Abstract
Programmed cell death-2 (PDCD2) protein is enriched in embryonic, hematopoietic, and neural stem cells, however, its function in stem/progenitor cell differentiation is unclear. We investigated the effects of PDCD2 knockdown on the development and differentiation of hematopoietic progenitor cells (HPC). CD34+ cells derived from normal human bone marrow and K562 leukemic cells were effectively transduced with short-hairpin RNA to knockdown PDCD2. Colony-forming assays were used to investigate the effects of PDCD2 loss on HPC clonogenic potential and on 12-O-tetradecanoyl-phorbol-13-acetate–and arabinofuranosylcytosine–induced terminal differentiation. In CD34+ clonogenic progenitors, PDCD2 knockdown decreased the total number of colony-forming units, increased the number of colony-forming units–granulocyte-erythroid-macrophage-megakaryocyte and burst-forming unit–erythroid primitive colonies, and decreased the number of burst-forming unit–erythroid mature colonies. Similar results were observed in K562 cells, suggesting that PDCD2 is important for HPC differentiation and/or survival, and for erythroid lineage commitment. Furthermore, 12-O-tetradecanoyl-phorbol-13-acetate–induced megakaryocytic differentiation and proliferation of K562 cells was not affected by PDCD2 knockdown. In contrast, arabinofuranosylcytosine-induced erythroid differentiation of K562 cells was significantly reduced with PDCD2 knockdown, with no effect on cell proliferation. The effects of PDCD2 knockdown were attributed to a cell cycle arrest at G0/G1, along with increased messenger RNA expression of early progenitor factors c-MYB and GATA-2, and decreased expression of erythroid factors GATA-1, EpoR, and γ-globin. We conclude that PDCD2 loss of function(s) impedes erythroid differentiation by inducing cell cycle arrest and increasing expression of early hematopoietic progenitor factors. These findings suggest that PDCD2 has a novel regulatory role in human hematopoiesis and is essential for erythroid development.
Defining the factors that regulate the developmental plasticity of hematopoietic progenitor cells and how multipotent progenitors commit to terminal cell fates is an important aspect of hematology. The hierarchical transition from hematopoietic stem cells (HSCs) to lineage-restricted progenitor cells involves progressive loss of self-renewal potential and modulation of proliferative ability and lineage commitment. During hematopoietic development, the differentiation potential occurs at the levels of transient primitive erythroblasts, multipotent progenitor cells (MPPs), and definitive HSCs [1]. Hematopoietic colony-forming cell (CFC) assays have been used extensively to assess hematopoietic progenitor content and to identify stimulatory and inhibitory growth factors [2,3]. For example, the sequence of differentiation stages for the erythroid lineage includes MPP, common myeloid progenitors that undergo progressive loss of lymphoid potential and segregation into the bipotent megakaryocytic-erythroid progenitor (MEP). When stimulated with growth factors, the erythroid-restricted progenitors differentiate into erythropoietin (Epo)-responsive earlier burst-forming unit–erythroid (BFU-E) and subsequent late colony-forming unit–erythroid (CFU-E). CFU-E then generate a cascade of erythroid precursors that progress through terminal erythroid differentiation. This erythroid precursor maturation is characterized by decreased cell size, hemoglobin accumulation, nuclear condensation, and expression of erythroid surface markers.
At the molecular level, hematopoietic MPPs undergo a progressive restriction of differentiation driven by lineage-restricting transcription factors (TFs) in response to cytokines, chemokines, and colony-stimulating factors [4]. Lineage-restricting TFs, such as SCL/TAL1, GATA-2, MYB, PAX5, PU.1, FLI1, or RUNX1, are critical for the specification and biological function of HSCs and MPPs. In addition, these essential hematopoietic TFs maintain MPPs, promote restriction of differentiation potential to produce unipotent progenitors, and may be required for lineage commitment to produce the 10 subtypes of mature blood cells. Although the expression of many of these factors is largely not hematopoietic-restricted, their roles in regulating hematopoietic differentiation have been established.
Clearly, many more important regulators of HPC differentiation remain to be identified. We hypothesized that PDCD2 plays important role(s) in directing lineage-specific differentiation of human HPCs for several reasons. Key functional evidence for a critical role of PDCD2 in stem cell development was recently reported in Drosophila by Minakhina and Steward, where the PDCD2 ortholog ZFRP8 was found to be essential for maintenance of hemocyte stem cells, but not their differentiated daughter cells [5]. Pdcd2 is enriched in mouse embryonic, neural, and hematopoietic stem cells [6], and is highly expressed in human embryonic stem cells compared with their differentiated derivatives [7]. Knockout mouse studies showed that Pdcd2 is essential for embryonic stem cell viability and self-renewal [8]. Mouse Pdcd2 has not been reported to bind to DNA as a transcriptional factor, but rather, physically interacts with host cell factor 1 [9], which regulates multiple phases of the cell cycle [10]. Cellular functions of human PDCD2 have not been fully elucidated. Human PDCD2 is located on chromosome 6q27 in a region involved in both translocations and deletions in leukemias and lymphomas [11–13], therefore, PDCD2 might also be required for hematopoietic cell fate decisions.
Leukemic cells frequently retain differentiation programs similar to those of normal hematopoietic progenitors. K562 leukemia cells, derived from a patient with chronic myeloid leukemia (CML) in erythroleukemia blast crisis [14], resemble bipotent MEPs, and have been extensively utilized to study erythroid and megakaryocytic differentiation. Arabinofuranosylcytosine (Ara-C) can be used to induce erythroid differentiation and subsequent hemoglobin synthesis in K562 cells [15]. In contrast, treatment of K562 cells with phorbol esters such as 12-O-tetradecanoyl-phorbol-13-acetate (TPA) leads to loss of erythroid properties and acquisition of megakaryoblastoid characteristics. These megakaryocytic features include synthesis and surface expression of glycoprotein IIIa with increased positivity of platelet peroxidase, increased cell volume, and DNA ploidy [16]. We used this cellular model to study the role of PDCD2 in MEP cell differentiation. In addition, we investigated the effects of lentiviral short-hairpin RNA (shRNA)–mediated knockdown of PDCD2 expression on colony-forming activities of normal human bone marrow CD34+ cells and K562 leukemia cells. We demonstrate that PDCD2 knockdown decreases survival of human HPCs; maintains MEPs and early erythroid progenitors; and reduces spontaneous and induced erythroid, but not megakaryocytic differentiation, suggesting that loss of PDCD2 interferes with progenitor cell commitment and/or terminal erythroid differentiation.
Materials and methods
Reagents
Cytosine β-D-arabinofuranoside (Ara-C), phorbol 12-myristate 13-acetate (PMA/TPA), and benzidine (4,4′-diaminobiphenyl: B3503) were purchased from Sigma (St Louis, MO, USA). Serum substitute for culturing human hematopoietic cells (BIT9500), and recombinant human stem cell factor were purchased from StemCell Technologies (Vancouver, BC, Canada). Recombinant human fms-like tyrosine kinase receptor-3 ligand (Flt3L) and recombinant human thrombopoietin were purchased from R&D Systems (Minneapolis, MN, USA). Recombinant fibronectin CH-296 (RetroNectin) was purchased from TaKaRa Bio-Clontech (Mountain View, CA, USA).
Cell culture and media
Human leukemia cell lines Reh (lymphocytic leukemia), ML1 (myeloblastic leukemia), K562 (erythroleukemia), HL-60 (promyelocytic leukemia), Jurkat (lymphoblastic leukemia), and 293T (virus producer packaging cell line) were generously provided by Dr. Ganesan (Cancer Institute of New Jersey, New Brunswick, NJ, USA). KG-1 (CD34 antigen-expressing myeloblastic leukemia) cells were purchased from ATCC (Manassas, VA, USA) and maintained in Iscove’s modified Dulbecco’s medium (IMDM; Lonza, Rockland, ME, USA) supplemented with L-glutamine, and 20% fetal bovine serum (FBS). K562, ML1, and Reh cells were maintained in RPMI 1640 medium (Mediatech Inc., Manassas, VA, USA) supplemented with 10% FBS. Jurkat cells were maintained in RPMI 1640 medium supplemented with 10 mM HEPES and 10% FBS. HL-60 and 293T were maintained in DMEM medium (Mediatech Inc.) supplemented with 10% FBS. Cultures were maintained at 37°C in an atmosphere of 5% CO2.
Lentiviral production and infection
To create the human GIPZ lentiviral shRNA targeting PDCD2 (pGIPZ-PDCD2) and GIPZ lentiviral nonsilencing control (pGIPZ), transfer vectors were purchased from Open Biosystems (Thermo Scientific, Huntsville, AL, USA). psPAX2 (packaging plasmid) and pMD2G (envelope plasmid) were purchased from AddGene (Cambridge, MA, USA). 293T cells were plated at 5.5 × 106 cells per 10-cm plate 1 day before transfection. Medium was changed to OPTIMEM reduced-serum medium (GIBCO-Invitrogen, Carlsbad, CA, USA) 30 min before transfection. Mixture of plasmid DNA consisting of transfer vector, packaging plasmid, and envelope plasmid were transfected using Open Biosystems Arrest-In Reagent (Thermo Scientific) according to manufacturer’s protocol. Cells were incubated with DNA/Arrest-In transfection complex for 5 to 6 hours, and then transfection media were aspirated and replaced with regular growth medium. The virus-containing media were collected after 48 hours and filtered through 0.22-μm filter. Viruses were concentrated by ultracentrifugation at 4°C for 1.5 hours at 24,500 rpm, and then reconstituted in IMDM medium, followed by determination of viral titers. The protocol for lentiviral transduction for primary human CD34+ hematopoietic stem/progenitor cells was modified from Millington et al. [17]. After sorting, CD34+ cells were kept in induction medium (IMDM, 20% BIT9500, 100 ng/mL stem cell factor, 100 ng/mL Flt3L, and 10 ng/mL thrombopoietin) for 48 hours before transduction. After the preinduction period, cells were split into three groups. The first group was kept in induction medium (control group), and the second and third groups were cultured in IMDM medium only (transduced groups). Cells were infected at multiplicity of infection of 40–100 in RetroNectin-coated plates by the spin-inoculation method (600g, 1.5 hours, 30°C). After 5 hours of incubation (5% CO2, 37°C), all of the necessary components of the induction medium were added. At 24 hours after infection, cells were monitored for green fluorescent protein (GFP) expression. CD34+ cells demonstrated a mean transduction efficiency ranging from 15% to 30%. Cells were counted and plated in colony assays. Similar procedures were done for K562 cells, but GFP+ cells were sorted at 48 hours post transduction and placed in liquid phase culture or clonogenic medium.
Antibodies, immunohistochemistry, and Western blot analyses
For Western blots, anti-PDCD2 polyclonal antibody (Protein-Tech Group, Chicago, IL, USA) (1:500) and anti–β-ACTIN (AC-15) monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) (1:1000) were used as primary antibodies and sheep anti-mouse IgG-horseradish peroxidase (Amersham-GE Healthcare, Piscataway, NJ, USA) (1:2000) and goat anti-rabbit IgG (H+L)-horseradish peroxidase (GIBCO-Invitrogen) (1:3000) were used. Immunohistochemistry was performed on formalin fixed paraffin-embedded tissue sections using anti-PDCD2 (1:100), anti-CD235a monoclonal antibody (1:500) (Pierce), and anti-CD41a (GPIIb/IIIa) (1:50) (a kind gift from Dr. B. Coller, Rockefeller University). Negative controls for immunohistochemistry included omitting the primary antibody during staining. For Western blotting, total protein lysates were extracted using 1× RIPA buffer (Boston BioProducts, Worcester, MA, USA) supplemented with mammalian protease arrest mix (786–331; GBiosciences, Maryland Heights, MO, USA), phosphatase inhibitor Cocktail (78420; Thermo Scientific, Inc.), 250 μM phenylmethylsulfonyl fluoride (PMSF), and 10 μg/mL aprotinin. Total proteins were estimated using a Bradford protein estimation kit as per manufacturer’s instructions. Total proteins at 40 μg each were separated on 8% to 16% Trisglycine precast gels (Lonza, Rockland, ME, USA), transferred to a nitrocellulose membrane (Amersham Hybond-ECL; GE Healthcare), and immunostained in the presence of 5% milk with anti-pdcd2 antibody, or anti–β-actin antibody after stripping. After three 15-min washes, secondary antibodies were applied in the presence of 5% milk and 2.5% bovine serum albumin. After an additional three 15-min washes, blots were developed with ECL Plus (Amersham GE Healthcare).
Purification of human bone marrow CD34+ cells
Bone marrow (BM) from healthy volunteers was collected in heparinized syringes according to University of Medicine and Dentistry of New Jersey institutional guidelines under an approved institutional review board protocol. Informed consent was obtained in all cases. Mononuclear cells were separated using density gradient centrifugation. Fresh normal bone marrow (NBM), not older than 10 hours, was diluted with 1 volume of RPMI 1640 medium and layered over Ficoll-Paque PLUS (GE Healthcare). Nearly 4 mL diluted BM were layered over 3 mL Ficoll in a 15-mL conical tube. Tubes were centrifuged at 400g for 30 min at 20°C in a swinging-bucket rotor without brakes. The upper layer and interphase were collected into a new 15-mL tube, washed twice with phosphate-buffered saline, and cells were then counted before labeling with magnetic bead–or fluorochrome-conjugated antibodies.
CD34+ cell purification was conducted using MACS cell sorter (Miltenyi Biotech, Auburn, CA, USA) and MACS CD34 Cell Isolation Kit using positive selection. Cells were then frozen in α–minimum essential medium with 20% FBS and 10% dimethyl sulfoxide, and were thawed when needed for prestimulation and transduction.
CFC assay for NBM and K562 leukemic cells
Assays for in vitro CFC for transduced and nontransduced CD34+ NBM cells and K562 cells were carried out in 1% methylcellulose medium supplemented with stem cell factor (50 ng/mL), granulocyte-macrophage colony-stimulating factor (10 ng/mL), interleukin-3 (10 ng/mL), and erythropoietin (3 U/mL) (Metho-Cult H4434; StemCell Technologies), which is formulated to support optimal growth of erythroid progenitors (CFU-E and BFU-E), granulocyte-macrophage progenitors (CFU-GM, CFU-G, and CFU-M) and multi-potential granulocyte, erythroid, macrophage and megakaryocyte progenitors (CFU-GEMM). CFC assays for CD34+ BM cells were performed by plating 6 × 103 to 1.7 × 104 cells/mL and for K562 cells from 103 to 1.7 × 104 cells/mL. Cultures were scored after 14 days for the presence of colonies (>40 cells). Erythroid colonies scoring were based on an estimate of the total number of cells in the colony, as well as the number of clusters making up the colony [2,3]. Scoring for primitive and mature BFU-E colonies was performed as described [18,19]. Benzidine staining of colonies was done as described previously [20].
Analysis of cellular differentiation
Erythroid differentiation was induced with 0.36 μM Ara-C. After 8 days, cells were cytospun for 5 min at 240 rpm, fixed with 100% methanol for 1 to 2 min, followed by standard Wright-Giemsa staining to determine the various stages of cell differentiation [21]. For hemoglobin content, cells were stained with benzidine solution as described previously [22]. Twenty microliters of 30% hydrogen peroxide mixed with 500 μL benzidine solution were added to an equal volume of cell suspension. The mixture was incubated at room temperature protected from light for 3 to 5 min. Cells were imaged and randomly selected fields were analyzed by ImageJ software (Treestar, Ashland, OR, USA). The staining threshold was set from 0 to 123 intensity to normalize background. Pixel densities from 0 to 500 were considered negative (cells without staining), 501 to 2000 were weak (light blue cells), and >2001 were strong (dark blue cells) staining. A total of at least 300 cells from two independent experiments were counted.
Megakaryocyte differentiation was induced with 10 nM TPA for 2 days. Differentiation was assessed by cellular morphology using standard Wright-Giemsa staining and with ploidy assay as a molecular feature of megakaryocytic differentiation [16] after propidium iodide staining. The ploidy classes were then determined after cytometric analysis.
Flow cytometry
For cell cycle analysis, cells were synchronized during a 24-hour culture in the absence of FBS (serum starvation) and subsequently serum stimulated for 72 hours in the presence of 10% FBS. A total of 4 × 104 cells were stained with saponin-propidium iodide solution (0.25% saponin, 50 μg/mL propidium iodide, 50 mg/mL RNAse A, 0.1% bovine serum albumin, 1 × phosphate-buffered saline) for 1 hour at room temperature.
To determine the purity of CD34+ cells after sorting, cells were stained with MACS anti-human CD34-phycoerythrin according to manufacturer’s recommendations, washed twice with phosphate-buffered saline, and stained with 2 μg/mL propidium iodide to exclude nonviable cells. Cells were analyzed using a FC500 Flow Cytometer (Beckman Coulter; Miami, FL, USA) with FlowJo software. At least 10,000 events were acquired to determine the proportion of positive cells compared with isotype control.
Quantitative real-time polymerase chain reaction
Total RNA was isolated by TRIzol Reagent (GIBCO-Invitrogen) according to the manufacturer’s protocol. Complementary DNA synthesis was performed with 400 to 800 ng total RNA using RevertAid Premium First Strand cDNA Synthesis kit with Ribolock RNAse inhibitor (Fermentas, Glen Burnie, MD, USA) according to manufacturer’s protocols. Quantitative real-time polymerase chain reaction (PCR) was performed in triplicate or quadruplicate, with Express SYBR GreenER Universal qPCR Supermix (Invitrogen), using the Step One Plus Real Time PCR instrument (Applied Biosystems). Quantitative real-time PCR primers were: γ-GLOBIN forward (CTTCAAGCTCCTGGGAAATGT) and reverse (GCAGAATAAAGCCTACCTTGAAAG), ε-GLOBIN forward (GCCTGTGGAGCAAGATGAAT) and reverse (GCGGGCTTGAGGTTGT), PDCD2 forward (CTGTGGAGCTGGGCTTCGCC) and reverse (CAGCAGGAAGGAGAGCGGGC), and GAPDH forward (AAATTGAGCCCGCAGCCTCCC) and reverse (CGGCTGGCGACGCAAAAGAA). Expression levels of γ-GLOBIN, ε-GLOBIN, and PDCD2 were estimated by minimal cycle threshold values (Ct) normalized to the reference expression of GAPDH in each sample.
Gene expression profile by semi-quantitative reverse transcription PCR
Total RNA and complementary DNA preparation has been prepared as described above. The G3PDH, EpoR, GATA-1, GATA-2, and c-MYB transcripts were amplified from the complementary DNA by conventional PCR using gene-specific primers Table 1. Each sample was tested in triplicate.
Table 1.
Oligonucleotides of forward and reverse primers for reverse transcription PCR
| mRNA | Forward primer | Reverse primer | Size of PCR product (bps) |
|---|---|---|---|
| G3PDH | 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ | 5′-CATGTGGGCCATGAGGTCCACCAC-3′ | 983 |
| c-MYB | 5′-GGCTGCCGCAGCCGGCTGAG-3′ | 5′-GAGCTTGTCCAGAAATATGGTC-3′ | 475 |
| GATA-1 | 5′-CAGGTACTCAGTGCACCAAC-3′ | 5′-TGGTAGAGATGGGCAGTACC-3′ | 334 |
| GATA-2 | 5′-CAGAAGAGCCGGCACCTGTT-3′ | 5′-GGGATGACTTCTCCTGCATG-3′ | 254 |
| EpoR | 5′-GGACACCTACTTGGTATTGC-3′ | 5′-GACGTTGTAGGCTGGAGTCC-3′ | 552 and 457 |
Statistical analysis
All analyses were done using SAS 9.2 (SAS Institute, Cary, NC, USA). Tukey’s adjustment was applied to all pairwise comparisons. Mixed models (proc glimmix procedure) were used to model benzidine treatment and cell cycle data. For colony-forming assay (Fig. 2 and 3), the numbers of colonies were normalized to the total number of plated cells. The analyses for different types of colonies were done using proc mixed procedure with cell (normalized number for each colony type) as a response and treatment (CONTROL, pGIPZ, and PDCD2) as a predictor. For the benzidine treatment (Fig. 5B) and cell cycle data (Fig. 4D and 5C), the model included percentage of cells (percent cells with different level of benzidine staining or in each phase of cell cycle) as a response and group (CONTROL, pGIPZ, and PDCD2 groups), treatment (with or without Ara-C), and parameters were grouped by treatment interaction as predictors.
Figure 2.
PDCD2 knockdown in CD34+ cells results in impaired hematopoietic progenitor cell growth and multilineage differentiation on methylcellulose. (A) Representative colonies (1, 2, 3, and 4) were prepared from cytospins. Morphology of colony number 1 (BFU-E(m) mature), 2 (CFU-GM), 3 (BFU-E(p) primitive), and 4 (CFU-GEMM) shown in light and GFP fluorescent images at 10× magnification. For colored images of these colonies, see Supplementary Figure E4 (online only; available at www.exphem.org). Morphologies were confirmed by Wright-Giemsa staining at magnifications of 100× and 400×. BFU-E(m) colonies consist mainly of erythroblasts, BFU-E(p) predominantly consist of proerythroblasts, CFU-GEMM are mixed-cell colonies (myeloblasts, granulocytes, pro- and erythroblasts), and CFU-GM are mainly represented by granulocytes. (B) Graph of total number of colonies in nontransduced (CONTROL) or transduced with either nonsilencing control (pGIPZ) or shRNA-PDCD2 (PDCD2) viruses. Numbers of colonies were counted and normalized to the number of plated cells, mean ± standard deviation (SD) were calculated in three independent experiments done in quadruplicate. (C) After transduction, CD34+ cells gave rise to 15% to 30% GFP+ colonies. The histogram shows only the percentage of GFP+ colonies derived from CD34+ cells in pGIPZ and PDCD2 groups (% mean ± SD of two independent experiments done in quadruplicate). Colonies were scored on day 14. * indicate p < 0.05 and were considered significant.
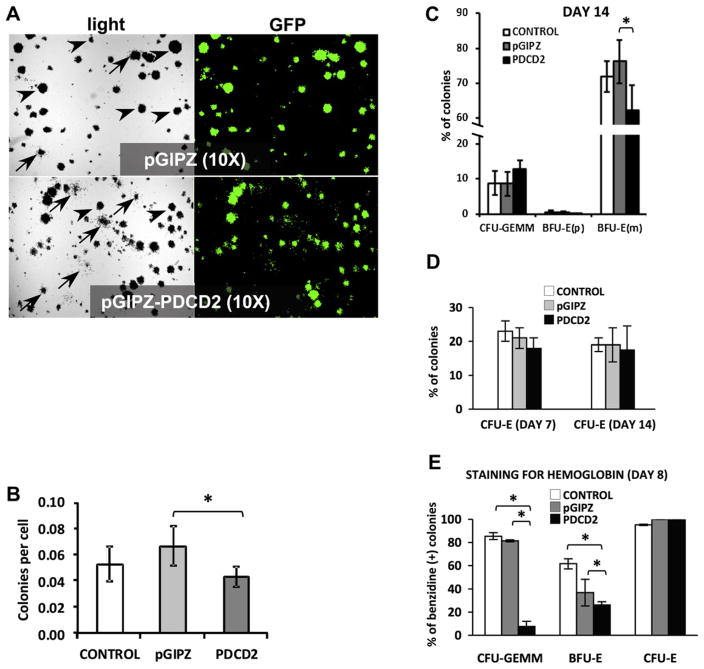
Figure 3.
PDCD2 knockdown in K562 leukemic cells results in impaired hematopoietic progenitor cells growth and multilineage differentiation on methylcellulose. (A) Representative light and GFP fluorescent images of lentiviral-transduced K562 cells that were sorted and cultured in methylcellulose medium (magnification of 10×). Arrowheads point to BFU-E(m) and arrows indicate CFU-GEMM colonies. (B) Graph of total number of colonies in nontransduced (CONTROL) or transduced with either control (pGIPZ) or shRNA-PDCD2 (PDCD2) viruses. Numbers of colonies were counted and normalized to the number of plated cells, mean ± standard deviation (SD) were calculated in three independent experiments done in quadruplicate. (C) Histogram showing the total percentage of different colonies produced by K562 cells in CONTROL (nontransduced) and treatment groups (transduced with pGIPZ-control vector and shRNA-PDCD2 expressing vector) (% mean ± SD were plotted from three independent experiments done in quadruplicate). (D) Histogram showing the percentage of CFU-E colonies produced at day 7 and day 14 from K562 cells in CONTROL (nontransduced) and treatment groups (% mean ± SD were plotted from three independent experiments done in quadruplicate). (E) Histogram showing the total percentage of benzidine-positive colonies produced by K562 cells (% mean ± SD, one experiment done in triplicate). Benzidine staining is a hemoglobin cytochemical stain. Colonies containing >10 positive (blue) cells were considered positive. * indicate p < 0.05, and were considered significant.
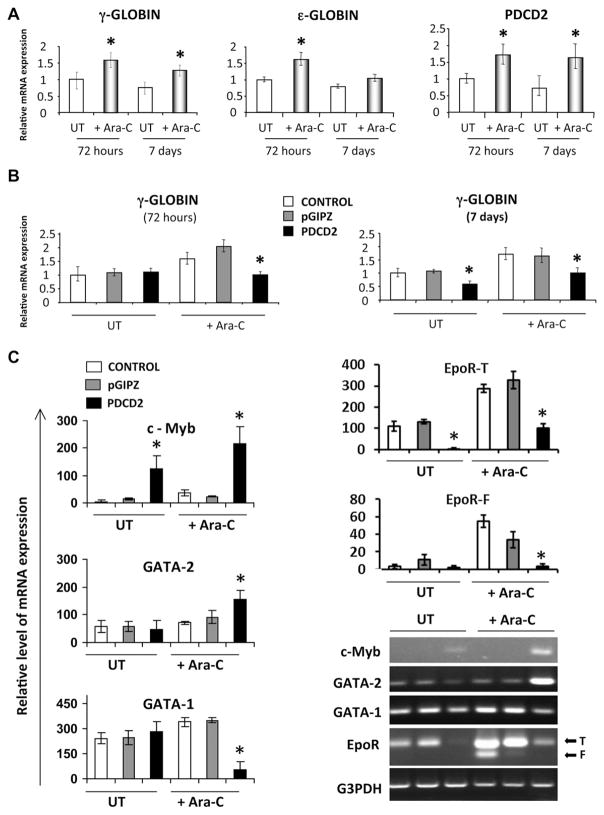
Figure 5.
PDCD2 knockdown impairs Ara-C–induced erythroid differentiation and alters cell cycle of K562 cells. K562 cells nontransduced (CONTROL) and transduced with a control (pGIPZ) and shRNA-PDCD2 (PDCD2) viruses were induced to differentiate into erythroid lineage by incubation with 0.36 μM of Ara-C for the number of days indicated. (A) Proliferation of cells was monitored each day for 3 days. The number of cells was quantified from three independent experiments and shown as a mean ± standard deviation (SD) (number of cells × 106). Differentiation of cells was examined by hemoglobin content using benzidine staining, and scored based on negative, weak, and strong benzidine staining. (B) Top; representative images showing negative, weak, and strong benzidine staining for hemoglobin. Bottom; histogram showing the distribution of benzidine-stained cells (% mean ± SD, 150 cells were counted in duplicate for each group in two independent experiments). ImageJ software was used to analyze the pixel density of each cell in the randomly selected areas. (C) Cell cycle analysis of transduced and nontransduced K562 cells before and 72 hours after stimulation with Ara-C. All cells in experimental groups were synchronized during a 48-hour culture in the presence of 1% FBS (serum starvation) and followed by subsequent serum stimulation for 8 days in the presence of 10% FBS. The distribution of cells in G0/G1, S, and G2/M phases was evaluated by measuring their DNA content by propidium iodide staining (% mean ± SD, five independent experiments). * p < 0.05 and were considered significant.
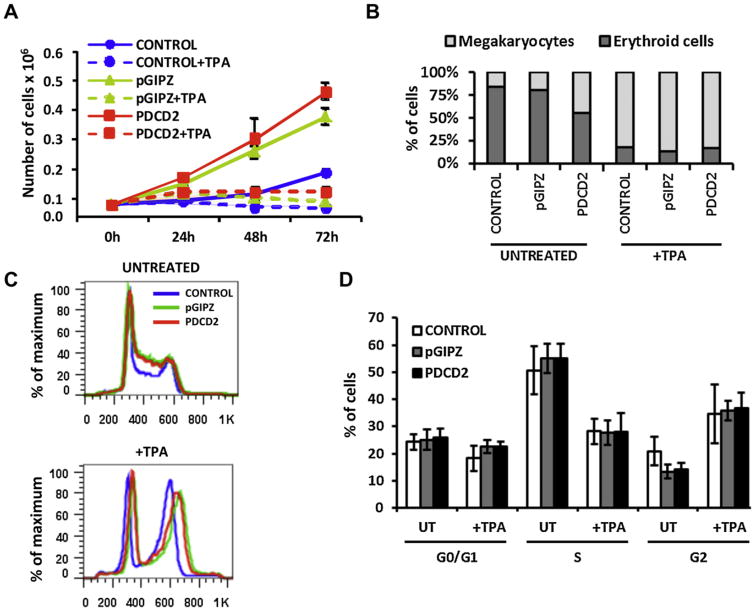
Figure 4.
PDCD2 knockdown does not affect TPA-induced megakaryocytic differentiation of K562 cells. K562 cells nontransduced (CONTROL) and transduced with a control lentiviral vector (pGIPZ) and one that encodes shRNA-PDCD2 (PDCD2) were induced to differentiate into the megakaryocytic lineage with TPA at a final concentration of 10 nM for the number of hours indicated. (A) Proliferation of cells was monitored each day for 3 days. The numbers of cells were quantified from three independent experiments and shown as the mean ± standard deviation (number of cells × 106). (B) The number of erythroid cells (proerythroblasts, intermediate cells, and mature erythroblasts) and megakaryocytes were counted (300 cells were counted for each group in two independent experiments, and mean cell numbers are shown). (C, D) Diagram demonstrating ploidy and cell cycle analyses of transduced and nontransduced K562 cells before and after stimulation with TPA. All cells in the experimental groups were synchronized during a 48-hour culture in the presence of 1% FBS (serum starvation) followed by serum stimulation for 48 hours in the presence of 10% FBS. (C) Ploidy analyses. The first peak represents cells in G0/G1 with 2N DNA content. The next peak represents cells in G2/M with 4N DNA content. Cells in between the 2N and 4N peak represent cells in S phase. (D) The distribution of cells in G0/G1, S, and G2/M phases was evaluated by measuring their DNA content by propidium iodide staining (% mean ± SD, three independent experiments). A p value <0.05 were considered significant, however, these experiments did show any statistically significant differences between groups.
Modeling was done separately for each cell type. All other data were statistically analyzed using Student’s t test (Excel software; Microsoft Corp., Redmond, WA, USA). Differences with p values ≤0.05 were considered statistically significant.
Results
PDCD2 is expressed in NBM and leukemia cell lines
To study the role of PDCD2 in hematopoiesis, we first performed immunohistochemical staining of NBM and found that PDCD2 protein was detected in multiple cell types at different levels (Fig. 1A). Antibody specificity was determined by omitting the primary antibody during immunohistochemical staining (Supplementary Figure E1; online only, available at www.exphem.org). Dual immunohistochemical staining indicated that PDCD2 is coexpressed with CD41a+ megakaryocytic cells (Fig. 1B) and CD235+ erythroid cells (Fig. 1C), respectively. Next, we examined PDCD2 protein levels in six different leukemic cell lines: Reh (acute lymphocytic leukemia), ML1 (myeloblastic leukemia), K562 (erythroleukemia blastic phase of CML), HL-60 (promyelocytic leukemia), Jurkat (lymphoblastic leukemia), and KG-1 (CD34+ myeloblastic leukemia). PDCD2 was detected in all six cell lines tested, with the highest level observed in K562 erythroleukemia cells (Fig. 1D).
Figure 1.
Detection of PDCD2 protein in human leukemia cell lines and normal human bone marrow. (A) Immunohistochemical (IHC) staining with anti-PDCD2 antibody of normal human bone marrow (arrows indicate positive PDCD2 staining) at magnifications of 40× and 100×. (B) Dual IHC with anti-PDCD2 (brown) and anti-CD41a (GPIIb/IIIa) (red) in NBM cells. Arrows indicate cells with coexpression of PDCD2 and CD41a. Insert: cytospin demonstrating the colocalization of PDCD2 and CD41a expression in a cell that is most likely of the megakaryocytic lineage. (C) Dual IHC with anti-PDCD2 (brown) and anti-CD235a (red) in NBM cells. Arrows indicate cells with coexpression of PDCD2 and CD235a. Note that cells with megakaryocytic phenotype (double arrow) only express PDCD2 in brown staining as expected, while erythroid cells have both brown and red staining. (D) Western blot showing the levels of PDCD2 protein in six leukemia cell lines. (E) Knockdown of PDCD2 with shRNA. Cells were harvested after 7 days post transduction to assess knockdown efficiency. Western blot analysis confirmed pdcd2 knockdown in shRNA-PDCD2 expressing K562 and 293T cell lines. Protein level was measured in total protein lysates isolated from nontransduced (CONTROL lane) and transduced (pGIPZ and PDCD2 lanes) K562 cells; and nontransfected (CONTROL lane) and transfected (pGIPZ and PDCD2 lanes) 293T cells. Blots were first probed with anti-PDCD2 antibody and then reprobed with anti–β-actin antibody.
Engineering of a lentiviral delivery system with high-efficiency RNA interference into hematopoietic progenitors
We generated a lentiviral shRNA delivery system for PDCD2 knockdown (Supplementary Figure E2A; online only, available at www.exphem.org). K562 cells were successfully transduced by nonsilencing control lentiviral vector (pGIPZ) and vector producing shRNA specific to the PDCD2 gene (pGIPZ-PDCD2). Transduced K562 cells were sorted at 48 hours post transduction for GFP expression (Supplementary Figure E2B; online only, available at www.exphem.org). We verified that >95% cells were effectively transduced by each of the lentiviral vectors on the basis of coexpression of GFP and functional shRNA. Knockdown of PDCD2 expression was successfully achieved in K562 and 293T cell lines, which was confirmed by Western blot analysis (Fig. 1E). The effects of PDCD2 shRNA on PDCD2 protein levels in K562 cells were examined over a 6-week period. Knockdown of PDCD2 began to subside at week 4, and expression returned almost to control levels by week 6 (data not shown). These results suggested that the effects of PDCD2 shRNA are time-dependent, and that knockdown of PDCD2 might interfere with hematopoietic cell proliferation and/or survival. The negative control shRNA viral vector transduction did not alter PDCD2 expression levels compared with nontransduced control. We concluded that our lentiviral system produces efficient and specific PDCD2 gene silencing. All of the experiments in this study were performed between weeks 1 and 3 post transduction to maintain consistency and reproducibility.
PDCD2 knockdown in CD34+ cells results in impaired HSC proliferation and differentiation
CD34+ cells were isolated from NBM of healthy volunteers under an approved institutional review board, and were transduced at multiplicity of infection of 40 to 100 with pGIPZ or pGIPZ-PDCD2 shRNA. Nontransduced cells served as additional controls. Cells were counted, and without sorting for GFP+ cells, were plated on methylcellulose semisolid medium supplemented with all necessary components to support optimal growth and differentiation of human hematopoietic progenitors (Fig. 2A and Supplementary Figure E3A; online only, available at www.exphem.org). After transduction, transduced cells gave rise to 15% to 30% GFP+ colonies. Colony types were distinguished based on previously published characteristics [2]. Scoring of erythroid colonies was based on the total number of cells in the colony, as well as the number of cell clusters making up the colony. This scoring method allowed us to identify erythroid progenitors derived from different stages based on fluorescence and Wright-Giemsa staining. Statistical analyses were performed in two steps: first, all formed colonies were counted and analyzed to examine the overall effect of viral transformation on the total population of cells. Second, only GFP+ colonies were scored and compared to identify the effects of PDCD2 gene knockdown vs control pGIPZ virus. In the first step, the total numbers of colonies (GFP+ and GFP+) produced by each group were normalized to the number of plated cells, and the mean and standard deviation were calculated. The normalized number of colonies per cell for CONTROL was 0.1017 ± 0.006; for pGIPZ was 0.1036 ± 0.004, and for PDCD2 was 0.0714 ± 0.003. There was a 30% statistically significant decrease in the total number of colonies in PDCD2 knockdown group (*p < 0.0001 when compared to CONTROL; *p = 0.006 when compared to pGIPZ) compared with control groups (Fig. 2B). This result suggests that PDCD2 is important to sustain HSC/HPC viability.
CFU-E are the smallest and most rapidly maturing erythroid colonies; they consist of one or two cell clusters containing 100 to 200 erythroblasts with distinctive reddish-orange hue due to their content of hemoglobin. BFU-E are a class of more primitive erythroid progenitors than CFU-E, and could be divided in two subgroups: mature (m) and primitive (p) [19]. BFU-E(m) are the immediate precursors of CFU-E, they are fully hemoglobinated and contain from 3 to 8 erythroblast clusters and ~100 to 500 cells (Fig. 2A, colony 1). BFU-E(p) are more primitive erythroid progenitors, which might not be fully hemoglobinated until day 18, and each contains 9 or more clusters of proerythroblasts and/or erythroblasts with >500 cells (Fig. 2A, colony 3). CFU-GM are late progenitors that are restricted to the granulocyte and/or macrophage lineages. CFU-GM colonies are relatively homogeneous and each consists of 50 to 100 cells (Fig. 2A, colony 2). CFU-GEMM are the most primitive multipotent progenitor colonies and contain progenitor cells that give rise to multiple lineages and appear with small numbers of granulocytes, macrophages, and/or megakaryocytes around the periphery of a spherical mass of hemoglobinated erythroid cells (Fig. 2A, colony 4). The morphology of each type of colony (1–4) was verified by Wright-Giemsa staining. All formed colonies were scored on day 14.
The distribution of all colony types in the three groups is shown in Supplementary Figure E3B; online only; available at www.exphem.org). To determine if PDCD2 knockdown has an impact on specific CFUs, we scored and compared all GFP+ colony types in pGIPZ and PDCD2 groups (Fig. 2C). There were only occasional GFP+ CFU-E colonies found, therefore, CFU-E colonies have not been scored in this experiment. There was a statistically significant increase in early progenitors (eightfold for CFU-GEMM; p = 0.016; twofold for BFU-E(p); p = 0.01) and a statistically significant decrease in late BFU-E(m) progenitors (1.3-fold; p = 0.006) in the PDCD2 group compared with the control pGIPZ group. The number of CFU-GM colonies was not significantly different among the groups. These results suggest that PDCD2 knockdown increases the numbers of hematopoietic multipotent progenitors and interferes with maturation of erythroid progenitors.
Effect of PDCD2 knockdown on colony-forming ability of K562 leukemic cells
We tested the effect of suppressing PDCD2 expression in K562 erythroleukemia cells. This cell line was chosen for multiple reasons. First, this cell line was derived from a CML patient in blastic phase; cells harbor the Ph1 chromosome and express BCR-ABL [14,23–25]. Second, K562 cells have been a useful model for studying differentiation along multiple hematopoietic lineages, including erythroid, granulocytic, and megakaryocytic lineages [20,26]. Third, the hemoglobins induced in K562 cells are embryonic and fetal, despite the origin of the cell line from an adult patient [27]. Finally, expression of PDCD2 protein was the highest among all tested leukemia cell lines (Fig. 1B).
Nontransduced (CONTROL) and lentiviral-transduced cells with control (pGIPZ) and shRNA-PDCD2 (PDCD2) cells were plated on methylcellulose medium similar to CD34+ fraction of cells from NBM (Fig. 3A). Total numbers of colonies produced by each group of cells were normalized to the number of plated cells, and mean and standard deviation were calculated (Fig. 3B) (CONTROL: 0.053 ± 0.013; pGIPZ: 0.067 ± 0.015; and pGIPZ-PDCD2: 0.043 ± 0.008). K562 cells with PDCD2 knockdown exhibited a significant decrease of ~36% in the number of colonies grown compared with the control virus group (p = 0.028). There was no significant difference between CONTROL and pGIPZ groups. These observations raised the possibility that PDCD2 inhibition might have a general effect on CML cells and does not exhibit specific effects on lineage differentiation of these cells. To address this issue, different types of colonies were scored in the three treatment groups (Fig. 3C). In the PDCD2 group, there was a 33% increase in the number of CFU-GEMM earliest multipotent progenitors without reaching a statistical significance (p = 0.089). In addition, a significant decrease of ~18% in BFU-E(m) colonies (p =0.006) and a statistically significant increase of 36% in CFU-E (p = 0.042) were observed (Fig. 3C). There were occasional BFU-E(p) colonies ranging from one to five per plate in each group, for which no statistical analysis were performed. CONTROL and pGIPZ groups did not show any statistically significant difference in all scored colonies.
It was previously reported that K562 cells in colony-forming assays demonstrate high rates of commitment (the decision to express a differentiated phenotype and to terminate proliferation in the absence of an inducer), and form large numbers of mixed colonies (based on number of cells accumulating hemoglobin in the colony) [28]. Because PDCD2 appears to be an important player in the differentiation process of early progenitors, and because accumulated hemoglobin is a good indicator of erythroid differentiation, we predicted that PDCD2 knockdown would affect the amount of hemoglobinated cells per colony. To test this hypothesis, we performed direct benzidine staining of colonies (Fig. 3E and Supplementary Figure E4; online only; available at www.exphem.org). In our study, three types of colonies were found, colonies containing only benzidine-positive cells, colonies containing only benzidine-negative cells, and those containing both cell types (mixed colonies). We divided these colonies into two groups based on benzidine staining: positive (≥10 stained cells/colony) and negative (<10 stained cells). All CFU-E colonies in pGIPZ and PDCD2 groups were 100% benzidine-positive, as were 95% of the colonies in the CONTROL group (95.2% ± 0.65%), confirmed by examining the morphology of these colonies. There was an 11-fold significant decrease in the number of benzidine-positive BFU-E colonies in the PDCD2 group (7.6% ± 4.43%) compared with the pGIPZ group (81.5% ± 0.97%) (p < 0.0001). Additionally, a 1.4-fold decrease in CFU-GEMM colonies without reaching statistical significance (PDCD2 group: 26% ± 2.91%; pGIPZ group: 36.8% ± 11.61%) was observed. These data are in agreement with our previous findings that PDCD2 knockdown leads to a decrease in erythroid differentiation of CFUs.
Effects of PDCD2 knockdown on TPA-induced megakaryocytic differentiation of K562 cells
Hematopoietic cell development is not only controlled by self-renewal of multipotent progenitor cells and their commitment to various hematopoietic lineages, but also is regulated by further differentiation and development of the committed cells. This prompted us to investigate the role of PDCD2 during enforced terminal lineage differentiation of K562 cells. Multipotent K562 cells can be chemically induced to differentiate to the megakaryocytic lineage using Ara-C. Nontransduced cells (CONTROL group) and transduced cells with a control virus (pGIPZ group) and shRNA-PDCD2–expressing virus (PDCD2 group) were treated with 10 nM TPA for 48 to 72 hours. To assess the effect of TPA on proliferation of nontransduced and transduced cells, cells were monitored for 72 hours (Fig. 4A). Transduced cells were proliferating at higher rates compared to nontransduced cells, which could be a direct viral effect on cell proliferation. At the same time, proliferation was inhibited in all groups after TPA induction, indicative of cells induced for terminal differentiation. Megakaryocytic differentiation of K562 cells induced by TPA was identified by three methods, cell morphology, cell polyploidy [29], and ploidy analyses using flow cytometry. Wright-Giemsa staining demonstrated a marked increase in cell size, extensive multinuclearity, and vacuolation after TPA induction in all groups (Supplementary Figure E5; online only, available at www.exphem.org). Percentages of erythroid (pro-erythroblasts, intermediaries, and mature erythroblasts) and megakaryocytic lineages were counted before and after TPA stimulation (300 cells were counted for each group in two independent experiments, mean percentages are shown) (Fig. 4B). There were no differences in cell ratio of megakaryocyte to erythroid before and after stimulation, with one exception, there was a significant decrease in the ratio of erythroid to megakaryocytic cells without TPA induction in the PDCD2 group (Fig. 4B) compared with treated cells. Because equal numbers of cells were examined, we deduced that PDCD2 knockdown has a specific effect on erythroid cells but not megakaryocytes because we did not observe any impact on induced megakaryocytic terminal differentiation by TPA in the PDCD2 group. Ploidy analysis by flow cytometry showed an identical pattern in all groups before and after TPA treatment, with a significant increase in 4N cells (increased ploidy indicative of megakaryocytic differentiation) in all groups after TPA induction (Fig. 4C). More detailed cell cycle analysis did not show any statistically significant differences in the different stages of the cell cycle between the groups before and after stimulation (Fig. 4D). Due to the observed effect of PDCD2 knockdown on the ratio between erythroid and megakaryocytic lineages, we decided to further investigate the role of PDCD2 in erythroid lineage differentiation.
PDCD2 knockdown leads to incomplete Ara-C–induced erythroid differentiation of K562 cells
To evaluate the effect of Ara-C on proliferation of nontransduced and transduced K562 cells, these cells were monitored for 1 week (Fig. 5A). Ara-C treatment decreased proliferation in the cells of the control groups (CONTROL and pGIPZ), as expected. However, in the PDCD2 group, Ara-C treatment showed only a partial suppression of proliferation at 72 hours (Fig. 5A), and this partial suppression of proliferation was significantly different from proliferation of the control groups (CONTROL and pGIPZ) at day 7 (Fig. 5A). This observation suggested that only a subset of K562 cells might be able to terminally differentiate into more mature erythroid lineages. To confirm this observation, nontransduced and transduced cells were treated with Ara-C for 8 days to achieve maximal stimulation and were then stained with benzidine, a hemoglobin-specific dye (Supplementary Figure E6; online only; available at www.exphem.org, and Fig. 5B). Cell differentiation was scored based on negative, weak, or strong benzidine staining. One hundred and fifty cells were analyzed and counted in duplicate using ImageJ software for each group in three independent experiments, and percentage of stained cells was calculated for each group. No untreated groups showed any differences in the number of benzidine-positive vs -negative cells. We did not observe any statistically significant differences in CONTROL and pGIPZ groups with or without Ara-C treatment. After induced erythroid differentiation with Ara-C, there was a fourfold significant decrease in number of cells with strong benzidine staining (p < 0.0001), and a 13-fold increase in number of cells with weak staining (p <0.0001) in the PDCD2 group. The number of negative cells did not change in the PDCD2 group compared with control pGIPZ group (Fig. 5B, lower panel). These data demonstrate a reduction in hemoglobin synthesis in K562 cells with PDCD2 knockdown, and point to a novel role for PDCD2 during erythroid differentiation.
To study the molecular mechanisms by which PDCD2 knockdown might regulate cell proliferation and erythroid differentiation, we performed cell cycle analysis during induction of erythroid differentiation with Ara-C (Fig. 5C). At 72 hours, knockdown of PDCD2 by itself had no significant effect on proliferation, but after Ara-C treatment, there was a significant increase in proportion of cells in G0/G1 in the control groups, but not with PDCD2 (37% vs 54% in control pGIPZ group; p value <0.001), and a significant increase in the proportion of cells in G2/M compartment (28% vs 10% in control pGIPZ group; p = 0.004). Ara-C–induced differentiation evidenced by accumulation of cells in G0/G1 and PDCD2 knockdown was associated with less accumulation of cells in G0/G1 compared with control pGIPZ group, suggesting that PDCD2 knockdown impedes terminal maturation of erythroid progenitors. As expected, no significant alterations of cell cycle profiles were observed between control groups (CONTROL and pGIPZ).
Gene expression changes with PDCD2 knockdown in Ara-C–induced erythroid differentiation of K562 cells
To determine whether PDCD2 might play a role in regulating the expression of erythroid- and/or progenitor cell-associated transcription factors, we first confirmed that treatment of K562 cells with Ara-C resulted in up-regulation of both γ-GLOBIN and ε-GLOBIN messenger RNA (mRNA), at 72 hours during early induction of erythroid differentiation and at 7 days when this induction is completed (Fig. 6A). The induction of γ-GLOBIN by Ara-C was highly statistically significant at both time points compared with ε-GLOBIN (Fig. 6A). Therefore, we used γ-GLOBIN as a molecular marker for induction of erythroid differentiation in our studies of the effects of PDCD2 knockdown. Next, we determined that PDCD2 mRNA levels are significantly up-regulated in K562 cells upon erythroid induction with Ara-C (Fig. 6A). The increased PDCD2 expression correlated with the up-regulation of γ-GLOBIN expression indicative of erythroid differentiation (Fig. 6A). In contrast, the levels of γ-GLOBIN mRNA determined by quantitative real-time PCR were significantly reduced with PDCD2 knockdown at both 72 hours and 7 days compared with control groups (CONTROL and pGIPZ) (Fig. 6B), confirming the failure of initiation and/or termination of erythroid maturation seen with PDCD2 knockdown. We next examined the expression of early erythroid (GATA-1 and EpoR) and hematopoietic progenitor cell markers (c-MYB, GATA-2) in K562 cells during Ara-C–induced differentiation (Fig. 6C). Reverse transcription PCR analyses were performed in three groups before and after stimulation with Ara-C treatment for 8 days in four independent experiments. Expression levels were normalized to G3PDH levels for each experiment, and mean ± standard deviation were plotted. There was a significant increase in mRNA expression of c-MYB and GATA-2 (nine- and twofold, respectively) and a sevenfold decrease in mRNA expression of GATA-1. These data are consistent with the reports describing an important role for c-MYB and GATA-2 in early hematopoiesis, while the expression of these factors diminishes during erythroid differentiation [30–32].
Figure 6.
PDCD2 knockdown impairs Ara-C–induced erythroid differentiation of K562 cells by altering gene expression of erythroid progenitor markers. (A) Q-PCR for γ-GLOBIN, ε-GLOBIN, GAPDH, and PDCD2 were done at 72 hours and after 7 days of induction with Ara-C. Data are shown as a ratio to GAPDH levels and are normalized from three independent experiments. (B) quantitative real-time PCR for γ-GLOBIN using complementary DNA from CONTROL, pGIPZ, and PDCD2 groups. (C) Semi-quantitative reverse transcription PCR analyses of c-MYB, GATA-2, GATA-1, EpoR (T = truncated, F = full-length), and G3PDH transcripts. Nontransduced cells (CONTROL, white bars) and transduced cells with either control (pGIPZ, gray bars) or shRNA-PDCD2 (PDCD2, black bars) lentiviral vectors were untreated (UT lane) or treated with Ara-C (Ara-C lane) for 7 days. ImageJ software was used to analyze band density in three independent experiments and normalized to G3PDH levels for each experiment, and mean ± SD were plotted. *p < 0.05 were considered significant.
K562 cells express two isoforms of erythropoietin receptor; a truncated (EpoR-T) and a full-length (EpoR-F) isoforms. The truncated (EpoR-T) isoform is longer as it contains the unspliced intron 7 [33]. Both EpoR isoforms were markedly up-regulated upon Ara-C induction, while PDCD2 knockdown was associated with a significant decrease in the expression of both EpoR isoforms with or without Ara-C induction (Fig. 6C). EpoR-T was more than 40 times decreased in PDCD2 group (p < 0.015 for both groups). In Ara-C–treated groups: EpoR-F was expressed only in control groups and showed a 10-fold decrease in PDCD2 group (p ≤ 0.023 for both groups), EpoR-T was expressed in all groups with more than threefold decrease in PDCD2 group (p ≤ 0.003 for both groups). These results support the previously published data that EpoR-T is predominant in immature erythroid progenitor cells, while EpoR-F is predominant in mature erythroid progenitors [34]. Because GATA factors and EpoR are involved in erythroid differentiation of K562 cells, the decreased expression of these factors in the PDCD2 group reflects an arrest in the erythroid maturation cascade of these cells even after Ara-C stimulation [32,35]. Taken together, these results suggest that PDCD2 knockdown inhibits erythroid differentiation of K562 cells by up-regulating TFs sustaining multilineage progenitors, while down-regulating erythroid-specific signals.
Discussion
The identification of novel proteins that regulate human stem cell and early progenitor cell-fate decision is a major goal in experimental and clinical hematology. In this study, we investigated the role of PDCD2 in lineage-specific differentiation of human HPCs. We demonstrated that PDCD2 influences the proliferation and differentiation of colony-forming units of HPCs, and is important for the terminal differentiation of erythroid progenitors.
Our work builds on earlier studies that demonstrated the involvement of PDCD2 in blood development. Using GFP-labeled homozygous mutant ZFRP8 clones in ZFRP8 heterozygous animals, Minakhina et al. provided the evidence that Drosophila hemocytes are established from a stem cell, and further, that the identity of these hemocyte stem cells is dependent on the function of ZFRP8, an ortholog of human PDCD2 [5]. More recently, our group utilized the zebrafish model to study pdcd2 function during embryonic hematopoiesis [36]. PDCD2 protein sequence comparisons revealed that PDCD2 proteins shares significant homology among all eukaryotes [36]. Loss of the zebrafish pdcd2 perturbs embryonic hematopoietic development with cell cycle defects and accumulation of erythroid progenitors during primitive hematopoiesis [36]. To validate these results in human cells, we took advantage of the CFC assay, which is a useful tool to define primarily quiescent progenitors, and could provide evidence for self-renewal capacity and/or multilineage differentiation potential. PDCD2 knockdown displayed particular selectivity for CFU-GEMM and BFU-E primitive colonies, which are representatives of early erythroid progenitors. Increased numbers of CFU-GEMM and BFU-E(p) colonies with PDCD2 knockdown might be explained by two views. One possibility is that loss of PDCD2 interferes with the ability of progenitor cells to terminally differentiate. The second view is that PDCD2 loss increases the ability of these progenitors to self-renew. It was recently demonstrated by England et al. that during definitive erythropoiesis, proerythroblasts have the capacity for extensive self-renewal divisions in vitro [37]. Their study challenges the traditional views of hierarchical hematopoietic differentiation and suggests that intermediate progenitors or transit amplifying cells might possess self-renewal features. Therefore, the two views of the effects of PDCD2 loss limiting differentiation or promoting stemness and self-renewal are not mutually exclusive, because self-renewal of stem and progenitor cells requires coordination of cell cycle progression and cell-fate choices [38]. In addition, PCDC2 might also be required for processing of self-renewal and/or differentiation signals, and loss of PDCD2 might deregulate the balance between these signals.
Early G1 phase of the cell cycle is a crucial period for cell-fate decision making in response to extracellular signals [38]. K562 cells are relatively resistant to the anti-metabolite Ara-C. When K562 cells are treated, Ara-C gets incorporated into their DNA, resulting in disruption of further DNA synthesis by activation of the ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) protein kinase function. Activation of ATM and ATR initiates downstream cellular response to DNA damage, and induces differentiation into erythrocytes without undergoing apoptosis [15,39]. In addition, induction of hemoglobin synthesis in K562 cells with Ara-C is dependent on cell cycle phase, with cells within late G1 and early S phases become sensitive to the effect of the agent and start to accumulate hemoglobin [15]. PDCD2 knockdown caused fewer cells to arrest at the G1 phase of the cell cycle, resulting in formation of colonies with limited abilities to accumulate hemoglobin. Flow cytometric analysis of shRNA-mediated knockdown of PDCD2 in K562 cells showed that treatment with Ara-C failed to significantly increase the percentage of cells in G0/G1 phase and cells accumulated in the G2 phase. It is possible that loss of PDCD2 prevented erythroid progenitors from reaching the late G1 phase and proceeding through terminal differentiation. However, prolonged exposure to Ara-C for up to 8 days did not improve the level of erythroid differentiation of K562 cells in the PDCD2 knockdown group, suggesting that these cells lost their ability to produce and/or accumulate hemoglobin. These data suggest that PDCD2 regulates cell cycle progression, and that perturbation of cell cycle distribution by itself does not necessarily lead to differentiation. Furthermore, PDCD2 effects appear to be specific to phases of the cell cycle that are necessary for erythroid differentiation. Further studies are required to clarify the role of PDCD2 and DNA damage during Ara-C–induced erythroid differentiation.
We have shown that K562 cells, upon treatment with TPA, showed acceleration in cell size increase, vacuolization, and polyploidy, which are unique features of megakaryocytes [26]. Interestingly, loss of PDCD2 expression did not interfere with these cellular changes leading to megakaryocytic differentiation, and no differences in cell cycle analyses were noticed. Accordingly, PDCD2 knockdown affected the ratio between megakaryocytic and erythroid cells toward decreased number of erythroid cells, suggesting that PDCD2 is essential for erythroid lineage differentiation from MEP, and that PDCD2 effects might correlate with levels of cell cycle activation during the progressive differentiation of these progenitors.
Our data suggest that PDCD2 is important for early stages of cell-fate decision making. There is currently a paucity of data describing the role of PDCD2 in embryonic and hematopoietic progenitor cell developmental processes. It was shown that mouse PDCD2 is a stem cell factor [6]. PDCD2 is also enriched in human embryonic stem cells compared with their differentiated lineage [7]. Knockout of PDCD2 in the mouse is embryonic lethal at preimplantation stages [8]. In addition, ZFRP8, the ortholog of PDCD2, is required for normal development of Drosophila hemocytes [40]. Importantly, few reports have presented evidence for deregulated PDCD2 expression in human leukemias and lymphomas. PDCD2 expression was shown to be repressed by the oncogenic transcriptional repressor protein BCL6 [41–43]. BCL-6 is required to retain erythroblasts in the cell proliferation stage in neonatal mice [44], however, whether these effects of BCL-6 on erythroid proliferation were dependent on PDCD2 function(s) was not determined. PDCD2 is localized in chromosome 6q27 in a region involved in both translocations and deletions in leukemias and lymphomas [11–13]. Collectively, these data and our results support the notion that PDCD2 plays an important role in maintaining stemness and/or fate decision of HPCs. Clearly, it will be critical to identify PDCD2 interacting partners that might be involved in regulating these critical functions.
To this end, we reasoned that if PDCD2 knockdown regulates the stemness of hematopoietic cells, it should lead to changes in gene expression profiles as well. MYB is an important regulator of cell differentiation at multiple decision forks in hematopoiesis [45]. Previous studies provided compelling evidence supporting a role for MYB as a regulator of the MEP lineage bifurcation, and a potent negative regulator of megakaryopoiesis. Studies showed that reduced MYB expression in MEP lineage–restricted progenitors increased megakaryocytic and reduced erythroid progenitors [46,47]. We found that the expressions of c-MYB along with GATA-2 are up-regulated in cells lacking PDCD2. c-MYB was previously shown to maintain ex vivo erythroblast proliferation [48], and constitutive activation of c-MYB blocks the maturation of murine erythroleukemia cells [49]. Whether PDCD2 loss had a direct impact on c-MYB ability to maintain human erythroid progenitors, or led to c-MYB up-regulation secondary to either failure of terminal differentiation or accumulation of undifferentiated progenitors remains to be investigated.
Conclusions
Several factors that regulate hematopoietic differentiation and progenitor cell fate determination such as c-MYB, GATA-2, HOX genes, and p21WAF1 have been implicated in leukemogenesis by arresting differentiation and promoting self-renewal [50]. We found that PDCD2 functions similarly to those factors by regulating HPC/MPP proliferation through maintaining an undifferentiated gene expression program in leukemic cells. The biochemical changes and molecular controls of PDCD2 expression that contribute to the specific sequence of events leading to aberrant differentiation during normal and malignant hematopoiesis will be important to uncover.
Supplementary Material
Acknowledgments
We thank Arthur Roberts for help with flow cytometry analyses, and Svetlana Minakhina, Ruth Steward, and Roger Strair for advice and critical reading of the manuscript.
Footnotes
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.exphem.2012.08.004.
Conflict of interest disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Funding disclosure
This work was supported by funds from the Cancer Institute of New Jersey (H.E.S) and support of the Robert Wood Johnson Foundation for the Child Health Institute of New Jersey (A.B.R).
References
- 1.Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- 2.Eaves CJ, Humphries RK, Eaves AC. In Vitro Characterization of Erythroid Precursor Cells and the Erythroid Differentiation Process. New York: Grune & Stratton; 1982. pp. 251–278. [Google Scholar]
- 3.Gordon MY. Human haemopoietic stem cell assays. Blood Rev. 1993;7:190–197. doi: 10.1016/0268-960x(93)90005-o. [DOI] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 7.Skottman H, Mikkola M, Lundin K, et al. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- 8.Mu W, Munroe RJ, Barker AK, Schimenti JC. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Dev Biol. 2010;347:279–288. doi: 10.1016/j.ydbio.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarr RB, Sharp PA. PDCD2 is a negative regulator of HCF-1 (C1) Oncogene. 2002;21:5245–5254. doi: 10.1038/sj.onc.1205647. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami T, Furukawa Y, Sudo K, et al. Isolation and mapping of a human gene (PDCD2) that is highly homologous to Rp8, a rat gene associated with programmed cell death. Cytogenet Cell Genet. 1995;71:41–43. doi: 10.1159/000134058. [DOI] [PubMed] [Google Scholar]
- 12.Merup M, Moreno TC, Heyman M, et al. 6q deletions in acute lymphoblastic leukemia and non-Hodgkin’s lymphomas. Blood. 1998;91:3397–3400. [PubMed] [Google Scholar]
- 13.Steinemann D, Gesk S, Zhang Y, et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer. 2003;37:421–426. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- 14.Lozzio BB, Lozzio CB, Bamberger EG, Feliu AS. A multipotential leukemia cell line (K-562) of human origin. Proc Soc Exp Biol Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- 15.Wanda PE, Walker MM. Hemoglobin induction by Ara-C in human erythroleukemic cells (K562) is cell-cycle dependent. Leuk Res. 1989;13:683–688. doi: 10.1016/0145-2126(89)90057-x. [DOI] [PubMed] [Google Scholar]
- 16.Rubin CI, French DL, Atweh GF. Stathmin expression and megakaryocyte differentiation: a potential role in polyploidy. Exp Hematol. 2003;31:389–397. doi: 10.1016/s0301-472x(03)00043-2. [DOI] [PubMed] [Google Scholar]
- 17.Millington M, Arndt A, Boyd M, Applegate T, Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS One. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory CJ, Eaves AC. Human marrow cells capable of erythropoietic differentiation in vitro: definition of three erythroid colony responses. Blood. 1977;49:855–864. [PubMed] [Google Scholar]
- 19.Eaves CJ, Eaves AC. Erythroid progenitor cell numbers in human marrow—implications for regulation. Exp Hematol. 1979;7(suppl 5):54–64. [PubMed] [Google Scholar]
- 20.Rowley PT, Ohlsson-Wilhelm BM, Farley BA. K562 human erythroleukemia cells demonstrate commitment. Blood. 1985;65:862–868. [PubMed] [Google Scholar]
- 21.Strobe W. Wright-Giemsa and nonspecific esterase staining of cells. In: Coligan J, editor. Current Protocols in Immunology. Hoboken, NJ: John Wiley & Sons, Inc; 1997. pp. A.3C.1–A.3C.3. [DOI] [PubMed] [Google Scholar]
- 22.Delgado MD, Quincoces AF, Gomez-Casares MT, et al. Differential expression of ras protooncogenes during in vitro differentiation of human erythroleukemia cells. Cancer Res. 1992;52:5979–5984. [PubMed] [Google Scholar]
- 23.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 24.Klein E, Ben-Bassat H, Neumann H, et al. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer. 1976;18:421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 25.Croce CM, Huebner K, Isobe M, et al. Mapping of four distinct BCR-related loci to chromosome region 22q11: order of BCR loci relative to chronic myelogenous leukemia and acute lymphoblastic leukemia breakpoints. Proc Natl Acad Sci U S A. 1987;84:7174–7178. doi: 10.1073/pnas.84.20.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leary JF, Ohlsson-Wilhelm BM, Giuliano R, LaBella S, Farley B, Rowley PT. Multipotent human hematopoietic cell line K562: lineage-specific constitutive and inducible antigens. Leuk Res. 1987;11:807–815. doi: 10.1016/0145-2126(87)90065-8. [DOI] [PubMed] [Google Scholar]
- 27.Benz EJ, Jr, Murnane MJ, Tonkonow BL, et al. Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc Natl Acad Sci U S A. 1980;77:3509–3513. doi: 10.1073/pnas.77.6.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowley PT, Ohlsson-Wilhelm BM. Demonstration of commitment by K562 human erythroleukemia cells. Prog Clin Biol Res. 1985;191:217–231. [PubMed] [Google Scholar]
- 29.Alitalo R. Induced differentiation of K562 leukemia cells: a model for studies of gene expression in early megakaryoblasts. Leuk Res. 1990;14:501–514. doi: 10.1016/0145-2126(90)90002-q. [DOI] [PubMed] [Google Scholar]
- 30.Rosson D, O’Brien TG. Constitutive c-myb expression in K562 cells inhibits induced erythroid differentiation but not tetradecanoyl phorbol acetate-induced megakaryocytic differentiation. Mol Cell Biol. 1995;15:772–779. doi: 10.1128/mcb.15.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci U S A. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- 33.Chiba S, Takahashi T, Takeshita K, et al. Selective expression of mRNA coding for the truncated form of erythropoietin receptor in hematopoietic cells and its decrease in patients with polycythemia vera. Blood. 1997;90:97–104. [PubMed] [Google Scholar]
- 34.Nakamura Y, Nakauchi H. A truncated erythropoietin receptor and cell death: a reanalysis. Science. 1994;264:588–589. doi: 10.1126/science.8160019. [DOI] [PubMed] [Google Scholar]
- 35.Rath AV, Schmahl GE, Niemeyer CM. Expression of transcription factors during sodium phenylacetate induced erythroid differentiation in K562 cells. Blood Cells Mol Dis. 1997;23:27–38. doi: 10.1006/bcmd.1997.0116. [DOI] [PubMed] [Google Scholar]
- 36.Kramer J, Granier CJ, Davis S, et al. PDCD2 controls hematopoietic stem cell differentiation during development. Stem Cells Dev. 2012 Aug 16; doi: 10.1089/scd.2012.0074. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.England SJ, McGrath KE, Frame JM, Palis J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood. 2011;117:2708–2717. doi: 10.1182/blood-2010-07-299743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 39.Takagaki K, Katsuma S, Kaminishi Y, et al. Role of Chk1 and Chk2 in Ara-C-induced differentiation of human leukemia K562 cells. Genes Cells. 2005;10:97–106. doi: 10.1111/j.1365-2443.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 40.Minakhina S, Druzhinina M, Steward R. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development. 2007;134:2387–2396. doi: 10.1242/dev.003616. [DOI] [PubMed] [Google Scholar]
- 41.Baron BW, Anastasi J, Thirman MJ, et al. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci U S A. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baron BW, Zeleznik-Le N, Baron MJ, et al. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:7449–7454. doi: 10.1073/pnas.0701770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baron BW, Hyjek E, Gladstone B, Thirman MJ, Baron JM. PDCD2, a protein whose expression is repressed by BCL6, induces apoptosis in human cells by activation of the caspase cascade. Blood Cells Mol Dis. 2010;45:169–175. doi: 10.1016/j.bcmd.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Asari S, Sakamoto A, Okada S, et al. Abnormal erythroid differentiation in neonatal bcl-6-deficient mice. Exp Hematol. 2005;33:26–34. doi: 10.1016/j.exphem.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol. 2008;20:247–256. doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M. Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol. 2006;26:7953–7965. doi: 10.1128/MCB.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandberg ML, Sutton SE, Pletcher MT, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Wessely O, Deiner EM, Beug H, von Lindern M. The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. EMBO J. 1997;16:267–280. doi: 10.1093/emboj/16.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke MF, Kukowska-Latallo JF, Westin E, Smith M, Prochownik EV. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ezoe S, Matsumura I, Satoh Y, Tanaka H, Kanakura Y. Cell cycle regulation in hematopoietic stem/progenitor cells. Cell Cycle. 2004;3:314–318. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.