potency was displayed by analogs of both ATP and GTP and was first observed with 3′-deoxy-, arabino-, and 2′-O-methyl-modified nucleoside triphosphates. The SAR was subsequently extended to 2′-C-methyl-ATP and 2′-C-methyl-GTP (11, 16, 23) with the result that in vitro potency was significantly improved.
Initial examination of the biological potential of 7-deaza-2′-C-methyl-adenosine focused on its toxicity and selectivity. This was of significant concern, since the closely related des-2′-methyl derivative of this lead compound, tubercidin, is very toxic (2, 29). Although 7-deaza-2′-C-methyl-adenosine did not result in detectable cytotoxicity in the replicon assay when assayed up to a 20 μM concentration for 24 h, additional testing at higher compound concentrations and for longer time periods was performed to further define the selective index. For comparative purposes, Jurkat cells, which are more sensitive to the cytotoxic effects of these compounds than Huh-7 cells, were incubated with up to 1 mM 2′-C-methyl-adenosine, 7-deaza-2′-C-methyl-adenosine, and tubercidin. The results revealed a striking difference in MTS cytotoxicity between 7-deaza-2′-C-methyl-adenosine (CC50 of >1 mM) and tubercidin (CC50 of ≪1 μM) (Table 7), which demonstrates the ability of the 2′-C-methyl modification to obviate cytotoxicity. A similar observation with other 2′-C modifications of tubercidin (13) has been reported. A second study examined the effect of the compound on the uptake and incorporation of [14C]thymidine into Huh-7 cells. These data paralleled the MTS cytotoxicity data, with measurable decreases in incorporation being detected only at very high micromolar (800 uM) concentrations of 7-deaza-2′-C-methyl-adenosine at the 72-h time point. In contrast, the des-2′-C-methyl derivative, tubercidin, exhibited demonstrable cytotoxicity at submicromolar concentrations, and 2′-C-methyl-adenosine was cytotoxic at 26 uM.
Tubercidin undergoes intracellular incorporation into nucleic acids and can yield cellular toxicity by inhibiting a number of metabolic processes, including RNA processing, nucleic acid synthesis, protein synthesis, and methylation of tRNA (2, 8, 29). It was therefore a concern that 7-deaza-2′-C-methyl-adenosine may be readily incorporated into RNA and could result in delayed cytotoxicity. Our data, shown in Table 8, confirm that tubercidin is readily incorporated into Huh-7 cellular RNA with an efficiency close to that for adenosine itself. In contrast, incorporation of 7-deaza-2′-C-methyl-adenosine into RNA occurred demonstrably less frequently than that of the two dideoxy control compounds, acyclovir and ddI. (It should be noted that the level of radiolabel incorporated in RNA for 7-deaza-2′-C-methyl-adenosine, ddI and acyclovir was very low and could be a result of radiochemical impurities in the nucleoside preparation.) This lack of incorporation displayed by 7-deaza-2′-C-methyl-adenosine provides further evidence that the polymerase-catalyzed incorporation of the corresponding monophosphate appears to be very specific for HCV NS5B polymerase. In the same experiment, an extremely small amount (below the limits of accurate quantitation) of radiolabel was found to be incorporated in DNA when [3H]7-deaza-2′-C-methyl-adenosine was placed on cells. This is consistent with previous reports on the selectivity of human DNA polymerases for 2′-deoxyribose nucleoside triphosphates (6, 17, 35) and that 2′-C-methyl-modified ribonucleoside diphosphates are not substrates for class I ribonucleotide reductase (22).
Adenosine and its corresponding phosphorylated species are involved in many cellular processes, and therefore, adenosine analogs have the potential to exhibit significant toxicity in vivo. Hence, in addition to the cellular analysis of cytotoxicity described above, the in vivo toxic potential of the lead compound was further evaluated. Tubercidin is lethal to mice, with an acute LD50 of ∼30 mg/kg, which is consistent with the previous data reported by Smith et al. (29). In contrast, the corresponding 2′-C-methyl derivative does not display observable toxicity when dosed at 2,000 mg/kg, the maximum dose tested. One of the main causes of the toxicity observed with tubercidin is believed to be its incorporation into cellular nucleic acids. Since significant incorporation into nucleic acids is not observed with 7-deaza-2′-C-methyl-adenosine and lethality for mice was not observed at concentrations >60-fold higher than the acute LD50 of tubercidin, it appears likely that the 2′-C-methyl-modified nucleoside is a very poor substrate for cellular enzymes involved in processes that lead to tubercidin-like toxicity.
The 7-deaza-2′-C-methyl-adenosine lead compound was tested against a diverse panel of viruses, and the data (Table 3) provide the following conclusions. First, antiviral selectivity of the compound is not limited to HCV, since it displays significant antiviral activity against several related and medically relevant plus-stranded-RNA viruses. This is not a surprising result, since sequence analyses demonstrate that RdRps are highly conserved among plus-strand-RNA viruses (20). Second, 7-deaza-2′-C-methyl-adenosine is a bona fide viral replication inhibitor, since it is active in true viral infectivity assays. Finally, the testing of the compound against this broad set of viruses also yielded a panel of toxicity data for seven different mammalian cell lines. Although these assays were of significant duration (≥3 days), 7-deaza-2′-C-methyl-adenosine yielded CC50s above the highest concentration tested, which, in most cases, was several hundred micromolar (Table 3). This is in contrast to the low-micromolar-concentration-toxicity data observed previously for 2′-C-methyl-adenosine in similar assays (23).
The cytotoxicity and antiviral selectivity data strongly suggest that the inhibition seen in the replicon assay is specific for HCV antiviral activity and is not a result of underlying toxicity. This is strengthened by correlation of 7-deaza-2′-C-methyl-adenosine resistance mutations which map to NS5B polymerase. Resistance selection with 2′-C-methyl-adenosine previously yielded a single serine-to-threonine active-site NS5B amino acid change at position 282 (23). It is not surprising that selection with 7-deaza-2′-C-methyl-adenosine yielded the same resistance mutation. As expected, 7-deaza-2′-C-methyl-adenosine (or its triphosphate) demonstrated significant shifts in inhibitory potency in assays with the resistant replicon (34-fold) and in enzyme assays with S282T NS5B RdRp (350-fold) (Table 2).
The incorporation of the modified nucleosides into the RNA hairpin primer/template substrate was examined in gel-based assays. The data in Fig. 1 demonstrate that the 7-deaza modification itself does not result in a chain-terminating nucleoside, since tubercidin monophosphate is readily incorporated and extended by the polymerase. In addition, the data demonstrate that a nucleoside with a single 2′-C-methyl-modified ribose does not result in complete chain termination (see Fig. 1, lanes for 2′-C-methyl-ATP). However, when the 7-deaza and 2′-C-methyl modifications are incorporated into one molecule, chain termination with the triphosphate of this analog is more efficient. Although the 7-deaza modification is a fairly conservative substitution, it is known to alter the glycosyl torsion angle, which can significantly shorten the glycosyl bond length (1). We speculate that this additional change in the molecule, combined with the 2′-C-Me, which has previously been suggested to sterically block the next incoming NTP (23), further disrupts the alignment of the 3′-OH for nucleophilic attack on the alpha-phosphorous of the next incoming NTP. The S282T mutant enzyme more readily extends the 2′-C-methyl-AMP-terminated primer, whereas both the WT and mutant enzymes are unable to extend the 2′-C-methyl-7-deaza-AMP-terminated primer under the conditions of this in vitro assay.
The pharmacokinetic parameters of 2′-C-methyl-7-deaza-adenosine were determined to further evaluate its potential as a therapeutic antiviral agent. 2′-C-methyl-adenosine was previously shown to lack oral bioavailability (23). In contrast, when rats were dosed with 2′-C-methyl-7-deaza-adenosine, a dramatic improvement in half-life and oral bioavailability was observed relative to results with 2′-C-methyl-adenosine. Excellent oral bioavailability and half-life were also observed in beagle dogs and rhesus monkeys. In addition, there was excellent exposure of compound in the rat liver, yielding a fairly consistent liver-to-plasma ratio of approximately 100 irrespective of dose and time. Thus, oral dosing of 7-deaza-2′-C-methyl-adenosine results in significant concentrations of the compound in the target organ, liver (30).
We speculated that the rapid plasma clearance observed for 2′-C-methyl-adenosine could be due to rapid deamination of the molecule by adenosine deaminase and/or cleavage by adenosine phosphorylase (5). The 7-deaza modification of tubercidin renders this compound inert to metabolism by adenosine deaminase and phosphorylase (5). It is therefore not surprising that the 7-deaza derivative of 2′-C-methyl-adenosine has better pharmacokinetic properties than 2′-C-methyl-adenosine itself.
In summary, a novel nucleoside analog inhibitor of HCV NS5B polymerase has been identified. The combination of the highly specific chain-terminating 2′-C-methyl-ribose modification and the chemically stable 7-deaza-adenosine has created a direct antiviral compound with promising therapeutic potential. The previously reported analog, 2′-C-methyl-adenosine, was a potent and specific inhibitor of NS5B but lacked appealing pharmacokinetics for further development. The incorporation of the 7-deaza motif has led to an improvement in potency against the enzyme and improved pharmacokinetics, while the 2′-C-methyl modification ameliorates toxicity that is associated with tubercidin. This combination has afforded an inhibitor that will be further investigated as a much-needed antiviral agent for HCV infection.
The genetic heterogeneity of HCV and the potential for the development of resistance will be important factors in the outcome for investigational compounds for treatment of HCV infection. Combination therapies have been successfully employed to reduce the emergence of resistance to HIV treatments and are likely to become an important part of HCV treatments. Therefore, it is expected that compounds like 2′-C-methyl-7-deaza-adenosine will ultimately be used as part of a combination therapy including existing therapeutic agent alpha interferon or ribavirin.
Acknowledgments
We thank Robert Sidwell and his team for advice and performing the non-HCV antiviral assays and B. Wolanski, Carrie A. Rutkowski, and Amy Simcoe for expert technical assistance.
REFERENCES
- 1.Abola, J., and M. Sundaralingam. 1973. Refinement of the crystal structure of tubercidin. Acta Crystallogr. B Struct. Crystallogr. Cryst. Chem. 29:697-703. [Google Scholar]
- 2.Acs, G., E. Reich, and M. Mori. 1964. Biological and biochemical properties of the analogue antibiotic tubercidin. Proc. Natl. Acad. Sci. USA 52:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, D. L., V. D. Stowell, K. L. Seley, V. R. Hegde, S. R. Das, V. P. Rajappan, S. W. Schneller, D. F. Smee, and R. W. Sidwell. 2001. Inhibition of measles virus replication by 5′-nor carbocyclic adenosine analogues. Antivir. Chem. Chemother. 12:241-250. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch, A., R. J. Leonard, and C. A. Nichol. 1967. On the mode of action of 7-deaza-adenosine (tubercidin). Biochim. Biophys. Acta 138:10-25. [DOI] [PubMed] [Google Scholar]
- 6.Bonnin, A., J. M. Lazaro, L. Blanco, and M. Salas. 1999. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J. Mol. Biol. 290:241-251. [DOI] [PubMed] [Google Scholar]
- 7.Boudou, V., L. Kerremans, B. De Bouvere, E. Lescrinier, G. Schepers, R. Busson, A. Van Aerschot, and P. Herdewijn. 1999. Base pairing of anhydrohexitol nucleosides with 2,6-diaminopurine, 5-methylcytosine and uracil as base moiety. Nucleic Acids Res. 27:1450-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco, L., and D. Vazquez. 1984. Molecular bases for the action and selectivity of nucleoside antibiotics. Med. Res. Rev. 4:471-512. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. S., R. L. Lafemina, D. L. Hall, A. L. Himmelberger, L. C. Kuo, M. Maccoss, D. B. Olsen, C. A. Rutkowski, J. E. Tomassini, H. An, B. Bhat, N. Bhat, P. D. Cook, A. B. Eldrup, C. J. Guinosso, M. Prhavc, and T. P. Prakash. August 2002. Preparation of nucleoside derivatives as inhibitors of RNA-dependent RNA viral polymerase. U.S. patent 6777395.
- 10.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 11.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. Recomm. Rep. 47:1-39. [PubMed] [Google Scholar]
- 13.Cory, A. H., V. Samano, M. J. Robins, and J. G. Cory. 1994. 2′-Deoxy-2′-methylene derivatives of adenosine, guanosine, tubercidin, cytidine and uridine as inhibitors of L1210 cell growth in culture. Biochem. Pharmacol. 47:365-371. [DOI] [PubMed] [Google Scholar]
- 14.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Division of Viral Hepatitis, National Center for Infectious Diseases, Centers for Disease Control and Prevention,. 2003. Viral hepatitis C: fact sheet. [Online.] www.cdc.gov/ncidod/diseases/hepatitis/c/fact.htm
- 16.Eldrup, A. B., C. R. Allerson, C. F. Bennett, S. Bera, B. Bhat, N. Bhat, M. R. Bosserman, J. Brooks, C. Burlein, S. S. Carroll, P. D. Cook, K. L. Getty, M. MacCoss, D. R. McMasters, D. B. Olsen, T. P. Prakash, M. Prhavc, Q. Song, J. E. Tomassini, and J. Xia. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 47:2283-2295. [DOI] [PubMed] [Google Scholar]
- 17.Gardner, A. F., and W. E. Jack. 1999. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 27:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274:57-71. [DOI] [PubMed] [Google Scholar]
- 19.Huffman, J. H., R. W. Sidwell, G. P. Khare, J. T. Witkowski, L. B. Allen, and R. K. Robins. 1973. In vitro effect of 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole, ICN 1229) on deoxyribonucleic acid and ribonucleic acid viruses. Antimicrob. Agents Chemother. 3:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 21.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 22.McFarlan, S. C., S. P. Ong, and H. P. Hogenkamp. 1996. Mechanism-based inhibition of ribonucleoside diphosphate reductase from Corynebacterium nephridii by 2′-C-methyladenosine diphosphate. Biochemistry 35:4485-4491. [DOI] [PubMed] [Google Scholar]
- 23.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain terminating ribonucleoside analogs which inhibit HCV replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 24.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 25.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 362:2095-2100. [DOI] [PubMed] [Google Scholar]
- 26.Shiffman, M. L. 2003. Natural history and risk factors for progression of hepatitis C virus disease and development of hepatocellular cancer before liver transplantation. Liver Transpl. 9:S14-S20. [DOI] [PubMed] [Google Scholar]
- 27.Sidwell, R. W., and J. H. Huffman. 1971. Use of disposable micro tissue culture plates for antiviral and interferon induction studies. Appl. Microbiol. 22:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 29.Smith, C. G., G. D. Gray, R. G. Carlson, and A. R. Hanze. 1967. Biochemical and biological studies with tubercidin (7-deaza-adenosine), 7-deazainosine and certain nucleotide derivatives of tubercidin. Adv. Enzyme Regul. 5:121-151. [DOI] [PubMed] [Google Scholar]
- 30.Szabo, E., G. Lotz, C. Paska, A. Kiss, and Z. Schaff. 2003. Viral hepatitis: new data on hepatitis C infection. Pathol. Oncol. Res. 9:215-221. [DOI] [PubMed] [Google Scholar]
- 31.Tomassini, J. E., E. Boots, L. Gan, P. Graham, V. Munshi, B. Wolanski, J. F. Fay, K. Getty, and R. LaFemina. 2003. An in vitro Flaviviridae replicase system capable of authentic RNA replication. Virology 313:274-285. [DOI] [PubMed] [Google Scholar]
- 32.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomei, L., Sergio Altamura, Linda Bartholomew, Antonino Biroccio, Alessandra Ceccacci, Laura Pacini, Frank Narjes, Nadia Gennari, Monica Bisbocci, Ilario Incitti, Laura Orsatti, Steven Harper, Ian Stansfield, Michael Rowley, Raffaele De Francesco, and Giovanni Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA dependent RNA polymerase. submitted to J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, G., M. Franklin, J. Li, T. C. Lin, and W. Konigsberg. 2002. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry 41:10256-10261. [DOI] [PubMed] [Google Scholar]