Summary
Bacteria sense and respond to their environment through the use of two-component regulatory systems. The ability to adapt to a wide range of environmental stresses is directly related to the number of two-component systems an organism possesses. Recent advances in this area have identified numerous variations on the archetype systems that employ a sensor kinase and a response regulator. It is now evident that many orphan regulators that lack cognate kinases do not rely on phosphorylation for activation and new roles for unphosphorylated response regulators have been identified. The significance of recent findings and suggestions for further research are discussed.
Introduction
Two-component regulatory systems are fundamental signal transduction systems in bacteria and they are also found in archaea and eukarya. The first component is a sensor kinase, often a membrane protein. The sensor kinase (HK) is autophosphorylated by intracellular ATP on a conserved histidine residue. Recent studies suggest that although many HKs are membrane proteins, they may be responding to cytoplasmic signals (Wang et al., 2012; Foo et al., 2015). Upon activation, a phosphoryl group is transferred from the HK to the N-terminal receiver domain of the second protein, a response regulator (RR). RRs are most often DNA binding proteins that activate or repress transcription upon phosphorylation at a conserved aspartate residue, although some output or effector domains have enzymatic activity or other functions. For example, RR FrzS has a C-terminal coiled-coil domain that is hypothesized to interact with a cytoskeletal motor (Mignot et al., 2005). In some systems, the HK can also dephosphorylate the phosphorylated RR, but this is less well-established (Kenney, 2010).
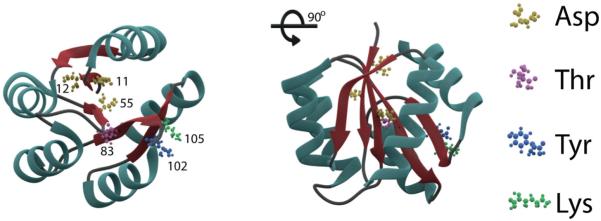
Substantial structural and biochemical characterization of RRs exists to enable a description of a series of molecular events involved in activation upon phosphorylation (see Hoch and Silhavy, 1995 for reviews). X-ray crystal structures of several receiver domains from homologous RRs have been solved, and they all reveal an (α/β)5 topology (summarized in Bourret, 2010). The five parallel β-strands form a hydrophobic core, surrounded by two α-helices on one side and three on the other. The receiver domains also share a group of conserved residues at the active site that are important for phosphorylation and signal propagation (Lee et al., 2001). These are (see Fig. 1): an aspartate residue that is the site of phosphorylation, Asp55 in OmpR, two additional acidic residues, Asp11 and Asp12 that coordinate a divalent metal ion (usually magnesium), these three aspartates form a catalytic or acidic triad, a lysine residue that interacts with the phosphoryl group, Lys105, an aromatic residue that functions as a rotamer, Tyr102, and a side-chain hydroxyl residue, Thr83, that also interacts with the phosphoryl group. Structural and functional analysis of activated response regulators has revealed the role of these conserved residues upon activation (Zhu et al., 1996; Mattison et al., 2002).
Fig. 1.
Conserved residues required for RR phosphorylation. The N-terminal receiver domain of OmpR is represented modelled on the CheY structure (PDB 4qyw). The side chains of conserved residues are depicted including: aspartate 11 and 12, aspartate 55 (site of phosphorylation), threonine 83, lysine 102 and tyrosine 105. The side chain of tyrosine 105 exists in both outward (depicted here) and inward positions. Alpha helices are illustrated in cyan, beta strands in red and loops are in grey.
In the presence of the HK or a small molecule phosphodonor such as acetyl phosphate or phosphoramidate, the threonine side-chain moves to form a charge-dipole interaction with the newly arrived phosphoryl moiety, a movement that is transmitted to the β4–α4 loop and is also tracked by the closely packed tyrosine side-chain. This ‘aromatic switch’, where the tyrosine moves from an exposed to a buried position, may be a general mechanism for intramolecular signalling in many RR proteins (Lee et al., 2001), although it does not appear to be valid for NtrC (Villali et al., 2014; Pontiggia et al., 2015; Vanatta et al., 2015). What remains less clear at present is how this conformational change is propagated to the C-terminal effector domain. Phosphorylation of the conserved aspartate residue often drives dimerization and modulation of DNA binding affinity. Many RRs can bind DNA without phosphorylation, including OmpR (Head et al., 1998), PhoP (Liu and Hulett, 1997), BvgA (Zu et al., 1996; Boucher et al., 1997), SsrB (Desai et al., 2016) etc., although phosphorylation increases the binding affinity and sometimes alters the specificity (Kenney, 2002). This minireview focuses on non-canonical activation by RRs including RRs that are not phosphorylated, lack the phosphorylated aspartate residue or possess other modifications and briefly summarizes the strategies employed. It is not intended to serve as an exhaustive review. For the purpose of this review, we refer to orphan RRs as those lacking cognate HKs, whereas pseudo-receivers or atypical RRs and non-canonical RRs are used interchangeably and indicate that either active site residues are lacking or activation is not via phosphorylation. In some cases, orphan RRs are also atypical.
RRs with distinct functions in unphosphorylated and phosphorylated states
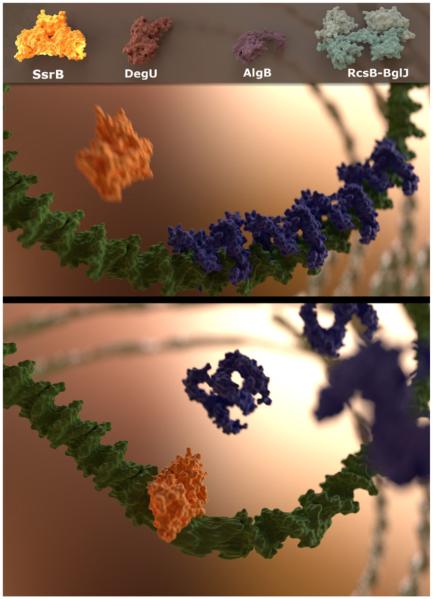
It has generally been assumed that phosphorylated RRs represented the active state, whereas the unphosphorylated state was inactive or inert. Recent studies have challenged this view. Some RRs make strong interactions between their N- and C-terminal domains, for example, MtrA and PrrA (Nowak et al., 2006; Friedland et al., 2007). Perhaps the best characterized of these is NarL, where N- and C-terminal interactions were first evident in the full-length crystal structure (Baikalov et al., 1996). Phosphorylation was required to reorganize the interface and promote dimerization and DNA binding. However, SsrB, a NarL homologue, was recently shown to bind and bend DNA in the absence of phosphorylation (i.e., when its kinase was not present or in a D56A mutant) (Desai et al., 2016), emphasizing that structural homology does not indicate functional homology (Fig. 2).
Fig. 2.
Unphosphorylated response regulators bind DNA and bend DNA to de-repress H-NS and relieve gene silencing. The upper panel shows a three dimensional reconstruction of SsrB (yellow) binding to the csgD (green) regulatory sequence (for details see Desai et al., 2016) on which the repressor, H-NS (blue), has formed a rigid nucleoprotein complex. Binding of SsrB introduces a bend in the csgD regulatory sequence, resulting in destabilization of the H-NS filament and a relief of transcriptional silencing, as shown in the lower panel. We propose that binding of other SsrB homologues (DegU, AlgR and RcsB-BglJ) to upstream gene sequences, may also lead to similar conformational changes and transcriptional activation by anti-silencing.
NarL homologue SsrB
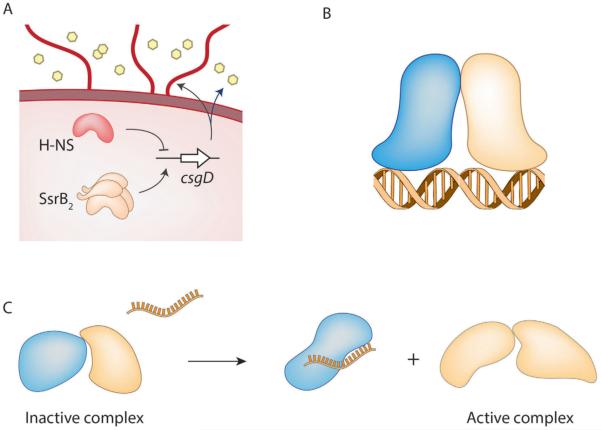
The two-component regulatory system, SsrA/B, present on the Salmonella Pathogenicity Island-2 (SPI-2), is responsible for the expression of a type three secretory system and the virulence effectors that are crucial for the survival of Salmonella enterica in macrophages. Activation of SPI-2 genes requires phosphorylation of SsrB at Asp56 by the tripartite sensor kinase, SsrA (Feng et al., 2004; Walthers et al., 2007). We recently discovered a novel role of unphosphorylated SsrB in the formation of biofilms, a multicellular lifestyle responsible for the establishment and maintenance of Salmonella in the carrier state in asymptomatic patients (Desai et al., 2016). In the absence of the HK SsrA, SsrB was sufficient to activate the transcription of csgD, which encodes for the master regulator of biofilms. In Escherichia coli and Salmonella, CsgD is required for the synthesis of cellulose and curli fimbriae that form the extracellular matrix of mature biofilms (Romling et al., 1998; Zogaj et al., 2001). Expression of csgD in a D56A SsrB mutant was similar to the wild type strain, providing the first evidence that phosphorylation of SsrB was not required for its positive role in regulating formation of biofilms. This was due to the ability of SsrB to activate csgD by relieving silencing by H-NS (Histone-like Nucleoid Structuring protein) (see Fig. 2). Single molecule Atomic Force Microscopy experiments showed that unphosphorylated SsrB was actually bound to the csgD regulatory region and introduced a bend of 82°. Further, such an SsrB-mediated conformational change in the promoter was responsible for displacing H-NS from discrete sub-regions on the DNA, enabling transcriptional activation. Unlike its subfamily member, NarL, SsrB does not require phosphorylation to bind DNA and mediate transcriptional activation via anti-silencing. However, for direct transcriptional activation of the SPI-2 genes, which requires an interaction with RNA polymerase, phosphorylation of SsrB is necessary (Walthers et al., 2007). Thus in Salmonella, the horizontally acquired RR, SsrB, has evolved to regulate the ancestral gene csgD, in a manner that is completely different from its classical function of regulating pathogenicity island genes (see Figs. 2 and 3A).
Fig. 3.
Strategies employed by RRs that do not involve phosphorylation.
A. Binding and bending DNA de-represses H-NS by driving it off the DNA or remodelling how it interacts with DNA. For example, unphosphorylated SsrB binds and bends the DNA and that relieves H-NS silencing at csgD.
B. Dimerization creates an active interface that is capable of DNA binding. This can involve homo- or hetero-dimerization.
C. RR sequestration via protection of the active surface (preventing dimerization) by small molecule binding or protein binding. A modification of this theme in the case of KaiA/KaiC involves KaiA protein binding to KaiC, stimulating KaiC autophosphorylation. For AmiR, AmiC binds and prevents AmiR interaction with RNA. Peptide binding to AmiC releases AmiR, allowing AmiR dimerization and activation.
NarL homologue RcsB
This same paradigm described above for unphosphorylated SsrB playing a role in activating Salmonella biofilms, may exist with RcsB (Fig. 3A). The RR RcsB, with the sensor kinase, RcsC, and the auxiliary regulatory protein, RcsA, form a complex phosphorelay signalling system involved in the synthesis of extracellular polysaccharide and some membrane and periplasmic proteins (see Majdalani and Gottesman, 2005 for a review). Unphosphorylated RcsB was reported to drive biofilm formation in Salmonella Typhimurium by activating expression of csgD, the master regulator (Latasa et al., 2012). If RcsB functions similarly to SsrB, then it would bind to the csgD promoter and de-repress H-NS. However, in this study (Latasa et al., 2012), no evidence of direct binding of unphosphorylated RcsB to the csgD regulatory region was provided, nor was there evidence provided for how unphosphorylated RcsB might mediate transcriptional activation of csgD. Instead, it was proposed by the authors that the presence of RscB~P leads to csgD repression through the production of RprA, a small regulatory RNA, leaving the open question as to what is the function of unphosphorylated RcsB?
OmpR subfamily archetype member OmpR
Recent evidence suggests that a variation on this paradigm is also relevant to OmpR-repression of genes required for neutralization during acid stress (Chakraborty et al., 2015). In the macrophage vacuole, the Salmonella cytoplasm acidifies via an OmpR-dependent repression of the cadC/BA operon. Expression of the cad genes would normally serve to neutralize the cytoplasm in response to acidification. OmpR binding and repression does not require phosphorylation (i.e., a D55A mutant is sufficient to repress the cadC/BA operon), although it does require the cognate HK EnvZ or the cytoplasmic domain of EnvZ, EnvZc. The assumption is that interaction of EnvZ with OmpR is sufficient to drive OmpR dimerization and DNA binding in the absence of phosphorylation (Chakraborty et al., 2016). A similar mechanism appears to function with the closely related OmpR homologue CpxR, where studies in Legionella pneumophila showed that a D53A CpxR mutant was no longer capable of transcriptional activation, but was still competent for repression (Feldhelm et al., 2015).
NarL homologue DegU
DegU is a RR in Bacillus subtilis belonging to the NarL subfamily. When phosphorylated by its atypical cytoplasmic HK DegS at conserved Asp55, it plays a central role in deciding cell fate by favouring biofilm formation. In the unphosphorylated form, DegU confers an ability to acquire foreign DNA via genetic competence (Dahl et al., 1992). Binding of unphosphorylated DegU enhanced auto-activation by stimulating binding of ComK to the upstream sites at comK (Hamoen et al., 2000). This cooperative effect of DegU on ComK binding to the comK regulatory region did not require direct protein–protein interactions and was presumed to occur through conformational changes in DNA (mediated by DegU-driven DNA bending?) (Hamoen et al., 2000). Thus, as with SsrB and OmpR, the unphosphorylated and phosphorylated forms of DegU control distinct pathways (Fig. 3A). However, precise experiments involving single molecule (Desai et al., 2016) or biochemical approaches (Will et al., 2014) are lacking to determine whether DegU, like SsrB, bends the DNA to increase the affinity of ComK binding.
LytR homologue AlgR
Another example of unphosphorylated RRs having distinct regulatory roles is found in the opportunistic pathogen Pseudomanas aeruginosa. It is known that phosphorylation of AlgR by the HK AlgZ is required to activate the fim genes for twitching motility (Whitchurch et al., 2002) and the hcn genes for hydrogen cyanide production (Cody et al., 2009). AlgR~P seems to be required to bind the fim and hcn promoters. On the other hand, transcriptional activation of the alg operon, which is required for the synthesis of the capsular polysaccharide, alginate, is directly mediated by two unphosphorylated response regulators, AlgB and AlgR (Ma et al., 1998; Leech et al., 2008). Pseudomonas strains carrying the D59N AlgB substitution and the D54N AlgR substitution retained the ability to form alginate-rich mucoid colonies. In this case, it is possible that unphosphorylated AlgR and AlgB are capable of binding to the algD promoter and with the help of co-activators such as IHF bring about conformational changes that result in transcriptional activation (see Okkotsu et al., 2014 for a review). IHF would then provide the DNA bending capability (Figs. 2 and 3A). However, the above predictions need to be tested experimentally using D59A AlgB or D54A AlgR strains, as de-amidation of Asn to Asp in CheY has been known to generate a wild type RR (Wolanin et al., 2003).
Orphan RRs mimic an active interface
In RRs that lack a conserved phosphorylated aspartate (sometimes referred to as atypical or pseudo-receiver domains), the most common substitution is glutamate (Maule et al., 2015). It remains enigmatic how some receivers are activated without phosphorylation, but many of these, including ChxR, HP1043, FrzS and AmiR are dimers in solution and appear to adopt an active conformation (Figs. 3B and 4), using a dimeric interface that is mostly similar to phosphorylated RRs (e.g., α4–β5–α5). NblR from Synechococcus elongatus sp PCC 7942 is a slight exception to these atypical RRs in that it retains an aspartate in the acidic pocket, but not the remaining residues required for phosphorylation (Ruiz et al., 2008). Although the active pocket and the YT pair of non-canonical RRs is often reorganized, much of the canonical post-phosphorylation mechanism appears to have been retained (Maule et al., 2015).
Fig. 4.
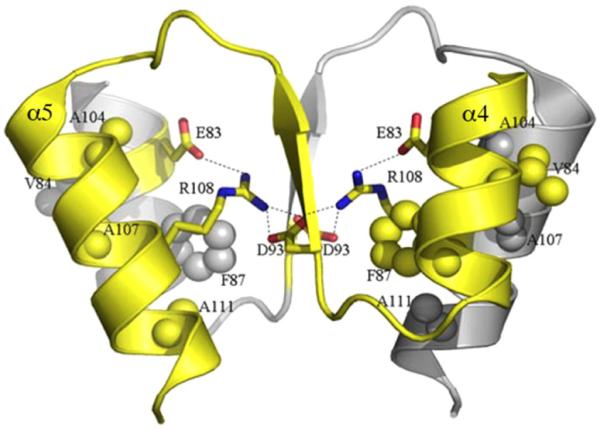
Structure of an active HP1043 RR dimer. Electrostatic and hydrophobic interactions of the dimeric interface are shown. A network of ionic interactions is formed between Glu83 (α4), Asp93 (β5) and Arg108 (α5). The core interactions of the hydrophobic patch consisting of Val84 (α4), Phe87 (α4), Ala104 (α5), Ala107 (α5) and Ala111 (α5) are also shown between the dimeric interface. Reprinted with permission from Hong et al., 2007).
OmpR homologue HP1043
Helicobacter pylori HP1043 is an orphan RR (i.e., no HK has been identified) that is essential for growth (Beier and Frank, 2000). It is also an atypical RR. In the unphosphorylated state, it is a symmetric dimer that binds to the target tlpB promoter DNA (Hong et al., 2007). The α4–β5–α5 dimer interface resembles that of the OmpR/PhoB subfamily (Figs. 3B and 4), although several conserved residues deviate from the typical interface (Hong et al., 2007). The stable interaction interface involves electrostatic and hydrophobic interactions and buries 800 Å2/monomer. Thus, the HP1043 dimer mimics the activated state, even in the absence of phosphorylation. The HP1043 dimer is structurally similar to the activated state of PhoB and ArcA (Toro-Roman et al., 2005; Arribas-Bosacoma et al., 2007) and the Tyr94 rotomer adopts the ‘active’ inward position. The fact that a stable dimer forms in solution probably constrains the structure, making it resolvable. This is unlike OmpR, in which the flexible linker presumably leads to crystals that diffract poorly.
An F87L mutant of HP1043, which substitutes a residue in α4 conserved in other RRs such as OmpR, is a monomer (Hong et al., 2007) (see Fig. 4). Surprisingly, the monomer was capable of DNA binding. Because actual binding data was not shown (only ‘+’ or ‘−’), it is not known whether the mutant possessed altered DNA binding affinity. One would expect that the monomer would exhibit lower binding affinity than the activated dimer. In any case, the HP1043 RR regulatory domain does not inhibit DNA binding activity of the C-terminal DNA binding domain and the two domains appear to function independently, for example, few chemical shift changes in the C-terminus were observed by NMR in the presence of DNA (Hong et al., 2007). This is unlike the archetype RR OmpR, for example, where even non-specific DNA resulted in many chemical shift changes (Rhee et al., 2008).
OmpR homologues GlnR and RamR
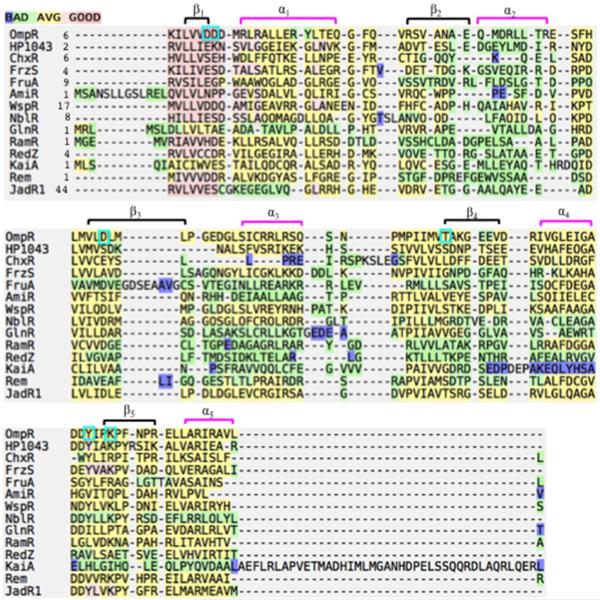
The GlnR protein of Actinomycetes is an OmpR subfamily member that coordinates expression of genes related to nitrogen metabolism. It also forms a homodimer through the α4–β5–α5 interface and has a conserved Asp residue in the active site (Figs. 3B and 4). It is considered an orphan RR since no cognate HK has been identified. Although Asp50 is conserved, the phosphorylation pocket is not conserved and is altered by charge interactions of Asp50 with Arg52 and Thr9 (Lin et al., 2014). Crystal structures of Amycolatopsis mediterranei GlnR or MTb GlnR indicated that except Asp50, conserved phosphorylation pocket residues were either not conserved or were not in the typical position. For example, conserved Mg-binding residues (OmpR residues Asp11 and Asp12, see Fig. 1) are replaced with Thr and Ala respectively. Furthermore, the ‘Y-T’ signalling residues Thr, Lys and Tyr (Fig. 5) are substituted with Val, Leu and Ile respectively (Lin et al., 2014). The GlnRD50A mutant is a monomer by gel filtration analysis, indicating that the conserved aspartate is required for dimerization. Residues Asp50, Arg52 and Thr9 are also conserved in GlnR proteins from Streptomyces coelicolor and MTb, but are not present in Gram-negative OmpR subfamily members, suggesting a common reaction mechanism. Thus, GlnR appears to be unphosphorylated, but the aspartate is critical for homodimerization and downstream DNA binding activity.
Fig. 5.
Alignment of RRs, highlighting the absence of key catalytic residues. Sequences of the N-terminal receiver domains of RRs or the first 120 amino acids in the case of AmiR, GlnR and RamR, were obtained from the UniProtKB database (www.uniprot.org/uniprot/) or the NCBI Gene database (http://www.ncbi.nlm.nih.gov/). Alignment of these sequences using the software Expresso (tcoffee.crg.cat) is shown. The ‘Expresso’ colour codes indicate the consistency in pairwise structural alignments. The group of conserved residues in the receiver domain of OmpR are boxed in turquoise, while the (α/β)5 topology is shown as black brackets for β-strands and magenta brackets for α-helices on top of the OmpR sequence.
RamR from S. coelicolor appears to be similar in this regard (Hudson and Nodwell, 2004; O’Connor and Nodwell, 2005). Attempts at phosphorylating the aspartate residue in vitro using phosphoramidate were unsuccessful and phos-tag analysis failed to demonstrate a phosphorylated GlnR from either A. mediterranei or S. coelicolor (Lin et al., 2014). The difficulty with a negative result is that the phos-tag barely detects PhoP~P (Lin et al., 2014) or OmpR~P (Adediran et al., 2014) in vivo, so a low level of GlnR~P might have simply gone undetected in these studies. However, the crystal structures confirm an inadequate phosphorylation pocket (Lin et al., 2014). Mutants that were unable to dimerize could not grow on minimal media in the presence of nitrate, nor were the GlnR gene targets activated, suggesting that phosphorylation-independent homodimerization was sufficient to activate GlnR (Fig. 3B).
Others
In Myxococcus xanthus, twitching motility is powered by type IV pili. The direction of movement is determined by the frequency of reversals along the long axis of the cell. Cell reversal correlates with RR FrzS accumulation at the new leading cell pole (Mignot et al., 2005). A combination of in vivo, crystallographic and NMR data suggest that the FrzS receiver domain retains structural and functional features of canonical receiver domains, but does not modulate switching or signalling via aspartate phosphorylation (Fraser et al., 2007). The FrzS receiver domain fold is highly similar to typical RRs, but the acidic triad (OmpR residues Asp11, 12, 55; see Fig. 5), the aspartate acid and the threonine residue in the active site are missing and the conserved Tyr102 is in an unusual conformation (Fraser et al., 2007). However, the α4–β5–α5 interface was maintained, and substitutions along this interface abolished M. xanthus S-motility (Fraser et al., 2007) (Figs. 3B and 5).
In a similar manner to FrzS, ChxR, a RR required for development in Chlamydia trachomatis, also appears to mimic the activated state in the absence of phosphorylation (Barta et al., 2014). ChxR lacks most of the conserved active site residues, containing a glutamic acid residue instead of aspartate (Koo et al., 2006), yet forms homodimers and binds to direct DNA repeats (Hickey et al., 2011a, 2011b).
FruA is a LysR family member from the social bacterium, Myxococcus xanthus, that controls cellular responses to C signalling during development. It may also fit this new paradigm of phosphorylation-independent dimerization (Fig. 3B). It appears to be an orphan, and is missing a couple of conserved aspartate residues, although dimerization has not been demonstrated (Mittal and Kroos, 2009).
The RR circadian clock protein KaiA from S. elongatus is proposed to function as a timing input-device. Its N-terminus is a pseudo-receiver domain in which the interface helix α4 is replaced with an unstructured, solvent-exposed loop that may serve an interfacing role (Williams et al., 2002). KaiA lacks the conserved aspartates for phosphorylation and magnesium binding (Fig. 5), these residues are instead replaced with Asn60, Glu12 and Ser13 respectively. The C-terminus of KaiA binds to KaiC, enhancing the KaiC rate of autophosphorylation (Williams et al., 2002). Phosphorylation of KaiA is not required for this effect (Fig. 3C).
Another active orphan RR is the Rem protein of S. meliloti. Rem is in a chemotaxis pathway and affects motility. It is expressed during exponential growth (Rotter et al., 2006) and is required for production of flagella. It has a glutamic acid in the active site. Potential phosphorylated aspartates Asp43, Asp45 and Asp47 were substituted with asparagine, but only combinations of multiple substitutions had an effect on swarming. The authors concluded that Rem was similar to HP1043. Dimerization was not examined, nor was the role of Glu50 in substituting for the phosphorylated Asp or a role in dimerization ever examined.
A question that arises from these discussions is what turns activated atypical receivers off? One mechanism is to only express the activated RR precisely when it is needed. For example, the RR Rem from S. meliloti is only expressed during exponential growth (Rotter et al., 2006). It has a glutamic acid (Glu50) in place of aspartate. Tight control appears to be achieved by synthesis, but whether proteolysis might play a role in the regulation of Rem has not been examined.
Regulation by sequestration
Some RRs are regulated by removal from their site of action by sequestration. AmiR is an orphan RR in Pseudomonas aeruginosa that lacks all of the conserved catalytic residues of RRs, including (OmpR residues are in parenthesis): Ser59 (Asp55), Asn 19 and Pro 20 (Asp 11, 12), Val86 (Thr83) and Gln108 (Lys105) (O’Hara et al., 1999). AmiR and AmiC constitute an amide-regulated transcription anti-termination system. AmiR is a coiled-coil dimer that binds RNA. AmiC binds amides at the interface of its N- and C-terminal domains (Pearl et al., 1994). In the absence of inducing amides, AmiR anti-termination activity is inhibited by interaction with AmiC (Fig. 3C). Addition of an inducing amide disrupts this complex in vitro. Once AmiR is liberated from AmiC, it multimerizes and binds to RNAs containing sequences upstream of an inverted repeat in the amidase promoter (Norman et al., 2000). Thus, AmiR activity is controlled by sequestration by AmiC, rather than phosphorylation (O’Hara et al., 1999).
A possible variation on the sequestering theme might occur during biofilm formation in Pseudomonas aeruginosa. The enzymatic output domain of the RR WspR~P is diguanylate cyclase activity, which is enhanced by the formation of higher order oligomers, visualized as distinct sub-cellular clusters. Cluster formation leads to synthesis of the exopolysaccharide PeI (Huangyutitham et al., 2013). WspR is canonical in that phosphorylation at Asp70 by the HK WspA is required to synthesize the second messenger, cyclic-di-GMP, and in turn, PeI. Binding of cyclic-di-GMP to the active WspR~P tetramer turns off its enzymatic activity. The question that arises is whether oligomerization of WspR~P tetramers affects the binding of cyclic-di-GMP, thus preventing auto-inhibition? This remains to be determined.
Heterodimer formation
Both typical and atypical RRs are capable of heterodimer formation. In this section, we highlight a few examples.
BldM/WhiI
Two RRs belonging to the FixJ/NarL subfamily in Streptomyces venezuelae form heterodimers to activate expression of group-II genes in the late stages of development (Al-Bassam et al., 2014). BldM and WhiI are orphan RRs that are also atypical, and heterodimer formation is an alternate way of regulating their DNA-binding activities. A BldM homodimer binds to group-I genes, whereas heterodimers of BldM/WhiI bind and activate group-II genes. WhiI alone does not activate transcription of group-II genes. Structural characterization of the BldM/WhiI heterodimers is lacking, and this would provide crucial information toward an understanding of the differences in activation and specificity mechanisms of heterodimers versus homodimers (Fig. 3B).
RcsB/BglJ and RcsB/GadE?
An example of heterodimerization of RRs occurs in the Gram-negative prototype, E. coli. The RR RcsB is part of a complex phosphorelay signalling system involved in the synthesis of extracellular polysaccharide and some membrane and periplasmic proteins (see above). In addition, unphosphorylated RcsB heterodimerizes with BglJ to activate the cryptic bgl operon by relieving H-NS silencing (Venkatesh et al., 2010) (Figs. 2 and 3A,B). RcsB/GadE heterodimers might also be activating the acid-responsive gadA promoter by anti-silencing (Castanie-Cornet et al., 2010). In contrast, RcsB homodimers activate environmentally-sensitive genes such as rpoS, osmB and osmC (Majdalani and Gottesman, 2005). Thus, homo and heterodimers of RRs can have different specificities.
Regulation by other covalent modifications
Post-translational modification of a RR at its conserved aspartate either by its cognate HK or small molecule phosphodonors such as acetyl phosphate modulates the activity of most RRs. However, to date, two additional covalent modifications have been discovered that also contribute to regulation of RR output activity.
Acetylation
In E. coli, the intracellular activity of RcsB is modified post-translationally by phosphorylation at Asp56, as well as by acetylation at Lys180 (Hu et al., 2013). Acetylation of Lys180 inhibited the ability of RcsB to bind to the rprA regulatory region, encoding a small regulatory RNA (Hu et al., 2013). However, a mutation in yfiQ, encoding the only known acetyl transferase in E. coli, did not abolish RcsB acetylation, suggesting perhaps a direct interaction of RcsB with acetyl-CoA. Although the precise mechanism by which acetylated RcsB controls gene expression is not known, it is interesting to speculate that acetylation of RcsB might prevent phosphoryl transfer from the RcsD histidyl phosphotransfer protein (HPT). It has not been established whether acetylation of the other six Lys residues in RcsB also affects its gene regulation functions (Hu et al., 2013).
Serine/threonine phosphorylation
Signal transduction pathways based on Serine/Threonine phosphorylation in Gram-positive bacteria show convergence with eukaryotic signalling pathways and also variable evolution of two-component regulatory systems across different phyla. The RR CovR in Group B Streptococci is a classic example where aspartate or threonine phosphorylation has opposing effects on its gene regulatory ability (Lin et al., 2009). Phosphorylation of CovR at Asp53 by the sensor HK CovS was required for DNA binding and activation of toxin gene expression. Phosphorylation of CovR at Thr65 by activation of the lone eukaryotic Ser/Thr kinase (eSTK) Stk1 drastically reduced its ability to bind DNA and activate gene expression, that is, phosphorylation at Thr65 of CovS reduced phosphorylation at Asp53. Reciprocally, Asp53 phosphorylation of CovR also reduced Stk1-driven Thr65 phosphorylation (Lin et al., 2009). Allosteric regulation of CovR activity by differential phosphorylation at Asp and Thr residues was also observed in Group A Streptococcus (Horstmann et al., 2014).
In contrast, in Bacillus subtilis, the activity of RR WalR is influenced by two separate classes of membrane-bound kinases, the cognate HK WalK, and the eSTK, PrkC (Libby et al., 2015). The WalR/K two-component system is essential and controls cell wall metabolism. Phosphorylation at Asp53 or Thr101 had similar effects on the downstream expression of WalR regulon genes. In the exponential growth phase, WalK phosphorylated WalR at the conserved Asp53, resulting in activation or repression of the WalR regulon genes involved in cell wall metabolism. However, when cells entered stationary phase, PrkC-dependent phosphorylation at Thr101 influenced the regulatory role of WalR (presumably as WalK activity was low). It is still not clear how the different forms of WalR~P compare in their ability to bind DNA or undergo conformational changes.
Small ligand binding
In the antibiotic-producing species Streptomycetes venezuelae (ISP5230), production of the broad-spectrum jadomycins is determined by a large gene cluster containing a regulatory gene specific for the pathway. JadR1 is an orphan RR in the OmpR subfamily and lacks the two N-terminal catalytic triad aspartate residues required for metal binding (replaced with Glu49 and Ser50). Instead, ligands of the late biosynthetic products, including jadomycin B (JdB) and aglycone (JdA), bind to the N-terminus of JadR1 (Fig. 3C). JdA binding enhanced JadR1 activity, whereas JdB binding inhibited its DNA binding activity (Wang et al., 2009). In another antibiotic pathway of S. coelicolor, the NarL homologue RedZ is also an orphan RR that is modified by undecylprodigisines. It lacks two aspartates for metal binding and the lysine of the conserved phosphorylation pocket (Guthrie et al., 1998). Thus, end product-mediated control of orphan RRs (through ligand binding) may be a widespread mechanism of regulation in antibiotic-producing bacteria, replacing or in some cases augmenting phosphorylation.
Final remarks
In summary, with more available structures and new assays and approaches for studying two-component regulatory systems, it is evident that there are many variations on the canonical theme. We are just beginning to appreciate that unphosphorylated RRs can evoke responses in pathways that are distinct from phosphorylated RRs and we presented some ways in which this has been shown to occur. Other orphan RRs can function without phosphorylation by mimicking an activated state, often via dimerization using the α4–β5–α5 interface.
Acknowledgments
We thank our RR colleagues who provided supporting information and clarification. We are grateful to Diego Pitta de Araujo and Cindy Zhang of the Science Communications team, MBI for figures. Supported by NIH R21-AI123640, VA5IOBX-00327 and a Research Centre of Excellence Grant for Mechanobiology from the MOE, Singapore.
Footnotes
The authors declare that they have no conflicts of interest.
References
- Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Moller J, Krogfelt KA, Kazmierczak K, et al. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun. 2014;82:670–682. doi: 10.1128/IAI.01149-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam MM, Bibb MJ, Bush MJ, Chandra G, Buttner MJ. Response regulator heterodimer formation controls a key stage in Streptomyces development. PLoS Genet. 2014;10:e1004554. doi: 10.1371/journal.pgen.1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Bosacoma R, Kim SK, Ferrer-Orta C, Blanco AG, Pereira PJ, Gomis-Ruth FX, et al. The X-ray crystal structures of two constitutively active mutants of the Escherichia coli PhoB receiver domain give insights into activation. J Mol Biol. 2007;366:626–641. doi: 10.1016/j.jmb.2006.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- Barta ML, Hickey JM, Anbanandam A, Dyer K, Hammel M, Hefty PS. Atypical response regulator ChxR from Chlamydia trachomatis is structurally poised for DNA binding. PloS One. 2014;9:e91760. doi: 10.1371/journal.pone.0091760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Frank R. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol. 2000;182:2068–2076. doi: 10.1128/jb.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher PE, Murakami K, Ishihama A, Stibitz S. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J Bacteriol. 1997;179:1755–1763. doi: 10.1128/jb.179.5.1755-1763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010;38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Mizusaki H, Kenney LJ. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol. 2015;13:e1002116. doi: 10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Winardhi RS, Yan J, Kenney LJ. Single cell analysis of acid/osmotic stress: a new view of pH regulation in prokaryotes. 2016 Submitted. [Google Scholar]
- Cody WL, Pritchett CL, Jones AK, Carterson AJ, Jackson D, Frisk A, Wolfgang MC, Schurr MJ. Pseudomonas aeruginosa AlgR controls cyanide production in an AlgZ-dependent manner. J Bacteriol. 2009;191:2993–3002. doi: 10.1128/JB.01156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- Desai SK, Winardhi RS, Periasamy S, Dykas MM, Jie Y, Kenney LJ. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. eLife. 2016;5:e10747. doi: 10.7554/eLife.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim YS, Zusman T, Speiser Y, Segal G. The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR’s function as a dual regulator and its connection to the effectors regulatory network. Mol Microbiol. 2016;99:1059–1079. doi: 10.1111/mmi.13290. [DOI] [PubMed] [Google Scholar]
- Feng X, Walthers D, Oropeza R, Kenney LJ. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol. 2004;54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- Foo YH, Gao Y, Zhang H, Kenney LJ. Cytoplasmic sensing by the inner membrane histidine kinase EnvZ. Prog Biophys Mol Biol. 2015;118:119–129. doi: 10.1016/j.pbiomolbio.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JS, Merlie JP, Jr., Echols N, Weisfield SR, Mignot T, Wemmer DE, Zusman DR, Alber T. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol Microbiol. 2007;65:319–332. doi: 10.1111/j.1365-2958.2007.05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland N, Mack TR, Yu M, Hung LW, Terwilliger TC, Waldo GS, Stock AM. Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry. 2007;46:6733–6743. doi: 10.1021/bi602546q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie EP, Flaxman CS, White J, Hodgson DA, Bibb MJ, Chater KF. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology. 1998;144:727–738. doi: 10.1099/00221287-144-3-727. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Van Werkhoven AF, Venema G, Dubnau D. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc Natl Acad Sci U S A. 2000;97:9246–9251. doi: 10.1073/pnas.160010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head CG, Tardy A, Kenney LJ. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- Hickey JM, Lovell S, Battaile KP, Hu L, Middaugh CR, Hefty PS. The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily. J Biol Chem. 2011a;286:32606–32616. doi: 10.1074/jbc.M111.220574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey JM, Weldon L, Hefty PS. The atypical OmpR/PhoB response regulator ChxR from Chlamydia trachomatis forms homodimers in vivo and binds a direct repeat of nucleotide sequences. J Bacteriol. 2011b;193:389–398. doi: 10.1128/JB.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. ASM Press; Washington, DC: 1995. [Google Scholar]
- Hong E, Lee HM, Ko H, Kim DU, Jeon BY, Jung J, et al. Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J Biol Chem. 2007;282:20667–20675. doi: 10.1074/jbc.M609104200. [DOI] [PubMed] [Google Scholar]
- Horstmann N, Saldana M, Sahasrabhojane P, Yao H, Su X, Thompson E, Koller A, Shelburne SA., 3rd Dual-site phosphorylation of the control of virulence regulator impacts Group A Streptococcal global gene expression and pathogenesis. PLoS Pathog. 2014;10:e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Basell K, et al. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol. 2013;195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangyutitham V, Guvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio. 2013;4:e00242–e00213. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Nodwell JR. Dimerization of the RamC morphogenetic protein of Streptomyces coelicolor. J Bacteriol. 2004;186:1330–1336. doi: 10.1128/JB.186.5.1330-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LJ. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol. 2002;5:135–141. doi: 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- Kenney LJ. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010;13:1–9. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo IC, Walthers D, Hefty PS, Kenney LJ, Stephens RS. ChxR is a transcriptional activator in Chlamydia. Proc Natl Acad Sci U S A. 2006;103:750–755. doi: 10.1073/pnas.0509690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa C, Garcia B, Echeverz M, Toledo-Arana A, Valle J, Campoy S, et al. Salmonella biofilm development depends on the phosphorylation status of RcsB. J Bacteriol. 2012;194:3708–3722. doi: 10.1128/JB.00361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Goss LA, Dworkin J. The eukaryotic-like Ser/Thr kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis. PLoS Genet. 2015;11:e1005275. doi: 10.1371/journal.pgen.1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, Kenney LJ, Rajagopal L. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol. 2009;71:1477–1495. doi: 10.1111/j.1365-2958.2009.06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Wang Y, Han X, Zhang Z, Wang C, Wang J, et al. Atypical OmpR/PhoB subfamily response regulator GlnR of Actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions. J Biol Chem. 2014;289:15413–15425. doi: 10.1074/jbc.M113.543504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Hulett FM. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Selvaraj U, Ohman DE, Quarless R, Hassett DJ, Wozniak DJ. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Byers N, Kenney LJ. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol. 2002;315:497–511. doi: 10.1006/jmbi.2001.5222. [DOI] [PubMed] [Google Scholar]
- Maule AF, Wright DP, Weiner JJ, Han L, Peterson FC, Volkman BF, Silvaggi NR, Ulijasz AT. The aspartate-less receiver (ALR) domains: distribution, structure and function. PLoS Path. 2015;11:e1004795. doi: 10.1371/journal.ppat.1004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Merlie JP, Jr., Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- Mittal S, Kroos L. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc Natl Acad Sci U S A. 2009;106:1965–1970. doi: 10.1073/pnas.0808516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RA, Poh CL, Pearl LH, O’hara BP, Drew RE. Steric hindrance regulation of the Pseudomonas aeruginosa amidase operon. J Biol Chem. 2000;275:30660–30667. doi: 10.1074/jbc.M000813200. [DOI] [PubMed] [Google Scholar]
- Nowak E, Panjikar S, Konarev P, Svergun DI, Tucker PA. The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis. J Biol Chem. 2006;281:9659–9666. doi: 10.1074/jbc.M512004200. [DOI] [PubMed] [Google Scholar]
- O’Connor TJ, Nodwell JR. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J Mol Biol. 2005;351:1030–1047. doi: 10.1016/j.jmb.2005.06.053. [DOI] [PubMed] [Google Scholar]
- O’Hara BP, Norman RA, Wan PT, Roe SM, Barrett TE, Drew RE, Pearl LH. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. Embo J. 1999;18:5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkotsu Y, Little AS, Schurr MJ. The Pseudomonas aeruginosa AlgZR two-component system coordinates multiple phenotypes. Front Cell Infect Microbiol. 2014;4:82. doi: 10.3389/fcimb.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L, O’hara B, Drew R, Wilson S. Crystal structure of AmiC: the controller of transcription anti-termination in the amidase operon of Pseudomonas aeruginosa. Embo J. 1994;13:5810–5817. doi: 10.1002/j.1460-2075.1994.tb06924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiggia F, Pachov DV, Clarkson MW, Villali J, Hagan MF, Pande VS, Kern D. Free energy landscape of activation in a signalling protein at atomic resolution. Nat Commun. 2015;6:7284. doi: 10.1038/ncomms8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter C, Muhlbacher S, Salamon D, Schmitt R, Scharf B. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol. 2006;188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz D, Salinas P, Lopez-Redondo ML, Cayuela ML, Marina A, Contreras A. Phosphorylation-independent activation of the atypical response regulator NblR. Microbiology. 2008;154:3002–3015. doi: 10.1099/mic.0.2008/020677-0. [DOI] [PubMed] [Google Scholar]
- Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J Mol Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanatta DK, Shukla D, Lawrenz M, Pande VS. A network of molecular switches controls the activation of the two-component response regulator NtrC. Nat Commun. 2015;6:7283. doi: 10.1038/ncomms8283. [DOI] [PubMed] [Google Scholar]
- Venkatesh GR, Kembou Koungni FC, Paukner A, Stratmann T, Blissenbach B, Schnetz K. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J Bacteriol. 2010;192:6456–6464. doi: 10.1128/JB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villali J, Pontiggia F, Clarkson MW, Hagan MF, Kern D. Evidence against the “Y-T coupling” mechanism of activation in the response regulator NtrC. J Mol Biol. 2014;426:1554–1567. doi: 10.1016/j.jmb.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. Embo J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Erova TE, Emery JA, Sargent JL, Harris JM, Semmler AB, et al. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J Bacteriol. 2002;184:4544–4554. doi: 10.1128/JB.184.16.4544-4554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will WR, Bale DH, Reid PJ, Libby SJ, Fang FC. Evolutionary expansion of a regulatory network by counter-silencing. Nat Commun. 2014;5:5270. doi: 10.1038/ncomms6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci U S A. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanin PM, Webre DJ, Stock JB. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry. 2003;42:14075–14082. doi: 10.1021/bi034883t. [DOI] [PubMed] [Google Scholar]
- Zhu X, Amsler CD, Volz K, Matsumura P. Tyrosine 106 of CheY plays an important role in chemotaxis signal transduction in Escherichia coli. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- Zu T, Manetti R, Rappuoli R, Scarlato V. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol Microbiol. 1996;21:557–565. doi: 10.1111/j.1365-2958.1996.tb02564.x. [DOI] [PubMed] [Google Scholar]