Summary
Fungal secondary metabolites (SMs) are extremely important in medicine and agriculture, but regulation of their biosynthesis is incompletely understood. We have developed a genetic screen in Aspergillus nidulans for negative regulators of fungal SM gene clusters and we have used this screen to isolate mutations that upregulate transcription of the non-ribosomal peptide synthetase gene required for nidulanin A biosynthesis. Several of these mutations are allelic and we have identified the mutant gene by genome sequencing. The gene, which we designate mcrA, is conserved but uncharacterized, and it encodes a putative transcription factor. Metabolite profiles of mcrA deletant, mcrA overexpressing, and parental strains reveal that mcrA regulates at least ten SM gene clusters. Deletion of mcrA stimulates SM production even in strains carrying a deletion of the SM regulator laeA, and deletion of mcrA homologs in Aspergillus terreus and Penicillum canescens alters the secondary metabolite profile of these organisms. Deleting mcrA in a genetic dereplication strain has allowed us to discover two novel compounds as well as an antibiotic not known to be produced by A. nidulans. Deletion of mcrA upregulates transcription of hundreds of genes including many that are involved in secondary metabolism, while downregulating a smaller number of genes.
Keywords: secondary metabolism, natural products, gene regulation, transcription, Aspergillus, fungi
Abbreviated summary
Using a new genetic screen, we have identified a conserved but uncharacterized gene that negatively regulates secondary metabolite production in the filamentous fungus Aspergillus nidulans. Deletion of the gene increases production of a large number of secondary metabolites including novel compounds and it alters the transcription of a large number of genes including many involved in secondary metabolite biosynthesis. Deletions of this gene and its homologs may be valuable tools for secondary metabolite discovery.
Graphical Abstract
Introduction
Fungal secondary metabolites (SMs) are a remarkably rich source of compounds with potential or actual medical value. They include, but are not limited to, antibiotics such as penicillin and cephalosporin, hypercholesterolaemic agents such as lovastatin, immunosuppressants such as cyclosporin, as well as anti-fungals such as echinocandin and derivatives [reviewed in (Martin, 1998; Palaez, 2004; Keller et al., 2005; Bok et al., 2006; Hoffmeister and Keller, 2007)]. It is not just the final products of SM pathways that are valuable, moreover. The final products suit the need of the producing organism, but intermediates in SM pathways may be more valuable to humans. Their chemistry is often more flexible than that of the final products, and it may be possible to modify intermediates to produce valuable compounds [e.g. (Somoza et al., 2012)].
Fungal SMs also include compounds that are important toxins. For example, the carcinogenic toxins aflatoxin and sterigmatocystin are produced by members of the genus Aspergillus [(Hajjar et al., 1989; Brown et al., 1996; Yu et al., 2004b; Yu et al., 2004a), reviewed in (Walton, 1996; Bhatnagar et al., 2002)]. They contaminate the food chain, causing cancer and food wastage and they sometimes occur at high enough concentrations to kill humans or animals directly (Cullen and Newberne, 1993). Other fungal secondary metabolites also contribute to human pathogenesis [e.g. (Gardiner et al., 2005; Fox and Howlett, 2008b; Hoffmeister and Keller, 2007; Kwon-Chung and Sugui, 2009)].
Extensive cloning and sequencing has revealed that, in fungi, genes that encode the proteins of a particular SM pathway are generally clustered together in the genome [reviewed in (Keller et al., 2005; Galagan et al., 2003; Hoffmeister and Keller, 2007; Brakhage and Schroeckh, 2011)]. The sequencing of fungal genomes has also revealed that in most cases the number of SM biosynthetic gene clusters (BGCs) in the genomes far outnumbers the number of SMs known to be produced by the species (Galagan et al., 2003; Nierman et al., 2005; Machida et al., 2005; Khaldi et al., 2010; Sanchez et al., 2012b; Andersen et al., 2013; Brakhage, 2013; Chiang et al., 2014; Yaegashi et al., 2014). This is because most fungal SM BGCs are not expressed under normal laboratory growth conditions. The full exploitation of the greater fungal secondary metabolome will require that we be able to obtain expression of heretofore-silent SM BGCs.
Given the importance of fungal SMs, the need for activating fungal SM clusters to exploit the fungal secondary metabolome and the need to prevent production of toxic fungal SMs such as aflatoxins, it is very important to understand the regulation of fungal SM genes. A great deal has been achieved (Keller et al., 2005; Yu and Keller, 2005; Keller et al., 2006; Perrin et al., 2007; Bok et al., 2006; Calvo, 2008; Hoffmeister and Keller, 2007; Fox and Howlett, 2008a; Hertweck, 2009; Georgianna et al., 2010; Yin and Keller, 2011; Sanchez et al., 2012b; Bayram and Braus, 2012; Brakhage, 2013). In particular, a great deal of progress has been made in understanding the veA/velB/laeA complex, a complex that is present in many fungi and is involved in the regulation of development and the transcription of a large number of SM gene clusters [reviewed in (Bayram and Braus, 2012)] and new components of this system are still being discovered (Ramamoorthy et al., 2013).
In spite of these advances and other advances involving replacement of native promoters with regulatable promoters, we still do not know how to activate the majority of the SM clusters even in a well-studied fungus such as Aspergillus nidulans (Yaegashi et al., 2014). There are clearly important aspects of fungal SM regulation that remain to be determined. Much of the previous work in identifying regulators of fungal SM has been directed toward identifying positive regulators, genes for which increased expression would lead to activation of silent SM clusters or greater expression of weakly expressed clusters. There is ample evidence that there are negative regulators of SM clusters, however, and that inactivation of such genes can result in the activation of SM clusters (Tsitsigiannis and Keller, 2006; Sprote and Brakhage, 2007; Reverberi et al., 2007; Hoffmeister and Keller, 2007; Bok et al., 2009; Reverberi et al., 2012). We have developed a genetic screen for identifying negative regulators in A. nidulans. Using a combination of genetics, molecular genetics and genomic sequencing, we have identified mutations that activate expression of a target non-ribosomal peptide synthetase (NRPS) gene. The mutations are in AN8694, a previously uncharacterized gene that has strong homologs in many fungi and is predicted to encode a zinc-finger transcription factor. We have deleted the gene and found that the deletion upregulates the production of several secondary metabolites, while inducing overexpression of the gene with a regulatable or strong constitutive promoter results in suppression of SM production. This gene is, thus, a potent, negative regulator of A. nidulans SM production. We assign the gene designation mcrA (multicluster regulator A) to AN8694. Deletion of mcrA in a multicluster deletion (genetic dereplication) strain we have constructed (Chiang et al., 2016) allowed us to identify two novel compounds and determine that they are produced by the cichorine biosynthetic pathway. We were also able to determine that A. nidulans produces felinone A, a recently discovered antibiotic (Du et al., 2014) and to propose a biosynthetic pathway for this compound. Deletion of mcrA homologs in Aspergillus terreus and Penicillium canescens alters SM production in these organisms, stimulating production of compounds not produced by wild-type strains cultured under the same conditions. These data suggest that deletions of mcrA homologs in fungi are promising tools for compound discovery. We have compared transcription profiles of a strain carrying the deletion of mcrA to its mcrA+ parental strain and found that McrA (the protein product of the mcrA gene) is involved in the regulation of hundreds of genes, including important SM BGCs.
Results
Isolation of mutations that activate expression of a silent non-ribosomal peptide synthetase gene
The strategy we developed to identify novel SM-activating mutations takes advantage of the fact that genes of SM BGCs are coordinately regulated such that transcription is activated for all the genes of the cluster at the same time. If one can select for a trans-acting mutation that activates expression of one gene in the cluster, the mutation is likely to activate expression of other genes in the cluster. Directly screening for mutations affecting production of SMs by analyzing mutagenized colonies with high performance liquid chromatography (HPLC) or mass spectroscopy (MS) would be a very time-consuming process so we developed a screen that allows us to select for mutations that upregulate a target cluster (Fig. 1).
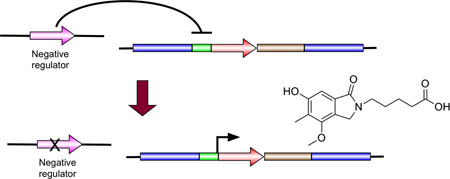
Fig. 1.
Strategy for selection of negative regulators of secondary metabolite gene clusters. A. Postulated negative regulation of AN1242 (which encodes a non-ribosomal peptide synthetase) by a gene separate from the AN1242 gene cluster. B. Replacement of the coding region of AN1242 by transformation with a linear molecule containing the coding region of the Aspergillus fumigatus, riboB gene (AfriboB) and a selectable marker (AfpyrG). The host strain, LO4389, carries mutations in the pyrG gene which confers pyrimidine auxotrophy and in the riboB gene which results in riboflavin auxotrophy. Transformation selection is for pyrimidine prototrophy conferred by AfpyrG. C. After transformation, the coding region of AfriboB is now under control of the AN1242 promoter. Since expression from the AN1242 promoter is normally repressed, AfriboB is not expressed at significant levels and the strain still requires riboflavin. D. The strain is then mutagenized. If expression of AN1242 is negatively regulated, a mutation in the negative regulator gene will allow activation of expression from the AN1242 promoter, driving transcription of the AfriboB coding sequence and, consequently, riboflavin prototrophy. Riboflavin prototrophs can be easily selected on media lacking riboflavin.
In a strain that carries the riboB2 allele, which confers a riboflavin requirement, we replaced the coding sequence for a key biosynthetic enzyme, a polyketide synthase (PKS) or NRPS, in the target SM cluster with the coding sequence of the Aspergillus fumigatus riboB gene (AfriboB) (Nayak et al., 2006) (Fig. 1B). This places AfriboB under control of the promoter of an SM cluster gene, and if the SM cluster is silent, transcription levels for the AfriboB coding sequence will be very low. The strain will, thus, require riboflavin for growth. Since AfriboB expression levels are predicted to be low, one cannot select transformants on the basis of riboflavin prototrophy and a second selectable marker (AfpyrG) was included in the construct to allow selection of transformants. If there is a negative regulator of transcription of the cluster, a mutation that inactivates the negative regulator will, in turn, activate transcription of the cluster (Fig. 1D). AfriboB transcription will consequently increase and the strain will no longer require riboflavin. Such mutants can, in principle, be selected easily on media lacking riboflavin.
In initial experiments we replaced seven NRPS genes and five PKS genes with the AfriboB coding sequence [using the Aspergillus Genome Database designations (http://www.aspgd.org/) NRPS = AN1242, AN1680, AN2064, AN2924, AN3495, AN4827 and AN5318; PKS = AN3612, AN6431, AN6791, AN8910 and AN9005]. In all but three of these (AN6791, AN2064 and AN5318) expression was low enough to prevent growth on glucose minimal medium (GMM) plates unsupplemented with riboflavin. Here we report results obtained with the NRPS gene AN1242. AN1242 has now been identified as the NRPS required for nidulanin A production (Andersen et al., 2013), although this was not known when we began this work.
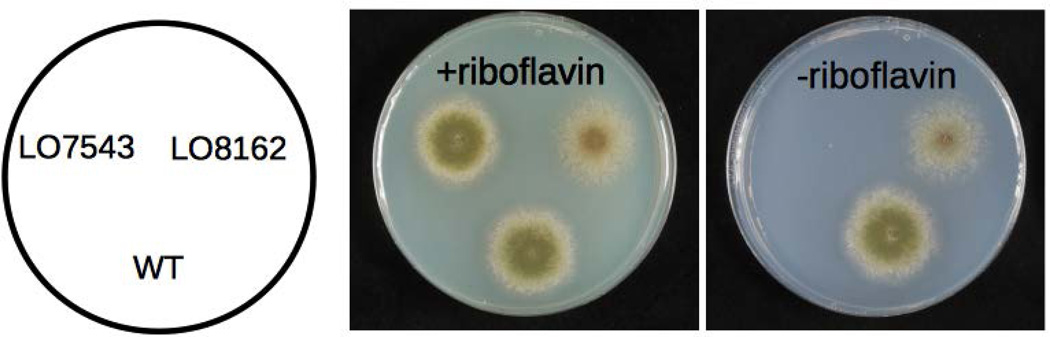
We replaced AN1242 with AfriboB, using the A. fumigatus pyrG gene (AfpyrG) as a selectable marker, in strain LO4389 (Fig. 1) creating strain LO7543 (Fig. 2). (Strains are listed in Table 1.) We then mutagenized with 4-nitroquinoline 1-oxide (NQO), which specifically causes base-pair substitutions (Downes et al., 2014; Bailleul et al., 1989). We used a range of NQO concentrations (0–3.5 µg ml−1) and selected mutants in which expression of AfriboB was activated on a medium lacking riboflavin (e.g. Fig. 2). We chose to analyze mutants obtained at the lowest NQO concentrations (0.5–1.0 µg ml−1; 38–44% kill), reasoning that since they had received the lightest mutagenesis they would carry the fewest extraneous mutations.
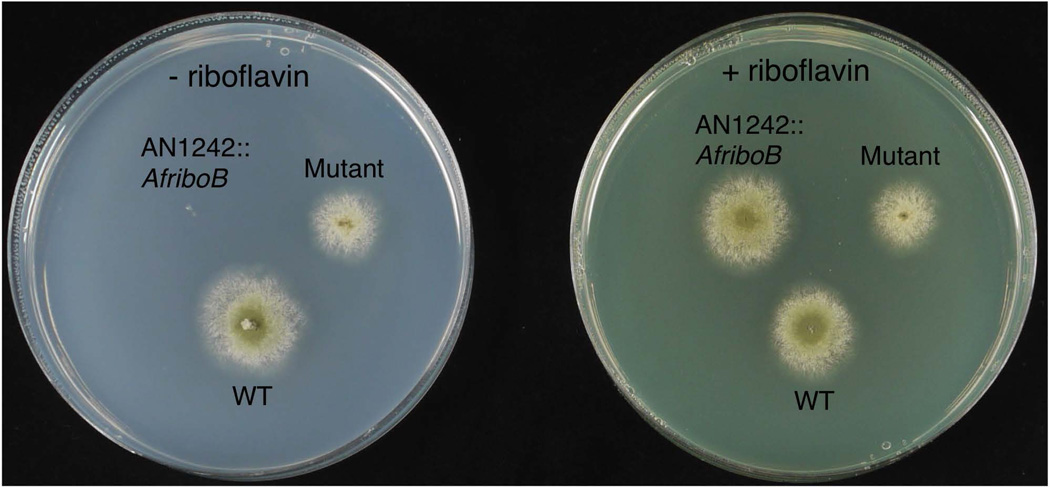
Fig. 2.
Growth of a wild-type (WT) strain, a strain (LO7543) in which the AN1242 coding region has been replaced with the AfriboB gene (AN1242::AfriboB), and a mutant (LO7668) which has been selected for the ability to grow in the absence of riboflavin. The AN1242::AfriboB strain, LO7543, does not grow in the absence of riboflavin but grows as well as the wild-type if riboflavin is added to the medium. The mutant grows in the absence of riboflavin.
Table 1.
A. nidulans strains used in this study.
| Strain | Genotype |
|---|---|
| LO1362 | pyroA4, riboB2, pyrG89, nkuA::argB |
| LO2804 |
pyroA4, riboB2, nkuA::argB (LO1362 transformed with a wild-type copy of A. nidulans pyrG) |
| LO4389 |
pyroA4, riboB2, pyrG89, nkuA::argB, sterigmatocystin cluster (AN7804- AN7825)Δ |
| LO7543 |
pyroA4, riboB2, pyrG89, nkuA::argB, sterigmatocystin cluster (AN7804- AN7825)Δ, AN1242:: AfriboB_AfpyrG (AN1242 coding sequence replaced) |
| LO7668 | A riboflavin prototophic mutant of LO7543 |
| LO8030 |
pyroA4, riboB2, pyrG89, nkuA::argB, sterigmatocystin cluster (AN7804- AN7825)Δ, emericellamide cluster (AN2545-AN2549)Δ, asperfuranone cluster (AN1036-AN1029)Δ, monodictyphenone cluster (AN10023-AN10021)Δ, terrequinone cluster (AN8513-AN8520)Δ, austinol cluster part 1 (AN8379- AN8384)Δ, austinol cluster part 2 (AN9246-AN9259)Δ, F9775 cluster (AN7906- AN7915)Δ, asperthecin cluster (AN6000-AN6002)Δ. |
| LO8111 | AN8694 (mcrA)::AfpyroA in LO8030 |
| LO8158- LO8159 |
pyroA4, riboB2, pyrG89, nkuA::argB, AN8694 (mcrA)::AfpyroA |
| LO8162 |
pyroA4, riboB2, pyrG89, nkuA::argB, sterigmatocystin cluster Δ, AN1242::AfriboB_AfpyrG (AN1242 coding sequence only replaced), AN8694 (mcrA)::AfpyroA |
| LO8166- LO8167 |
pyroA4, riboB2, pyrG89, nkuA::argB, AfpyroA-alcA(p)AN8694 [promoter of AN8694 (mcrA) replaced with the alcA promoter] |
| LO8936 -LO8937 |
pyroA4, riboB2, pyrG89, nkuA::argB, AfpyrG-gpdA(p)AN8694 (mcrA) [promoter of AN8694 (mcrA) replaced with the gpdA promoter] |
| LO9345 | AN6448(pkbA = cicF)::AfpyrG in LO8111 |
| LO10255- LO10257 |
pyroA4, riboB2, pyrG89, nkuA::argB, sterigmatocystin cluster Δ, AN1242::AfriboB_AfpyrG (AN1242 coding sequence only replaced), AN8694 (mcrA)::AfpyroA, yA::ptrArmcrA (mcrA deleted and reinserted at the yA locus using pyrithiamine resistance as a selectable marker) |
| LO10260- LO10262 |
pyroA4, riboB2, pyrG89, nkuA::argB, AN8694 (mcrA)::AfpyroA, laeA::AfpyrG (deletion of both mcrA and laeA) |
| LO10269- LO10271 |
pyroA4, riboB2, pyrG89, nkuA::argB, AN8694 (mcrA)::AfpyroA, AtpyrG- gpdA(p)laeA (laeA promoter replaced with the gpdA promoter in a mcrA deletion strain |
| LO10279- LO10281 |
pyroA4, riboB2, pyrG89, nkuA::argB, laeA::AtpyrG (laeA deletion) |
| LO10285- LO10287 |
pyroA4, riboB2, pyrG89, nkuA::argB, AtpyrG-gpdA(p)laeA (laeA driven by the gpdA promoter) |
All strains carry veA1. Note that since some of the molecular genetic manipulations are novel, there is no standard nomenclature for them.
Characterization of mutant alleles and identification of activating mutations
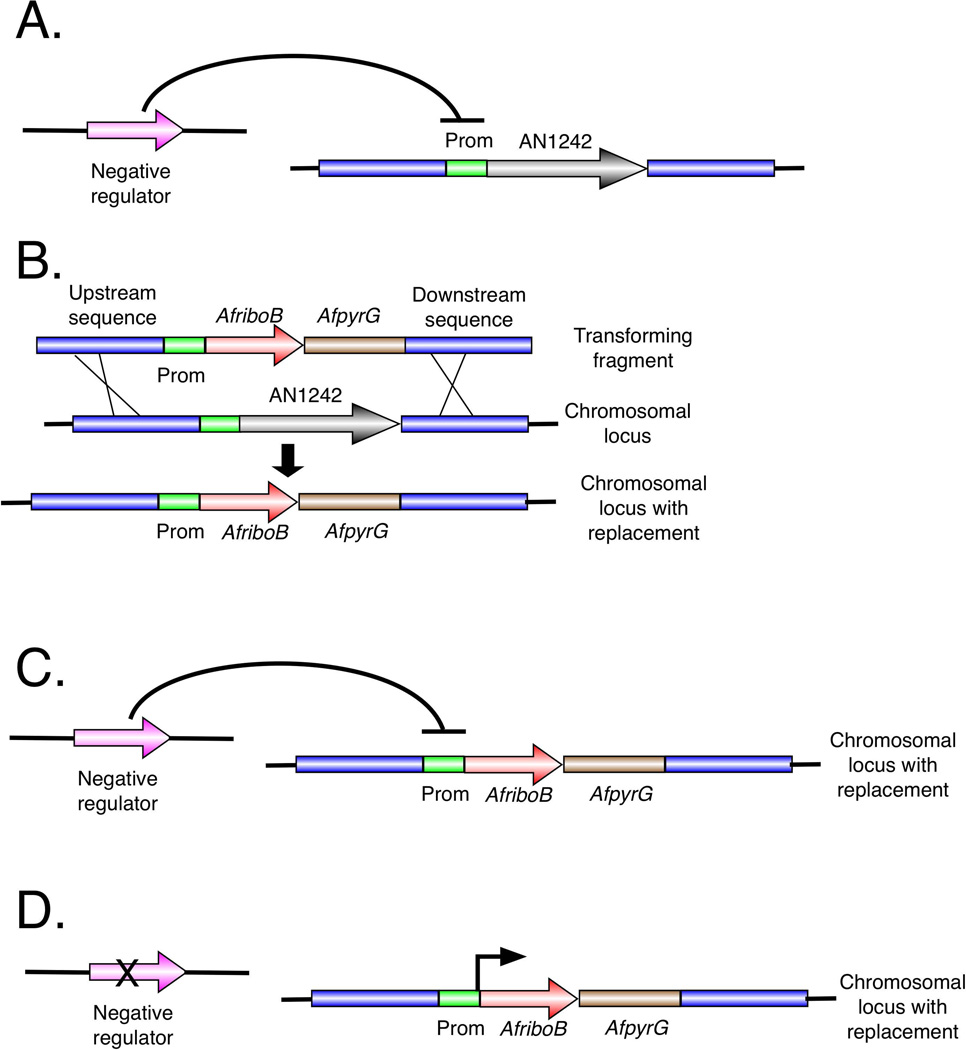
Activating mutations can be in the cluster (e.g. mutations in the AN1242 promoter), or in a gene that is distant from the cluster and regulates the cluster in trans. Mutations that act in trans are strongly predicted to activate the entire cluster and are likely to be more informative with respect to SM regulation than mutations in the promoter of the target gene. We devised a simple way to determine if activating mutations were distant from the cluster (Fig. 3). We constructed a second strain, a tester strain, that, like the originally mutated strain, has the AfriboB coding sequence under control of the promoter of our target gene. We used a different selectable marker, however, for transformation, AfpyroA [which complements the pyridoxine requirement conferred by the A. nidulans pyroA4 mutation (Nayak et al., 2006)]. Importantly for subsequent identification of the mutation by sequencing, the tester strain and the mutated strain are derived by transformation from the same parent and, thus, should carry very few sequence differences [e.g. single nucleotide polymorphisms (SNPs)].
Fig. 3.
Mapping activating mutations after the initial screen. A. Determination of the distance of the activating mutation from the AN1242 cluster. A mutant strain (top line) carries the AfriboB coding sequence replacing the AN1242 coding sequence and therefore under control of the AN1242 promoter (Prom). The strain also carries an activating mutation (*) that increases transcription from the AN1242 promoter and, thus, expression of AfriboB. This mutant strain is crossed to a strain that is isogenic except that it does not carry an activating mutation and the replacement of the AN1242 coding sequence has been carried out using AfpyroA as a selectable marker (making it a pyridoxine prototroph). If a crossover occurs between the activating mutation and AfriboB under control of the AN1242 promoter (i.e. the AN1242 locus), progeny that are prototrophic for both riboflavin and pyridoxine will result. The frequency of progeny that are prototrophic for riboflavin and pyridoxine, thus, reflects the distance between the activator mutation and the AN1242 gene cluster. B. Determination if two activating mutations are allelic. Two strains carrying separately isolated trans-activating mutations are crossed. One carries AfpyrG as a selectable marker and the other carries AfpyroA. Both are riboflavin prototrophs because they both carry activating mutations. The pyridoxine prototrophs come from crosses such as the one shown in A. Crossing over between the two activating mutations will result in progeny that carry no activating mutation and, thus, are riboflavin auxotrophs. If the activating mutations (*) are allelic, the frequency of riboflavin auxotrophs will be very low.
We crossed 20 mutants selected for riboflavin prototrophy to the tester strain. The fraction of the progeny that are prototrophic for both riboflavin and pyridoxine will be proportional to the distance between the activating mutation and the target SM gene (Fig. 3A). One mutation was closely linked to the target cluster with only a single recombinant among hundreds of progeny and a second was linked with a recombination frequency of approximately 5.5%. Eighteen were clearly transactivators, however, with recombination frequencies >21%. We chose five transactivators at random and crossed them to each other to determine if the activating mutations are in the same gene (Fig. 3B). In these crosses, the frequency of riboflavin auxotrophs will be proportional to the distance between the two mutations. We found that the recombination frequencies in all pairwise crosses of the five transactivating mutations were less than 1%. All five of these transactivating mutations are thus, in all likelihood, allelic.
The fact that all five of these transactivating mutations are allelic strongly suggested that the mutant gene is important for regulation of AN1242 and we, consequently, identified the gene by genomic sequencing. A challenge in identifying mutations by genomic sequencing is that if one compares a mutant strain to a wild-type strain, there are likely to be sequence differences including SNPs that are unrelated to the mutation of interest. Since the strains in which we isolated our activating mutations were from the same parent as the tester strain, with the necessary genetic markers introduced by transformation rather than crossing, SNPs would be minimal except for those that occur during mutagenesis. Because even minimal mutagenesis might create irrelevant mutations in addition to those that activate AN1242, however, we developed a scheme to allow us to filter the irrelevant mutants from our data. For a given transactivating mutation, we chose 10 segregants that carried the activating mutation from the crosses of mutants to the tester strain and made a single barcoded genomic DNA library. We next chose 10 segregants that did not carry the activating mutation and made a second barcoded library. The two libraries were sequenced in a single sequencing run to >100X coverage. We applied bioinformatics filters to filter out SNPs or other alterations that occurred within both groups, leaving only SNPs that were specific to the segregants that carried the activating mutation.
Using this approach, we attempted to identify two separately isolated but allelic (recombination frequency <1%) mutations that transactivate AN1242. Gratifyingly, our analysis of sequence data revealed a single putative transactivating mutation in each case and each mutation was in the predicted coding region of the same gene, AN8694. In one case the mutation was a G to T transversion of nucleotide 621 (with the A of the start codon being nucleotide 1) of AN8694 resulting in the replacement of an arginine with a leucine. In the second case there was a G to A transition at nucleotide 759 resulting in a cysteine to tyrosine change. AN8694 is on chromosome III whereas AN1242 is on chromosome VIII. The facts that there was a single consistent difference between the mutant and wild-type strains in each case, and that independently isolated mutations were in the same gene, provided strong evidence that these mutations are, indeed, transactivating mutations for AN1242 and, thus, that AN8694 plays an important role in the regulation of transcription of AN1242.
AN8694 is a previously uncharacterized gene. The only mention of AN8694 that we could find in the literature is that its transcription is downregulated by deletions of velB and vosA (Ahmed et al., 2013). The gene model in the AspGD database predicts that its product is a 399 amino acid protein and our RNAseq data (not shown) are consistent with this gene model. The product of AN8694 is predicted to be a zinc-finger transcription factor with a GAL4-like Zn2Cys6 binuclear cluster DNA binding domain extending from amino acids 156 to 196. The predicted product of AN8694 also, notably, contains proline rich regions including several polyproline regions. A BLAST search with AN8694 revealed that many species of fungi have a single, strong homolog of AN8694 (Data for the 99 top hits are shown in Dataset S1). The homology was strong not just among species of Aspergillus, but across many genera of ascomycetes revealing that AN8694 encodes a protein that is highly conserved. Among species of Aspergillus the identity generally extended over the entire molecule (>95% coverage). There were five hits among Aspergillus species that showed less coverage. In some cases, the reduced coverage may be due to errors in annotation. For example, the A. terreus homolog [designated ATET_7219 in AspGD and ATEG_07219 in the fungal genome datablase (Fungidb.org) and the National Center for Biotechnology information (http://www.ncbi.nlm.nih.gov/)] has only 74% coverage because the product of this gene is annotated to lack the 93 N-terminal amino acids of the predicted product of AN8694. However, a TBLASTN search of the A. terreus genome with the product of AN8694 reveals reading frames with strong identity to aa 5–93 in the sequences upstream of the annotated ATET_7219 (using the AspGD designation for consistency) start codon, suggesting that the start codon for the A. terreus gene is misannotated. A number of species from other genera showed >90% coverage. In a number of cases the homologous proteins in other species appeared to lack the N-terminal 140 amino acids or so present in the predicted A. nidulans protein, but in nearly all cases the region starting with the DNA binding domain and extending to the C-terminus was very highly conserved.
AN8694 is a negative regulator of the production of nidulanins and other secondary metabolites
Since most random mutations cause loss or reduction of function rather than enhanced function, the two mutations we identified in AN8694 are likely to reduce or eliminate the activity of AN8694. If so, it follows that AN8694 is a negative regulator of expression of AN1242 (as predicted from the design of the screen). To determine if this was the case, we deleted AN8694 in strain LO7543, the strain we originally mutated that carries AfriboB replacing the coding sequence of AN1242. We designated the resulting strain LO8162. LO8162 grew on medium lacking riboflavin, confirming that deletion of AN8694 activates expression of AN1242 and, thus, that AN8694 is a negative regulator of transcription of AN1242 (Fig. 4). Finally, to add an additional layer of redundancy to the proof that AN8694 is a regulator of AN1242, we reinserted AN8694 at the yA locus creating strains LO10255–10257. LO10255-LO10257 required riboflavin (Fig. S1). Deletion of AN8694, thus, activates the AN1242 promoter and reinsertion of AN8694 at another locus in the deletion strain inactivates the promoter.
Fig. 4.
Growth of a WT strain (FGSC4), LO7543, a strain carrying a replacement of the coding region of AN1242 with the coding sequence of the AfriboB gene and LO8162, which is a transformant of LO7543 in which AN8694 has been deleted. The strains are growing on minimal medium supplemented or unsupplemented with riboflavin. In the absence of riboflavin, LO7543 does not grow, but LO8162, carrying a deletion of AN8694, grows, revealing that expression of AfriboB at the AN1242 locus has been activated.
While some regulators of fungal SM production are specific for particular clusters (e.g. cluster-specific transcription factors that are associated with some SM gene clusters), others (e.g. LaeA) regulate several or many gene clusters (Bok and Keller, 2004; Keller et al., 2006; Brakhage, 2013). Regulators that control the expression of multiple SM gene clusters are often called global regulators. (This term seems to imply that the regulator gene controls all SM gene clusters, but no regulator identified to date has been shown to regulate more than a fraction of SM clusters.) Because AN8694 mutants were originally selected to activate AN1242 on glucose minimal medium (GMM) plates, we expected that on the same medium AN8694Δ would activate expression of the entire nidulanin A cluster resulting in nidulanin A production. We also wanted to determine if it would activate production of other compounds as well. We deleted AN8694 in strain LO1362 [=TN02A7 (Nayak et al., 2006)] in which all SM clusters are intact, creating strain LO8158. We examined the effects of deletion of AN8694 as well as overexpression of this gene on SM production in a variety of media. SMs were extracted from plates and media as detailed in the Materials and Methods and analyzed by HPLC-diode array detector (DAD)-MS. Previous efforts from our laboratories and others have resulted in the purification and structural determination of many A. nidulans SMs (Yaegashi et al., 2014), and we were able to identify these previously discovered compounds by comparing their HPLC retention time, UV-Vis absorption, and mass spectra.
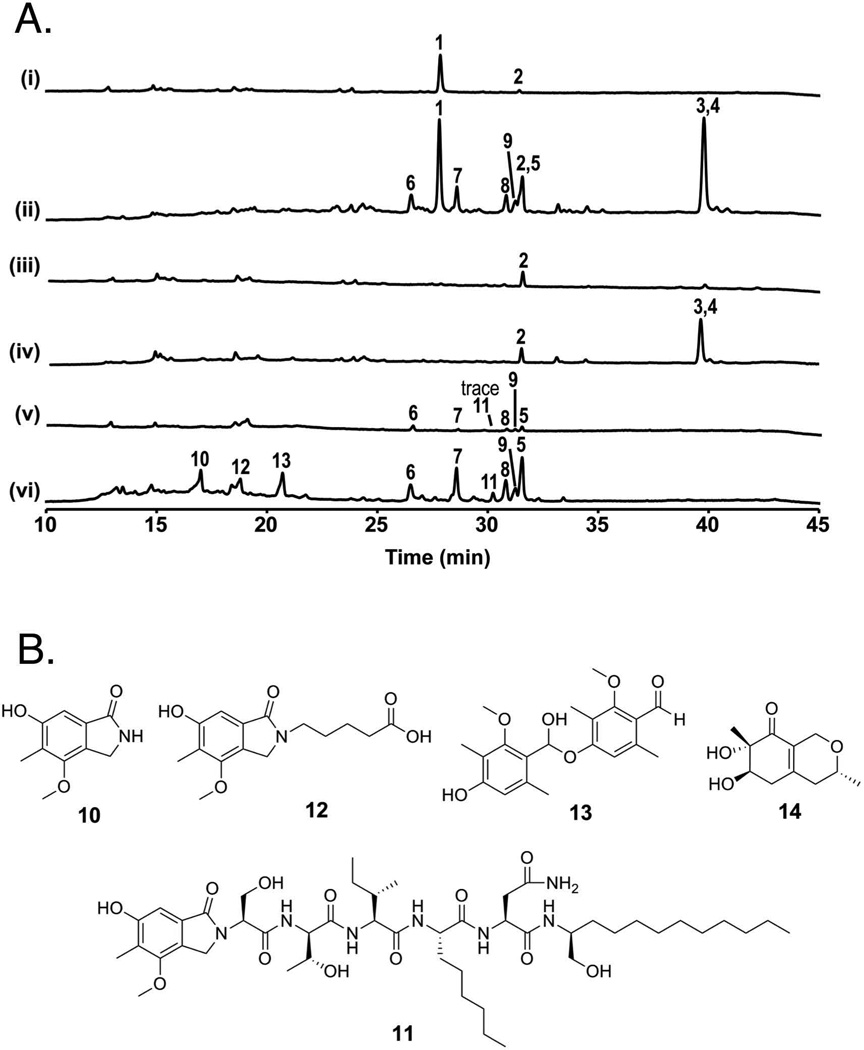
We first examined the metabolites produced in the AN8694Δ strain LO8158 relative to its parental strain LO1362 cultivated on GMM plates (Fig. 5A, i and ii). A major metabolite, sterigmatocystin (1), and a trace amount of terrequinone (2) were identified in the parental strain LO1362. In addition to compounds 1 and 2, seven additional metabolites (3 – 9) were detected in the AN8694Δ strain LO8158. Compounds 3 and 4 were identified as shamixanthone and emericellin from the prenyl xanthone pathway by comparing the spectral data previously reported by our groups (Sanchez et al., 2011). Compounds 5 – 9 were identified as nidulanin A and its derivatives, based on the extracted-ion chromatogram (EIC) reported by Andersen et al. (Andersen et al., 2013) as well as MS/MS fragmentation (Fig. S2, confirming that AN8694 is a negative regulator of the nidulanin A biosynthesis pathway.
Fig. 5.
A. HPLC paired profile scans of parental strains and AN8694 deletion strains. [UV-Vis total scan (200 – 600 nm)] (i) Parental strain LO1362, (ii) LO8158, the AN8694 deletion strain created from (i). (iii) parental strain LO7543, (iv) LO8162, the AN8694 deletion strain created from (iii), (v) parental strain LO8030, (vi) LO8111, the AN8694 deletion strain created from (v). All strains were grown on GMM plates. 1 is sterigmatocystin, 2 is terrequinone, 3 is shamixanthone, 4 is emericellin, 5 is nidulanin A, 6 – 9 are nidulanin A derivatives (see Fig. S1), 10 is cichorine, 11 is aspercryptin, 14 is felinone A. Note that 2 and 5 were co-eluted at 31.6 min; 3 and 4 were co-eluted at 39.6 min. B. Chemical structures of compounds 10 – 14 identified from LO8111.
We next compared LO7543, the strain that was originally mutated, to LO8162, which is identical except that it carries AN8694Δ (Fig. 5A, iii and iv). Since a key nidulanin A biosynthesis gene is replaced in both strains and the entire sterigmatocystin cluster is deleted in both strains, no nidulanins or sterigmatocystin were detected in either strain. As seen for LO1362 and LO8158, shamixanthone (3) and emericellin (4) were upregulated by AN8694Δ. Reinsertion of the AN8694 gene at the yA locus of LO8162 (creating strains LO10255-LO10257 returned SM production to that seen with LO7543 (Fig. S3), demonstrating that the absence of AN8694 was the cause of enhanced production of SMs in LO8162. Taken together, our data so far indicated that AN8694 is a negative regulator of the biosynthesis of prenyl xanthones (3 and 4) and nidulanins (5 – 9).
Deletion of AN8694 results in the upregulation of several SMs under different culture conditions
In many cases, microorganisms produce different SMs under different growth conditions (Bode et al., 2002). We were consequently interested in determining if deletion of AN8694 affected SM production in a variety of culture conditions. Note that all SM profiles were repeatable and based on analysis of two or more sister transformants although in most cases data from a single, representative strain are shown. When grown on lactose minimal medium (LMM) plates, both the parental strain LO1362 and LO8158 (AN8694 deleted in LO1362) produced austinol (15), dehydroaustinol (16), and intermediates from the sterigmatocystin pathway (17 – 21) (Fig. S4A i and ii) so AN8694 does not appear to regulate production of these compounds on LMM plates. On the other hand, F9775B (22), F9775A (23), shamixanthone (3), emericellin (4), and epishamixanthone (24), were upregulated in AN8694Δ mutants, LO8158 and LO8162 (AN8694Δ in strain LO7543 which carries the AfriboB gene coding sequence replacing the coding sequence of AN1242), on LMM plates (Fig. S4A, i – iv). In yeast extract, agar, glucose (YAG) plates, prenyl xanthones (3, 4, and 24), nidulanins (5 – 9), cichorine (10), asperthecin (27), emodic acid (28), emodin (29), and an unknown (30) were identified (Fig. S4B, i – iv). However, there was no consistent pattern of upregulation or downregulation by AN8694Δ on YAG plates. We also examined secondary metabolites produced in liquid media (Table S2). When cultured in liquid GMM, as on GMM plates, prenyl xanthones and nidulanins were upregulated in AN8694Δ mutants. In addition, F9775A and B were also upregulated in liquid GMM (Table S2).
Deletion of AN8694 in a genetic dereplication strain results in the discovery of novel compounds
We have previously developed methods for deleting entire A. nidulans SM clusters (Chiang et al., 2013) and we have used these methods to engineer a strain (LO8030) in which the SM clusters responsible for the biosynthesis of the following major SMs are deleted: sterigmatocystin, the emericellamides, asperfuranone, monodictyphenone, terrequinone, F9775A and B, asperthecin and both portions of the split SM cluster that produces austinol and dehydroaustinol (Chiang et al., 2016). These deletions greatly reduce the SM background allowing SMs from other clusters to be detected more easily. In addition, deletion of major SM BGCs may free up CoA precursors to allow greater production of low level SMs and we noted that nidulanins (5 – 9) were produced at low, but detectable, levels in LO8030 (Fig. 5A, v). It also produced a trace amount of 11, a compound we have discovered recently and designated aspercryptin (Chiang et al., 2016). We have also discovered the BGC responsible for aspercryptin biosynthesis (AN7873-AN7884) and designated it atn (Chiang et al., 2016). Aspercryptin incorporates a cichorine (10) moiety and we have characterized and designated the cluster responsible for cichorine biosynthesis as cic (AN6443 - AN6449) (Sanchez et al., 2012a).
We deleted AN8694 in LO8030 creating strain LO8111. Compounds 5 – 11 were upregulated in LO8111 (Fig. 5A, v and vi). AN8694 is, consequently, a negative regulator of cichorine and aspercryptin as well as the nidulanins. In addition, two peaks (12 and 13) were consistently detected in LO8111 grown on GMM that were not detectable or produced in low quantities in the LO8030 parent (Fig. 5A, v and vi). In order to characterize the chemical structures of 12 and 13, we isolated both compounds from a large-scale culture of LO8111. The structures of 12 and 13, elucidated by their spectroscopic data, are shown in Fig. 5B. Both are new compounds (for details of purification and structure elucidation, see supporting information). From their chemical structures, 12 and 13 are likely to be metabolites produced by the cichorine biosynthesis pathway (Sanchez et al., 2012a). Indeed, deletion of AN6448, the PKS responsible for the biosynthesis of cichorine (10), eliminated the production of 11 as well as 12 and 13 (Fig. S5), demonstrating the requirement of AN6448 for the biosynthesis of 12 and 13. We propose biosynthesis pathways of 12 and 13 in Fig. S6A.
Although not detected by HPLC-DAD-MS, a minor compound, felinone A (14), a recently discovered antibiotic (Du et al., 2014), was also isolated from large-scale cultures of LO8111. We have characterized all the non-reducing polyketide synthase (NR-PKS) products produced by A. nidulans (Ahuja et al., 2012). Comparison of the carbon skeleton of 14 with those of other NR-PKS products suggests that 14 is a product of the AN7903, dba, cluster and we propose a biosynthetic pathway for felinone A in Fig. S6B. Two products have previously been reported for the dba cluster (Ahuja et al., 2012; Gerke et al., 2012) and felinone A is a third product, not previously known to be produced by A. nidulans.
Upregulation of AN8694 results in downregulation of secondary metabolites
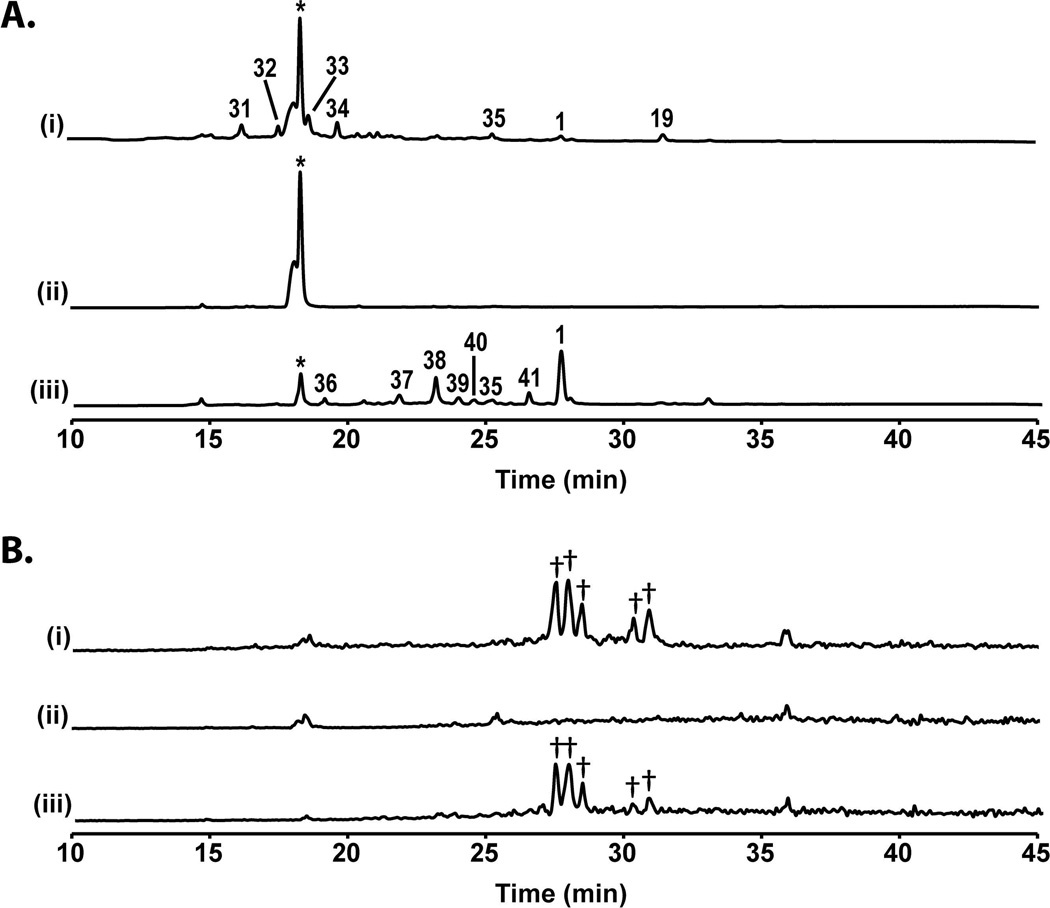
Since AN8694 is a negative regulator of the production of several secondary metabolites, we reasoned that upregulation of this gene might reduce production of SMs that are normally produced in culture. We placed AN8694 under control of the regulatable alcA promoter [alcA(p)] (Doy et al., 1985; Felenbok, 1991) in the LO1362 background. alcA(p) is repressed by glucose and strongly induced by a number of compounds including cyclopentanone. We analyzed the effects of overexpression of AN8694 on SM production using five closely related strains, LO1362, two transformants of LO1362 that carry alcA(p)AN8694 (LO8166 and LO8167) and two transformants of LO1362 that carry AN8694Δ (LO8158 and LO8159). We grew the strains in liquid LMM and induced with 10 mM cyclopentanone. After 48 hours we harvested the cultures, evaluated AN8694 expression by quantitative RTPCR and extracted SMs with ethyl acetate and analyzed SM production by HPLC-DAD-MS. Quantitative RTPCR indicated that AN8694 transcription was upregulated approximately 5 X in LO8166 relative to the parental strain LO1362 Fig. S7). While this is not a huge upregulation, it must be remembered that AN8694 inhibits SM production at its normal transcription levels and a 5 X upregulation over a normally inhibitory level is likely to cause further inhibition. Results with SM production are shown in Fig. 6. Using a DAD to detect compounds containing chromophores, LO1362 produced approximately 8 detectable compounds under these conditions including sterigmatocystin (1), 2,4-dihydroxy-3-methyl-6-(2-oxopropyl)benzaldehyde (from the AN7903, dba cluster (Ahuja et al., 2012; Gerke et al., 2012)) (34), microperfuranone (35), and four unknowns (19 and 31 – 33). Only one peak was visible in the alcA(p)AN8694 strains and this peak was identified as lumichrome, which we believe to be metabolized from the riboflavin added to the culture as a nutritional supplement. In contrast, the AN8694Δ strains produced sterigmatocystin (1), microperfuranone (35), and sterigmatocystin intermediates (36 – 41) under the same conditions. Total ion chromatogram (TIC) revealed that the production of emericellamides (Chiang et al., 2008) was also essentially eliminated by induction of alcA(p)AN8694 (Fig. 6B). These data indicate that overexpression of AN8694 results in a reduction of SM production. We should note, however, that alcA(p)AN8694 strains did not grow well after induction so some of the effects of alcA(p)AN8694 induction might be related to reduced growth.
Fig. 6.
Overexpression of AN8694 suppresses SM production. A. HPLC UV-Vis total scan (200 – 600 nm) profiles of parental strain LO1362, (i), alcA(p)AN8694 strain LO8166 (ii), and AN8694Δ strain LO8158 (iii) grown in LMM medium under inducing conditions. 1 is sterigmatocystin, 34 is dihydroxy-3-methyl-6-(2-oxopropyl)benzaldehyde, 35 is microperfuranone, 19 and 31 – 33 are unknowns, 36 – 41 are sterigmatocystin intermediates. *: lumichrome B. Total ion chromatogram (TIC) of A. †: emericellamides A and C – F.
We also placed AN8694 under control of the gpdA promoter [gpdA(p)] (Punt et al., 1990), which is a constitutive promoter that drives transcription at high levels (Punt et al., 1991). The gpdA(p)AN8694 strains were partially restricted for growth. Growth was sufficient, however, to allow us to look at the effects of AN8694 over expression on media containing glucose. In liquid GMM, the emericellamides were consistently downregulated whereas F9775A (23) and F9775B (22) were upregulated (Table S3). In liquid LMM, sterigmatocystin (1), terrequinone (2) and the emericellamides were consistently downregulated. On solid medium, gpdA(p)AN8694 caused downregulation of prenyl xanthones (3 and 4), nidulanins (5 – 9), and emodins (28 and 29). Sterigmatocystin (1) and emericellamides were downregulated on GMM and LMM (Table S3). Again, these data are consistent with AN8694 being a negative regulator of several SM gene clusters.
In summary, our data indicate that AN8694 is involved in the regulation of the SM biosynthetic pathways that produce nidulanins (5 – 9), sterigmatocystin (1), the prenylated xanthones (3, 4 and 24), asperthecin (11), cichorines (10, 12 and 13), F9775A and B (23 and 22), the emericellamides, and possibly terrequinone (2), asperthecin (27) and microperfuranone (35). In most, but not all, cases, deletion of AN8694 increases compound production and overexpression reduces compound production. The effects of AN8694Δ and AN8694 overexpression differ on various media suggesting that AN8694 may connect secondary metabolism to primary metabolism. In view of these findings we designate AN8694 mcrA (multi-cluster regulator A).
mcrA is a master transcription regulator
mcrA regulates several SM BGCs and this implies that it is involved in the regulation of transcription of many genes involved in SM biosynthesis. Secondary metabolism is intimately connected to a number of other processes in the cell, however, such as primary metabolism and development (Bayram and Braus, 2012; Bayram et al., 2012; Katz et al., 2013; de Souza et al., 2013; Brakhage, 2013; Garzia et al., 2013). We wished to determine, therefore, if the effects of mcrA on gene expression extended beyond genes involved in secondary metabolism. We carried out RNAseq analysis on LO1362 and its daughter mcrA deletant strain LO8158 in triplicate from material grown in liquid GMM. Samples were collected after 24 hours of incubation. Note that this is a single growth condition and our metabolite profile data indicate that expression of SM BGCs differs on different media. Note also that metabolite profiles were obtained with material grown in liquid culture for four days or on plates for five days. Transcription profiling on a variety of media and on samples taken at a range of incubation times will be required to determine the full effects of mcrA on transcription. Such analysis is beyond the scope of this study, but we would anticipate that with different growth conditions mcrAΔ would cause alterations of transcription beyond those we see with a single growth condition and time of incubation.
Combined data from three separate RNAseq experiments are listed in Dataset S2. Surprisingly, edgeR analysis (Robinson et al., 2010) revealed that deletion of mcrA significantly altered the transcription of 1352 genes (p<0.05), more than 10% of the genes of A. nidulans. These data reveal that mcrA is a master regulator of transcription. 623 genes were upregulated at least 5X in the mcrAΔ strain relative to the parent. Among these, transcripts from 112 genes were detected in the mcrAΔ strain that were never detected in the parental strain in more than 4 × 107 reads. In many cases these only produced a small number of reads in the mcrAΔ strain, indicating that their expression is stimulated by mcrAΔ but not to high levels. In other cases, both the fold increase and the final transcript abundance were large. For example, AN8781, a conserved but uncharacterized open reading frame, was upregulated 20X in the mcrAΔ strain and averaged more than 12,000 reads per 107 total reads in three separate experiments.
SM biosynthesis genes were among the genes most strongly upregulated. AN1242 showed a 510X increase in the deletant relative to the parent. AN11934, which is adjacent to AN1242 and almost certainly involved in nidulanin A synthesis (Andersen et al., 2013), showed an increase of 386X). AN11080, a prenyl transferase that is required for nidulanin A synthesis (Andersen et al., 2013) was upregulated 20X. In addition, although it has not been previously noted, we found that AN8483, which is adjacent to AN11080 in the genome, is co-regulated with the other nidulanin biosynthesis genes and we, therefore, suspect that it is involved in nidulanin biosynthesis. Other SM biosynthetic genes showed large increases as well. Based on the SM BGCs defined by Inglis et al. (Inglis et al., 2013), at least five core SM biosynthetic genes from 4 BGCs showed a > 40X increase (Table S4) and core biosynthetic genes from 13 SM BGCs showed a > 5X increase in transcription. (Note that Inglis et al. considered AN11080 to be a core biosynthetic gene, but its function appears to be to prenylate nidulanins.) Of these, eight were expressed at relatively low levels even in the mcrAΔ strain (less than 100 reads/107 total reads, mean of three experiments). These genes are, thus, regulated in part by mcrA but additional factors must be involved in the regulation of their transcription. For example, our metabolite profile data suggest that transcription of some of these BGCs will be higher after longer incubation periods.
It is relatively easy to identify core SM biosynthetic genes such as PKSs and NRPSs by homology, but determining which genes, in addition to these core biosynthetic genes, make up particular BGCs is more difficult. However, in instances in which the genes that make up a BGC have been determined experimentally, there is clear evidence that mcrA coordinately regulates the genes of the cluster. For example, we have previously determined that AN2545, AN2547, AN2548 and AN2549 are required for emericellamide biosynthesis (Chiang et al., 2008). These genes were upregulated 48X, 61X, 196X and 41X respectively in the mcrAΔ strain, whereas adjacent genes were not upregulated significantly (Dataset S2). The nidulanin A cluster showed similar coordinate regulation, as mentioned above, as did the aspernidine cluster (Yaegashi et al., 2013) and the aspercryptin cluster (Andersen et al., 2013; Chiang et al., 2016). In instances in which the upregulation of the core biosynthetic gene was barely above our 5X threshold and the number of RNAseq reads was low, co-regulation was absent or less obvious (e.g. monodictyphenone and F9775A, B). Fewer genes were downregulated in the AN8694 deletion (see Dataset S2). Ninety-nine were downregulated at least 5X and these did not include any core SM biosynthetic genes.
We looked over the genes regulated by mcrA to see if there were obvious patterns that might give clues as to the mechanism of action of mcrA. We did not find Gene Ontology (GO) terms to be particularly informative in this regard, but we have included descriptions of known or putative functions of these genes from the AspGD web site in Dataset S2. For convenience we have grouped genes with similar putative functions together in Dataset S3. Bearing in mind that many of the descriptions may be imprecise or incomplete and there are many genes with no putative function, we believe the groupings and descriptions are informative. As expected, many of the genes that are upregulated by mcrA are involved directly in secondary metabolism and others in primary metabolism. There are also numerous putative cytochrome P450 genes and transporter genes that are likely involved in primary or secondary metabolism. For the most part genes that encode cytoskeletal elements or cell cycle regulatory proteins are absent from the list of mcrA regulated genes except for AN0453 which is a cyclin involved in conidiation. Several additional genes regulated by mcrA are involved in conidiation as well. Importantly, deletion of mcrA upregulates at least six transcription factors associated with secondary metabolism gene clusters. These include AN1599 [ent-pimara-8(14),15 diene cluster (Bromann et al., 2012)] AN2025, AN2036, AN7896 (DHMBA cluster (Gerke et al., 2012), AN10060, AN10021 (monodictyphenone, shamixanthone (Chiang et al., 2010; Sanchez et al., 2011) as well as AN7819 which is a putative regulatory gene within the sterigmatocystin gene cluster. Consistent with the fact that some SMs are downregulated under some conditions by mcrAΔ, we noted that mcrAΔ downregulated two transcription factors associated with SM gene clusters, AN9132 and AN3502. These data reveal that one of the mechanism of actions of mcrA is to regulate regulators of secondary metabolism. The mcrA deletion upregulates at least 12 additional putative transcription factors and it is possible that some of these may stimulate secondary metabolism even though they are not tightly linked to SM clusters and it is worth noting that mcrAΔ upregulates AN5874 a putative laeA-like methyl transferase that could have a role in chromatin level gene regulation.
Deletion of mcrA homologs in Aspergillus terreus and Penicillium canescens alters production of secondary metabolites
Because mcrA is conserved in ascomycetes, we hypothesized that deletion of mcrA homologs in species of fungi other than A. nidulans would lead to alterations of SM profiles. As discussed above, we identified an mcrA homolog in A. terreus strain NIH2624 (ATET_7219) using a BLASTP search of the NCBI protein database. As discussed above, we suspect that ATET_7219 is incorrectly annotated and that the product of ATET_7219 is actually longer and aligns with the entire length of McrA. We also identified an mcrA homolog in Penicillium canescens, CE25191_2989, in the Joint Genomes Institute database (http://genome.jgi.doe.gov/cgi-bin/dispGeneModel?db=Penca1&id=159801). This gene, too, is shorter than mcrA as annotated with aa 103 of mcrA aligning with aa 1 of P. canescens. However, the RNA reads in the JGI database suggest to us that the P. canescens gene is longer than annotated. We deleted each of these genes and we found that the deletions altered the metabolite profiles of each of these genes as anticipated (Fig. S8). As in A. nidulans, some metabolites were reduced whereas more metabolites appeared or increased. Although it is beyond the scope of this manuscript, we are actively working to identify the new compounds that appear in the deletions of the mcrA homologs. These data indicate that deletions of mcrA homologs will be a useful tool to elicit secondary metabolite production in ascomycetes.
Interactions with the veA/velB/laeA network
The veA/velB/laeA network is a very important regulator of secondary metabolism, and it is still yielding secrets even though it has been studied extensively over many years. It is likely that fully elucidating mcrA functions and interactions will require similar long-term efforts. We curious, however, as to the relationship, if any, between mcrA and laeA. The deletion of mcrA resulted in a 2.4 X upregulation of laeA (AN0807), a value that is statistically significant but rather small. Deletion of mcrA caused no significant change in expression of veA (AN1052) or velB (AN0363). These data suggest that mcrA probably does not exert its effects by regulating laeA, veA, or velB. It has been shown, however, that transcription of mcrA (AN8694) is downregulated by deletion of velB (Ahmed et al., 2013) so mcrA appears to have some connection to this network.
LaeA is a positive regulator of secondary metabolism. Deletion of laeA reduces the production of several secondary metabolites and overexpression increases SM production (Bok and Keller, 2004; Bayram and Braus, 2012). We, therefore, created strains that carried mcrAΔ as well as laeAΔ or laeA overexpressed (oelaeA) by replacing its native promoter with the inducible and repressible alcA promoter (Doy et al., 1985; Waring et al., 1989; Felenbok et al., 2001) or the strong constitutive gpdA promoter (Punt et al., 1988; Punt et al., 1991). Construction of these strains caused us to note an effect of laeAΔ on growth of mcrAΔ strains. Strains carrying mcrAΔ exhibit nearly normal rates of radial growth so colony diameter is only slightly inhibited relative to parental strains (Fig. 4, S1). We noted, however, that hyphal density within mcrAΔ colonies (as well as our original mcrA mutants) appeared reduced relative to parentals, giving the colonies a thin appearance with fewer conidia than normal. The apparent reduction in conidiation may simply be due to the reduced hyphal density. This difference was barely noticeable on complete medium (YAG), but obvious on minimal media. Deletion of laeA exacerbated this defect causing a thin colony phenotype and reduction of conidiation even on complete medium (Fig. S10). Overexpression of laeA driven by alcA(p) or gpdA(p) restored growth to the levels seen with laeA+, mcrAΔ strains (not shown). We next asked whether overexpression of laeA would activate the AN1242 promoter by placing laeA under control of the gpdA promoter in strain LO7543 which is mcrA+ and in which the AN1242 coding sequence is replaced by AfriboB. gpdA(p)laeA did not result in sufficient activation of the AN1242 promoter to convey riboflavin prototrophy, (not shown).
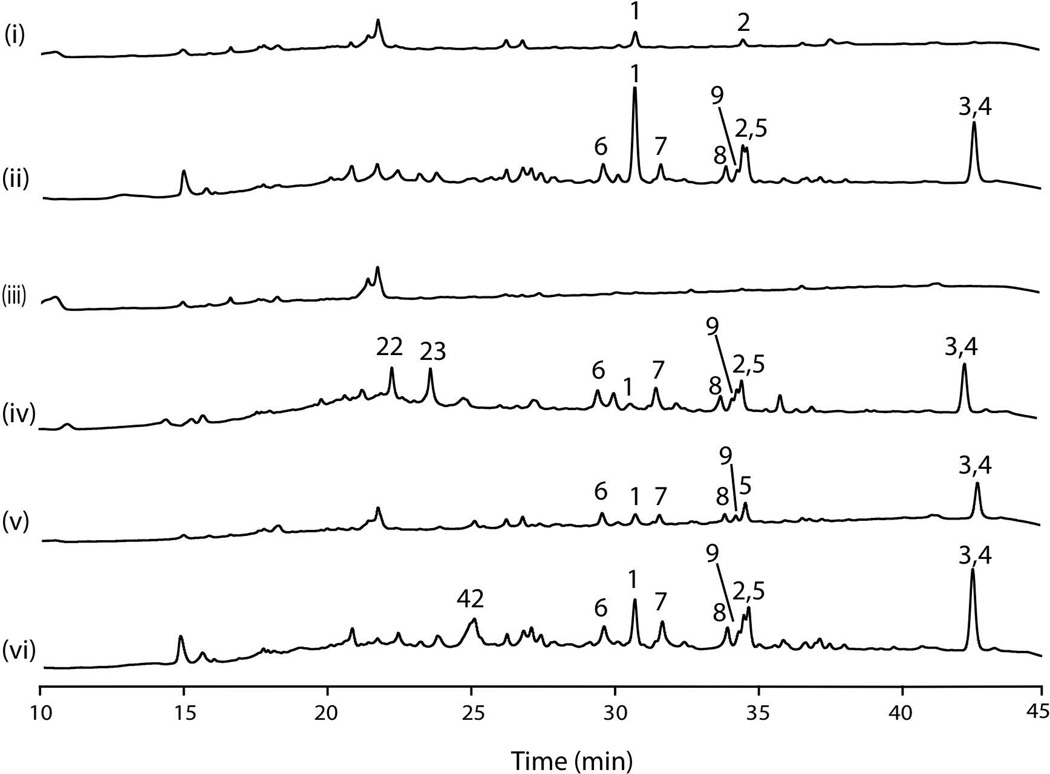
We next examined the effects of mcrAΔ along with laeAΔ or overexpression of laeA on SM production. These experiments were carried out on GMM plates under conditions identical to those used to obtain the data shown in Fig. 5. We first confirmed that SM production of the parental strain L01362 is modest under these conditions (Fig. 7 i). We also examined production of SMs in the alcA(p)mcrA strain LO8166. On GMM alcA(p) is repressed by glucose resulting in downregulation of mcrA and, as anticipated, SM production was boosted similar to the mcrA deletion (compare Fig. 7ii to Fig. 5ii). We deleted laeA in LO1362 creating the sister transformants LO10279-LO10281. We confirmed that, as expected, the deletion abolished almost all SM production in each of the three sister transformants and results for LO10279 are shown in Fig. 7iii. Next we created laeA, and mcrA double deletions strains by deleting laeA in the mcrAΔ strain, LO8158, creating sister transformants LO10260-LO10262. Interestingly, we found that for many SMs mcrAΔ overrode the inhibitory effects of laeAΔ. (All three strains gave similar results. Results for LO10260 are shown in Fig. 7iv). Austinol, dehydrooaustinol, nidulanin A, nidulanin intermediates, terrequinone, shamixanthone and emericellin were all produced in the laeAΔ, mcrAΔ strain. There was one notable exception. Sterigmatocystin was produced in the double deletion strain, but at much lower levels than in the mcrAΔ, laeA+ strains (Fig. 5 ii) or mcrA repressed, laeA+ strains (Fig. 7 ii). This result suggests that laeA plays a particularly important role in sterigmatocystin production that is not overridden by mcrAΔ. Surprisingly, two compounds, F9775A and F9775B were produced in laeA, mcrA double deletion strains at much higher levels than in the mcrAΔ, laeA+ or mcrA repressed laeA+ strain.
Fig. 7.
Effects of alteration of mcrA and laeA expression on SM production. HPLC UV-Vis total scan (200–600 nm). Growth was on GMM plates and extraction and HPLC conditions were identical to those used for Fig. 5. i. parental strain LO1362 (laeA+mcrA+). ii. LO8166 (laeA+alcA(p)mcrA). The glucose in GMM plates represses the alcA promoter resulting in repression of mcrA transcription and an SM profile similar to the mcrA deletion strain AN8158 (Fig. 5 ii). iii. LO10279 (laeAΔ, mcrA+). Deletion of laeA eliminates most SM production. iv. LO10260 (laeAΔ, mcrAΔ). Deletion of mcrA stimulates the production of SMs even if laeA is deleted. [Compare to laeA+mcrAΔ strains (Fig. 5 ii) or laeA+mcrA repressed strains (ii).] Sterigmatocystin (1) is a notable exception. However, F9775 A and B (23 and 22) are produced in this strain whereas they were not detected in laeA+mcrAΔ strains under the same conditions. v. LO10285 (gpdA(p)laeA, mcrA+). Upregulation of laeA driven by the strong constitutive gpdA promoter results in enhanced SM production relative to the parental strain (i). vi. L010269 (gpdA(p)laeA, mcrAΔ). Deletion of mcrA combined with overexpression of laeA results in the production of a new compound (42). For traces iii-vi three separate genetically identical strains (sister transformants listed in Table 1) were analyzed and in each case the results were very similar for the three strains. Representative traces are shown. 1 is sterigmatocystin, 2 is terrequinone, 3 is shamixanthone, 4 is emericellin, 5 is nidulanin A, 6 – 9 are nidulanin A derivatives (see Fig. S1), 23 is F9775A, 22 is F9775B, 42 is an unknown compound. Emericellamides were detected by mass spectroscopy in all samples but they are not UV active and thus not visible in these traces.
Overexpression of laeA results in enhanced SM production (Bok and Keller, 2004) and putting laeA under control of the strong constitutive promoter of the gpdA gene (Punt et al., 1988; Punt et al., 1991) in the parental strain LO1362, creating strains LO10285-LO10287, resulted in enhanced SM production as expected (Results for LO10285 are shown in Fig. 7v). Since both mcrAΔ and overexpression of laeA produce SM production, we created strains carrying both mcrAΔ and gpdA(p)laeA by putting laeA under control of gpdA(p) in LO8158 creating strains LO10269–10271. We hoped that SM production would be doubly enhanced. SM production was strong in the double mutant strains, but not dramatically greater than in the mcrAΔ parent (Fig. 7vi). However, one novel compound was produced (42) in these strains under these conditions and it is possible that additional new compounds will be produced in this strain under other conditions.
Discussion
We have developed a genetic screen that allows mutations in negative regulators of secondary metabolite gene clusters to be isolated through a simple nutritional selection. By replacing the coding sequences of target SM genes with the AfriboB coding sequence in a strain that is auxotrophic for riboflavin, we can select for mutations that activate transcription of the gene by selecting for the ability to grow on medium unsupplemented with riboflavin. We have used the screen to isolate negative regulators of the nidulanin A cluster. The levels of transcription of AfriboB from promoters of genes from eight of 11 other clusters were sufficiently low, moreover, to allow selection of riboflavin prototrophs. It should, thus, be possible to apply the approaches reported here to isolate mutations in negative regulators of these and other SM clusters, and this may be a valuable general strategy for isolating negative regulators of secondary metabolism.
The strain construction strategy that we report here is doubly advantageous in that it facilitates the determination of whether a mutation is cis-activating or trans-activating, and it allows the mutation to be identified in a single sequencing run using progeny from a single cross. In fact, we were able to identify two different allelic mutations in a single run and, given the huge amounts of sequence available from a single run with current generation sequencers, it should be possible to identify several mutations, allelic and/or non-allelic, in a single sequencing run. These strategies should facilitate rapid and cost-effective identification of additional negative regulators of secondary metabolism.
Our approach has allowed us to identify a previously unstudied negative regulator of secondary metabolism that we designate mcrA. Our secondary metabolite profiles, obtained on a variety of media, reveal that mcrA plays a role in the regulation of bioactive compounds such as nidulanins, sterigmatocystin, the prenylated xanthones, aspercryptin, cichorines, F9775A and B, the emericellamides, and possibly terrequinone and microperfuranone. Sterigmatocystin, notably, is a precursor of aflatoxins (Brown et al., 1996) and both sterigmatocystin and the aflatoxins are extremely toxic and carcinogenic and are major public health and agricultural problems (Fujii et al., 1976; Williams et al., 2004). It is also noteworthy that the effects of the mcrA deletion on production of secondary metabolites differ on different growth media. This, and the fact that mcrAΔ alters expression of genes involve in primary metabolism, may indicate that McrA plays a role in connecting primary metabolism with secondary metabolism. In addition, mcrAΔ allowed us to discover two new compounds synthesized by the cichorine biosynthetic pathway and to discover that A. nidulans produces felinone A and an additional, apparently novel, but as yet unidentified, compound was produced in the gpdA(p)laeA, mcrAΔ strains.
Homology searches indicate that McrA, the protein product of mcrA, is likely a transcription factor with a Gal4 DNA binding domain. Transcription profiling revealed that mcrA is a master regulator, affecting the transcription of hundreds of genes. The mcrA gene is conserved in ascomycetes, and there are only one or two strong homologs per species, suggesting that mcrA homologs are conserved master regulators. It follows that deletion of mcrA homologs in other fungi may activate silent secondary metabolite BGCs and that deletions of mcrA homologs may be very valuable in discovering compounds produced by heretofore-silent SM gene clusters. Our initial experiments with A. terreus and P. canescens confirm that this is the case.
Our original screen was for a regulator of nidulanin A biosynthesis and our transcriptional profiling confirms that mcrA is a potent regulator of the nidulanin A biosynthetic genes. Although mcrA regulates many genes, secondary metabolite BGCs are prominent among them with five core SM biosynthetic genes from four BGCs being upregulated more than 50X by the mcrA deletion on a single medium (liquid GMM). It is worth noting that the upregulation of transcription of BGCs on liquid GMM did not result in a detectable upregulation of the product of the BGC in all instances. This could indicate that there is posttranscriptional regulation of SM biosynthesis or accumulation in some cases so that an increase in transcription may not result in an accumulation of product. Alternatively, upregulation of transcription could simply result in an increase in compound production that is too low to be detected under the conditions we have employed.
mcrAΔ stimulates SM production regardless of whether laeA is present, absent or overexpressed and this suggests that McrA, in general, acts independently of LaeA. There are clearly connections to the veA/velB/laeA system, however. Deletion of velB has been reported to downregulate AN8694, which we have now designated mcrA (Ahmed et al., 2013) and this might be expected to stimulate SM production. However, the velB deletion is not reported to stimulate SM production (Bayram et al., 2008). Deletion of mcrA causes a modest upregulation of laeA and deletion of laeA exacerbates the weak growth defect caused by mcrAΔ. mcrAΔ causes only a small stimulation of sterigmatocystin production if laeA is deleted so for this metabolite laeAΔ is epistatic to mcrAΔ. mcrAΔ and oelaeA collaborate to produce a new compound not found in the single mutants and, finally, F9775 A and B are produced in mcrAΔ, laeAΔ strains but not in either single mutant. These findings show that mcrA and the veA/velB/laeA system are connected, but they are not easily integrated into a simple, linear signaling model. Some of the findings are consistent with mcrA acting downstream of veA/velB/laeA. In this case, mcrA would be proximal to SM production and deletion of mcrA would cause SM production regardless of veA/velB/laeA signaling. This model does not, however, explain the aforementioned effects of laeAΔ on growth of mcrAΔ strains, nor the epistatic effect of laeAΔ on sterigmatocystin production nor the stimulation of F9775 A and B production in the double deletion strain.
In this regard, we call mcrA a master SM regulatory gene because mutation or deletion of this gene alters the production of a number of secondary metabolites. However, it may be more appropriate to think of regulatory networks than master regulators. These networks would integrate various signals (light, carbon source, nitrogen source, chemical environment, physical contact with predatory or prey organisms, developmental stage and perhaps many other things) and activate production of SMs as appropriate. Key regulators such as LaeA and McrA would be at nodes of these networks and alteration of these proteins would, thus, affect production of many SMs. We also note that since data to date suggest that LaeA and McrA together regulate fewer than half of the A. nidulans SM gene clusters, other important SM regulators likely await discovery.
Experimental Procedures
Mutagenesis
Mutagenesis procedures were modified from those of Holt and May (Holt and May, 1996). 5 × 109 spores of LO7543 were washed in 20 ml P-buffer (0.1M Potassium phosphate, pH 7.0) and resuspended in 10 ml P-buffer. 1.0 ml aliquots (5 × 108 spores) were transferred to 15 ml tubes and placed on ice. NQO (Sigma #N8141) was maintained as a 1.0 mg ml−1 stock solution in dimethylformamide. NQO was added to the tubes at 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5 µg ml−1 (dimethylformamide was normalized to 100 µl per sample); the tubes were vortexed briefly and incubated in a 32°C New Brunswick G76 gyratory shaker/water bath, shaking at 200 rpm, for 30 min. The reactions were quenched by addition of 4 ml 5% Sodium thiosulfate (filter-sterilized) to each tube. Serial dilutions were made and spread on GMM + pyridoxine + riboflavin) to generate a kill curve. GMM is 10 g L−1 D-glucose, 6 g L−1 NaNO3, 0.52 g L−1 KCl, 0.52 g L−1 MgSO4·7H2O, 1.52 g L−1 KH2PO4, 15 g L−1 agar, and 1 ml L−1 trace element solution (Vishniac and Santer, 1957). Each sample was also spread on GMM + pyridoxine to select for mutants capable of growing in the absence of ribflavin (ribo+ mutants). Plates were incubated at 37°C for 3 days. A total of 456 ribo+ mutants were generated from 1.83 × 109 spores that were spread on the MM + pyridoxine plates. Colonies for further analysis were chosen from the 0.5 and 1.0 µg ml−1 NQO samples (38.5 and 44.5% kill respectively).
Molecular Genetic Methods
Replacement of the coding sequences of core SM biosynthetic genes was carried out as previously described using fusion PCR to create transforming fragments (Nayak et al., 2006; Szewczyk et al., 2006; Oakley et al., 2012). Correct gene integration in each transformant was verified by at least three diagnostic PCR amplifications using different primer pairs. Deletion of entire SM clusters was carried out as previously described (Chiang et al., 2013). Most clusters were deleted using the loop out recombination procedure. Correct deletion of entire clusters was verified by diagnostic PCR amplifications using primers outside of the ends of the clusters.
Molecular genetic manipulations in A. terreus and P. canescens including protoplasting and gene targeting were carried out as previously described (Guo et al., 2014 ; Yaegashi et al., 2015). Deletions were confirmed by diagnostic PCR. The diagnostic PCR strategy is shown in Figure S9.
Genome sequencing
DNA was prepared from pooled hyphae using the method of Dunne and Oakley (1988) with minor modifications followed by purification over two, sequential CsCl gradients. Purified DNA was sheared and fragments 300–400 bp were ligated to barcode sequences using using an Illumina TruSeq DNA LT sample preparation Kit. Sequencing was 100 bp single read on a HiSeq 2500 sequencer.
Genotype calling
Short reads were aligned to reference sequence, A. nidulans FGSC A4 (AspGD version s09-m02-r02 (Cerqueira et al., 2014)), via BWA (Li and Durbin, 2009) using default parameters. Read groups were added using Picard (http://picard.sourceforge.net). Genotype calls were made through GATK package (McKenna et al., 2010): IndelRealligner executed with default parameters; UnifiedGenotyper executed with the following non-default parameters: “-nt 32 -dt NONE -glm BOTH -G Standard --output_mode EMIT_ALL_SITES -A AlleleBalance --computeSLOD”; VariantFiltration executed using the expression “((DP - MQ0) < 5) || ((MQ0 / (1.0 * DP)) >= 0.5) || AB > 0.1”. Resulting VCF files were uploaded into PolyDB (https://github.com/gustavo11/polydb). An initial set of candidate mutations was defined by selecting all variable positions between wild-type and mutant that were neither tagged as LowConfidence nor LowQual and that were located in regions with read coverage (DP field in the VCF file) with more than 50 reads on both wild-type and mutant samples. Reads supporting each candidate mutation were manually inspected using JBrowse (Skinner et al., 2009). Only candidates with more than 95% of the aligned reads in agreement with the called genotype were retained and included in the final set.
RNAseq samples and their analysis
1 × 106 spores of LO1362 and LO8158 were grown in 150 ml GMM liquid medium supplemented with riboflavin (2.5 mg L−1), pyridoxine (0.5 mg L−1), uracil (1 g L−1) and uridine (10 mM) in 1 L flasks at 37°C with shaking at 100 rpm for 24 hours. To obtain triplicate sets of data from each strain, three independent cultures of each strain were prepared and harvested. After harvesting by Miracloth filtration, cells were frozen with liquid nitrogen and disrupted to frozen powder with a CoolMill TK-CM20S (Tokken Inc., Japan). Total RNAs were purified from the frozen powder with NucleoSpin RNA plus kit (Machery-Nagel GmbH & Co., Germany). The RNA samples were analyzed with a 2100 Bioanalyzer (Agilent Technologies Inc., CA, U.S.A.) with RNA6000 nano reagent (Agilent Technologies Inc.) to confirm no obvious deterioration. To prepare sequencing libraries, Poly(A)-RNAs were purified from the total RNAs with NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Inc., MA, U.S.A.), and then directional RNA libraries of the poly(A)-RNAs were made with NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Inc.) according to the manufacturer’s instructions. The libraries were sequenced with the Illumina MiSeq platform (Illumina Inc., CA, U.S.A.) with MiSeq reagent kit v3 (150 cycles; Illumina Inc.). Short read data were aligned to reference sequence, A. nidulans FGSC A4 (AspGD version s10-m03-r26 (Cerqueira et al., 2014)), via HISAT (Kim et al., 2015). Mapped reads were visualized and analyzed with SeqMonk software (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and CLC genomics workbench software (CLC Bio., Denmark). Exact tests of differential expression of the two data sets were carried out with the EdgeR package (Robinson et al., 2010). RNAseq raw reads have been deposited at the NCBI Gene Expression Omnibus (GEO) (accession number GSE85927).
Quantitative RT-PCR
1 × 106 spores of LO1362 and LO8166 were grown in 100 mL LMM liquid medium supplemented with riboflavin (2.5 mg/L), pyridoxine (0.5 mg/L), uracil (1 g/L) and uridine (10 mM) in 300 ml flasks at 37°C with shaking at 100 rpm for 18 hours. After adding cyclopentanone to the cultures to a final concentration of 10 mM, cells were cultured further for 2 days and then harvested. To obtain triplicate sets of data from each strain, three independent cultures of each strain were prepared. After harvesting by Miracloth filtration, cells were frozen with liquid nitrogen and disrupted to a frozen powder with a Multi-beads Shocker (YASUI KIKAI Co., Ltd., Japan). Total RNAs were purified from the frozen powder with a NucleoSpin RNA plus kit (Machery-Nagel GmbH & Co., Germany). The RNA samples were analyzed with 2100 Bioanalyzer (Agilent Technologies Inc., CA, U.S.A.) with RNA6000 nano reagent (Agilent Technologies Inc.). Reverse transcription was carried-out with ReverTra Ace (TOYOBO Co., Ltd., Japan) according to manufacturer’s instructions with 0.5 to 1 µg of total RNAs as the template and with oligo-dT as the primer. For quantitative RT-PCR, we used a KAPA SYBR FAST qPCR kit for LightCycler 480 (KAPA BIO, U.S.A.) along with a LightCycler 480 (Roche Diagnostics, U.S.A.). The primers used for AN8694 (mcrA) were 5’-GGAGACGAAAGATCCGATGC-3’ and 5’-CGACGAGGTCTATGATCCTG-3’, and those used for actA were 5’-CGTTATCGACAATGGTTCGG-3’ and 5’-CGTGCTCAATGGGGTATCTG-3’. Purified PCR products of each cDNA were used as standard samples for calibration. AN8694 (mcrA) mRNA was quantified relative to actin [actA, AN6542] mRNA using the software of the real time PCR system. Fold change was further calculated with each triplicate value as the ratio of AN8694 mRNA amount in the expression-induced strain to that in the wild-type strain.
Fermentation and HPLC-DAD-MS analysis
For agar plate cultures, A. nidulans strains were incubated at 37°C on GMM [as above except that Hutner’s trace element solution (Hutner et al., 1950) was used at 1ml L−1], LMM (15 g L−1 D-lactose, 6 g L−1 NaNO3, 0.52 g L−1 KCl, 0.52 g L−1 MgSO4·7H2O, 1.52 g L−1 KH2PO4, 15 g L−1 agar, and 1 ml L−1 Hutner’s trace element solution), or YAG (5 g L−1 yeast extract, 20 g L−1, D-glucose, 15 g L−1 agar, and 1 ml L−1 Hutner’s trace element solution) plates were supplemented with riboflavin (2.5 mg L−1), pyridoxine (0.5 mg L−1), or uracil (1 g L−1) and uridine (10 mM) when necessary. Plates were inoculated with 1.0 × 107 spores per 10-cm plate. After 5 days, three plugs (7-mm diameter) were cut out and transferred to a 7-ml screw-cap vial. The material was extracted with 3 ml of methanol followed by 3 ml of 1:1 dichloromethane-methanol, each with a 1-hr sonication. The extract was transferred to a clean vial and the solvent was evaporated to dryness by TurboVap LV (Caliper LifeSciences). The residues were re-dissolved in 0.3 ml of DMSO:MeOH (1:4) and 10 µl was injected for LC-DAD-MS analysis as described previously (Bok et al., 2009). MS/MS was conducted with a normalized collision energy of 35 and isolation width of 2 m/z.
For liquid culture, 3 × 107 spores were grown in 30 ml GMM or LMM liquid medium (recipes same as above except no agar added) in 125 ml flasks at 37°C with shaking at 180 rpm. After 4 days, culture medium and hyphae were collected by filtration. Culture medium was extracted with the same volume of EtOAc. In order to extract the most acidic phenolic compounds, the water layer was extracted with the same volume of EtOAc again after acidification (pH = 2). Both EtOAc extracts were then evaporated by TurboVap LV. The residues were re-dissolved in 0.5 ml of DMSO:MeOH (1:4) and analyzed by LC-DAD-MS as described above. The hyphae collected were soaked in 30 ml of methanol overnight to extract metabolites not secreted out to the liquid medium. The methanol extract was then collected by filtration and the solvent was evaporated by TurboVap LV. The residues were re-dissolved in 1 ml of DMSO:MeOH (1:4) and analyzed by LC-DAD-MS as described above.
For alcA(p) induction, 10 mM of cyclopentanone was added to the LMM liquid culture 18 hr after inoculation. Culture medium and hyphae were collected 48 hr after cyclopentanone induction by filtration, followed by extraction and HPLC-DAD-MS analysis as described. Fermentation and LC-MS analysis for A. terreus and P. canescens are listed in the captions to Fig. S8.
Supplementary Material
Acknowledgments
We are grateful for funding support from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (P01-GM084077), the H. L. Snyder Medical Foundation, and the Irving S. Johnson Fund of the University of Kansas Endowment. Research reported in this publication was made possible in part by the services of the KU Genome Sequencing Core Laboratory, which is supported by NIGMS under award number P20GM103638. We would also like to acknowledge the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine for use of the MiSeq sequencer and related equipment. JRW and GCC were funded by the National Institute of Allergy and Infectious Diseases at the US National Institutes of Health (R01 AI077599). TA was funded in part by JSPS Grant-in-Aid for Scientific Research (C) Grant number 42619003.
Footnotes
The authors declare no conflict of interests.
Author contributions
C. Elizabeth Oakley carried out the genetic screen, mapping crosses, much of the strain construction, purified DNA for sequencing and participated in the editing of the manuscript. Manmeet Ahuja constructed some of the strains used. Wei-Wen Sun carried out some of the metabolite analyses and prepared some of the figures. Ruth Entwistle carried out some replacements of core secondary metabolism biosynthetic genes with the riboB coding sequence. Tomohiro Akashi carried out the transcription analyses. Gustavo Cequeira carried out sequence analysis to identify AN8694 as the gene harboring the mutations that activated expression from the AN1242 promoter. Jennifer Russo supervised the sequence analysis. Clay Wang supervised the metabolite analysis and helped edit the manuscript. Yi-Ming Chiang carried out some metabolite analyses, prepared portions of the manuscript, prepared some figures and helped edit the manuscript. Berl Oakley conceived and directed the genetic screen and wrote the majority of the manuscript.
References
- Ahmed YL, Gerke J, Park HS, Bayram Ö, Neumann P, Ni M, et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biol. 2013;11:e1001750. doi: 10.1371/journal.pbio.1001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans . J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci U S A. 2013;110:E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Daubersies P, Galiegue-Zouitina S, Loucheux-Lefebvre MH. Molecular basis of 4-nitroquinoline 1-oxide carcinogenesis. Jpn J Cancer Res. 1989;80:691–697. doi: 10.1111/j.1349-7006.1989.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O, Bayram OS, Ahmed YL, Maruyama J, Valerius O, Rizzoli SO, et al. The Aspergillus nidulans MAPK Module AnSte11-Ste50-Ste7-Fus3 Controls Development and Secondary Metabolism. PLoS Genet. 2012;8:e1002816. doi: 10.1371/journal.pgen.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- Bhatnagar D, Yu J, Ehrlich KC. Toxins of filamentous fungi. Chem Immunol. 2002;81:167–206. doi: 10.1159/000058867. [DOI] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bok J-W, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang Y-M, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, Schroeckh V. Fungal secondary metabolites - strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Bromann K, Toivari M, Viljanen K, Vuoristo A, Ruohonen L, Nakari-Setala T. Identification and characterization of a novel diterpene gene cluster in Aspergillus nidulans . PLoS One. 2012;7:e35450. doi: 10.1371/journal.pone.0035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans . Proc Natl Acad Sci U S A. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42:D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y-M, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, et al. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y-M, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CCC, Oakley BR. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y-M, Wang CCC, Oakley BR. In: Analyzing fungal secondary metabolite genes and gene clusters. Osbourn A, Goss R, Carter G, editors. Hoboken, N. J: John Wiley & Sons; 2014. pp. 175–193. [Google Scholar]
- Chiang YM, Ahuja M, Oakley CE, Entwistle R, Asokan A, Zutz C, et al. Development of Genetic Dereplication Strains in Aspergillus nidulans Results in the Discovery of Aspercryptin. Angew Chem Int Ed Engl. 2016;55:1662–1665. doi: 10.1002/anie.201507097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, et al. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen JM, Newberne PM. In: Acute hepatotoxicity of aflatoxins. DL E, JD G, editors. London: Academic Press; 1993. pp. 1–26. [Google Scholar]
- de Souza WR, Morais ER, Krohn NG, Savoldi M, Goldman MH, Rodrigues F, et al. Identification of metabolic pathways influenced by the G-protein coupled receptors GprB and GprD in Aspergillus nidulans. PLoS One. 2013;8:e62088. doi: 10.1371/journal.pone.0062088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes DJ, Chonofsky M, Tan K, Pfannenstiel BT, Reck-Peterson SL, Todd RB. Characterization of the mutagenic spectrum of 4-nitroquinoline 1-oxide (4-NQO) in Aspergillus nidulans by whole genome sequencing. G3 (Bethesda) 2014;4:2483–2492. doi: 10.1534/g3.114.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doy CH, Pateman JA, Olsen JE, Kane HJ, Creaser EH. Genomic clones of Aspergillus nidulans containing alcA, the structural gene for alcohol dehydrogenase and alcR, a regulatory gene for ethanol metabolism. DNA. 1985;4:105–114. doi: 10.1089/dna.1985.4.105. [DOI] [PubMed] [Google Scholar]
- Du FY, Li XM, Zhang P, Li CS, Wang BG. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Mar Drugs. 2014;12:2816–2826. doi: 10.3390/md12052816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne PW, Oakley BR. Mitotic gene conversion, reciprocal recombination and gene replacement at the benA, β-tubulin locus of Aspergillus nidulans . Mol Gen Genet. 1988;213:339–345. doi: 10.1007/BF00339600. [DOI] [PubMed] [Google Scholar]
- Felenbok B. The ethanol utilization regulon of Aspergillus nidulans: the alcA-alcR system as a tool for the expression of recombinant proteins. J Biotechnol. 1991;17:11–17. doi: 10.1016/0168-1656(91)90023-o. [DOI] [PubMed] [Google Scholar]
- Felenbok B, Flipphi M, Nikolaev I. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog Nucleic Acid Res Mol Biol. 2001;69:149–204. doi: 10.1016/s0079-6603(01)69047-0. [DOI] [PubMed] [Google Scholar]
- Fox EM, Howlett BJ. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 2008a;11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Fox EM, Howlett BJ. Biosynthetic gene clusters for epipolythiodioxopiperazines in filamentous fungi. Mycol Res. 2008b;112:162–169. doi: 10.1016/j.mycres.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Fujii K, Kurata H, Odashima S, Hatsuda Y. Tumor induction by a single subcutaneous injection of sterigmatocystin in newborn mice. Cancer Res. 1976;36:1615–1618. [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa . Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Waring P, Howlett BJ. The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology. 2005;151:1021–1032. doi: 10.1099/mic.0.27847-0. [DOI] [PubMed] [Google Scholar]
- Garzia A, Etxebeste O, Rodriguez-Romero J, Fischer R, Espeso EA, Ugalde U. Transcriptional changes in the transition from vegetative cells to asexual development in the model fungus Aspergillus nidulans. Eukaryot Cell. 2013;12:311–321. doi: 10.1128/EC.00274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, et al. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol. 2010;11:213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J, Bayram O, Feussner K, Landesfeind M, Shelest E, Feussner I, Braus GH. Breaking the silence: protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl Environ Microbiol. 2012;78:8234–8244. doi: 10.1128/AEM.01808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CJ, Sun WW, Bruno KS, Wang CC. Molecular genetic characterization of terreic acid pathway in Aspergillus terreus. Org Lett. 2014;16:5250–5253. doi: 10.1021/ol502242a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar JD, Bennett JW, Bhatnagar D, Bahu R. Sterigmatocystin production by laboratory strains of Aspergillus nidulans . Mycol Res. 1989;93:548–551. [Google Scholar]
- Hertweck C. Hidden biosynthetic treasures brought to light. Nat Chem Biol. 2009;5:450–452. doi: 10.1038/nchembio0709-450. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Holt CL, May GS. An extragenic suppressor of the mitosis-defective bimD6 mutation of Aspergillus nidulans codes for a chromosome scaffold protein. Genetics. 1996;142:777–787. doi: 10.1093/genetics/142.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutner SH, Provasoli L, Schatz A, Haskins CP. Some approaches to the study of the role of metals in the metabolism of microorganisms. Proc Am Philos Soc. 1950;94:152–170. [Google Scholar]
- Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ME, Braunberger K, Yi G, Cooper S, Nonhebel HM, Gondro C. A p53-like transcription factor similar to Ndt80 controls the response to nutrient stress in the filamentous fungus, Aspergillus nidulans. F1000Res. 2013;2:72. doi: 10.12688/f1000research.2-72.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N, Bok J, Chung D, Perrin RM, Shwab EK. LaeA, a global regulator of Aspergillus toxins. Med Mycol. 2006;(44 Suppl):83–85. doi: 10.1080/13693780600835773. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Sugui JA. What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Med Mycol. 2009;1(47 Suppl):S97–S103. doi: 10.1080/13693780802056012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, et al. Genome sequencing and analysis of Aspergillus oryzae . Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Martin JF. New aspects of genes and enzymes for beta-lactam antibiotic biosynthesis. Appl Microbiol Biotechnol. 1998;50:1–15. doi: 10.1007/s002530051249. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans . Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus . Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Edgerton-Morgan H, Oakley BR. Tools for manipulation of secondary metabolism pathways: rapid promoter replacements and gene deletions in Aspergillus nidulans . Methods Mol Biol. 2012;944:143–161. doi: 10.1007/978-1-62703-122-6_10. [DOI] [PubMed] [Google Scholar]
- Palaez F. In: Biological activities of fungal metabolites. An Z, editor. New York: Marcel Dekker; 2004. pp. 49–92. [Google Scholar]
- Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt PJ, Dingemanse MA, Jacobs-Meijsing BJ, Pouwels PH, van den Hondel CA. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene. 1988;69:49–57. doi: 10.1016/0378-1119(88)90377-0. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RD, Pouwels PH, van den Hondel CA. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene. 1990;93:101–109. doi: 10.1016/0378-1119(90)90142-e. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Zegers ND, Busscher M, Pouwels PH, van den Hondel CA. Intracellular and extracellular production of proteins in Aspergillus under the control of expression signals of the highly expressed Aspergillus nidulans gpdA gene. J Biotechnol. 1991;17:19–33. doi: 10.1016/0168-1656(91)90024-p. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, Dhingra S, Kincaid A, Shantappa S, Feng X, Calvo AM. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS One. 2013;8:e74122. doi: 10.1371/journal.pone.0074122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi M, Gazzetti K, Punelli F, Scarpari M, Zjalic S, Ricelli A, et al. Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl Microbiol Biotechnol. 2012;95:1293–1304. doi: 10.1007/s00253-012-3985-4. [DOI] [PubMed] [Google Scholar]
- Reverberi M, Zjalic S, Punelli F, Ricelli A, Fabbri AA, Fanelli C. Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit Contam. 2007;24:1070–1075. doi: 10.1080/02652030701553244. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Entwistle R, Corcoran D, Oakley BR, Wang CCC. Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans . Med Chem Commun. 2012a;3:997–1002. doi: 10.1039/C2MD20055D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang Y-M, et al. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans . J Am Chem Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Somoza AD, Keller NP, Wang CC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012b;29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza AD, Lee KH, Chiang YM, Oakley BR, Wang CCC. Reengineering an azaphilone biosynthesis pathway in Aspergillus nidulans to create lipoxygenase inhibitors. Org Lett. 2012;14:972–975. doi: 10.1021/ol203094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprote P, Brakhage AA. The light-dependent regulator velvet A of Aspergillus nidulans acts as a repressor of the penicillin biosynthesis. Arch Microbiol. 2007;188:69–79. doi: 10.1007/s00203-007-0224-y. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans . Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Keller NP. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol Microbiol. 2006;59:882–892. doi: 10.1111/j.1365-2958.2005.05000.x. [DOI] [PubMed] [Google Scholar]
- Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD. Host-selective toxins: agents of compatibility. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Williams JH, Phillips TD, Jolly PE, Stiles JK, Jolly CM, Aggarwal D. Human aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. Am J Clin Nutr. 2004;80:1106–1122. doi: 10.1093/ajcn/80.5.1106. [DOI] [PubMed] [Google Scholar]
- Yaegashi J, Oakley BR, Wang CC. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J Ind Microbiol Biotechnol. 2014;41:433–442. doi: 10.1007/s10295-013-1386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegashi J, Praseuth MB, Tyan SW, Sanchez JF, Entwistle R, Chiang YM, et al. Molecular Genetic Characterization of the Biosynthesis Cluster of a Prenylated Isoindolinone Alkaloid Aspernidine A in Aspergillus nidulans. Org Lett. 2013;15:2862–2865. doi: 10.1021/ol401187b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegashi J, Romsdahl JY-MC, Wang CCC. Genome mining and molecular characterization of the biosynthetic gene cluster of a diterpenic meroterpenoid, 15-deoxyoxalicine B, in Penicillium canescens . Chem Sci. 2015;6:6537–6544. doi: 10.1039/c5sc01965f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Keller NP. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49:329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Bhatnagar D, Cleveland TE. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus . FEBS Lett. 2004a;564:126–130. doi: 10.1016/S0014-5793(04)00327-8. [DOI] [PubMed] [Google Scholar]
- Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004b;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Keller N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.