Highlights
-
•
Host proteins must distinguish viral RNA from self RNA to trigger immune responses.
-
•
5′ RNA cap is key specificity determinant of host signaling.
-
•
Viruses use a variety of mechanisms to evade host RNA sensors.
-
•
RNA cap synthesis and recognition present novel therapeutic targets.
Abstract
Cytosolic recognition of viral RNA is important for host innate immune responses. Differential recognition of self vs non-self RNA is a considerable challenge as the inability to differentiate may trigger aberrant immune responses. Recent work identified the composition of the RNA 5′, including the 5′ cap and its methylation state, as an important determinant of recognition by the host. Recent studies have advanced our understanding of the modified 5′ RNA recognition and viral antagonism of RNA receptors. Here, we will discuss RIG-I and IFIT proteins as examples of host proteins that detect dsRNA and ssRNA, respectively.
Current Opinion in Structural Biology 2016, 36:133–141
This review comes from a themed issue on Nucleic acids and their protein complexes
Edited by David MJ Lilley and Anna Marie Pyle
For a complete overview see the Issue and the Editorial
Available online 23rd February 2016
http://dx.doi.org/10.1016/j.sbi.2016.01.009
0959-440X/Published by Elsevier Ltd.
Introduction
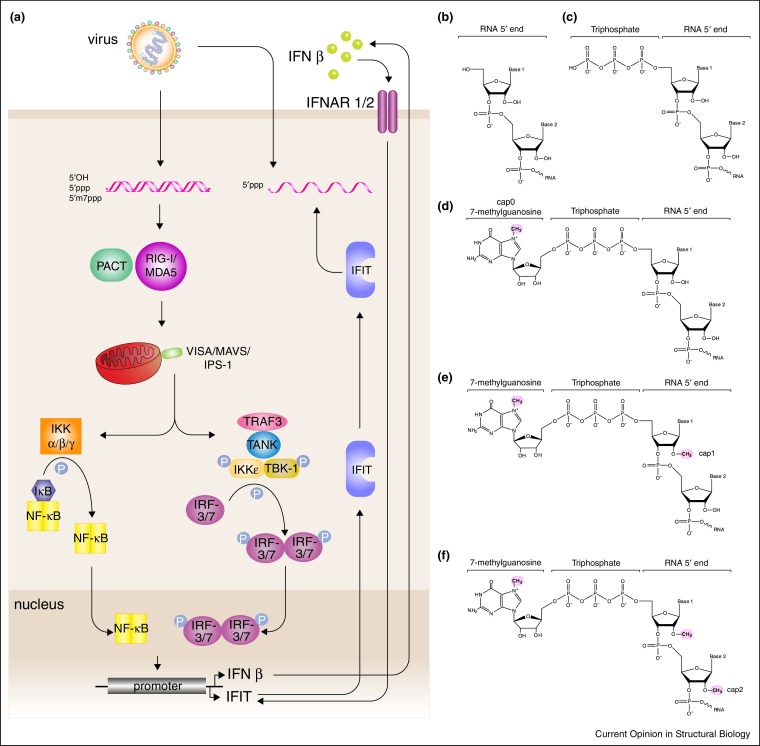
The innate immune system serves as a first line of defense against viral infections. Germline encoded pattern recognition receptors (PRRs) detect pathogens and promote innate immune responses, including activation of type I interferons (IFNs) and stimulation of antiviral genes by recognizing a variety of pathogen associated molecular patterns (PAMPs) such as viral RNA (Figure 1 ). In addition to limiting infections, innate immunity is required to activate humoral responses and to develop long-term protection via adaptive immune responses [1]. Dysregulation of IFN signaling is detrimental to the host, resulting in events such as cytokine storms during infections [2] or autoimmune disorders [3, 4]. Recent studies, discussed below, highlight the need for self/non-self-recognition of the RNA 5′ modifications to discriminate among the viral RNAs that are present within the cytosol during infection.
Figure 1.
Signaling pathways that respond to 5′ modified RNA. (a) RLRs and IFIT family proteins recognize 5′ modified dsRNA and ssRNA, respectively. Chemical structure of 5′ modifications, (b) 5′OH, (c) 5′ppp, (d) 5′ cap0, (e) 5′ cap1, and (f) 5′ cap2.
Retinoic acid inducible gene-I (RIG-I) like receptors (RLRs), which include RIG-I, melanoma differentiation associated factor gene 5 (MDA-5), and laboratory of genetics and physiology 2 (LGP2), are PRRs that contain super family 2 (SF2) RNA helicase domains [5, 6]. RLRs contain a DEX/DH box ATPase core formed by helicase domain 1 (Hel1) and 2 (Hel2), a helicase insertion domain (Heli) and an RNA binding domain known as the C-terminal domain (CTD, also called repressor domain (RD)) [7•] (Figure 2 ). The tandem caspase activation and recruitment domains (CARDs) are present at the N-terminus of RIG-I and MDA-5, which LGP2 lacks. The N-terminal CARD domains engage in protein–protein interactions with other CARD domain containing proteins, most notably with mitochondrial associated antiviral signaling molecule (MAVS, also known as IPS-1, VISA, CARDIF) and therefore is an important signaling component [8]. Unlike in RIG-I, the CARDs do not interact with the Hel2i or other domains within MDA-5 [9, 10]. Therefore, despite high sequence conservation and similar domain organization, there are number of differences in MDA-5 and RIG-I regulation.
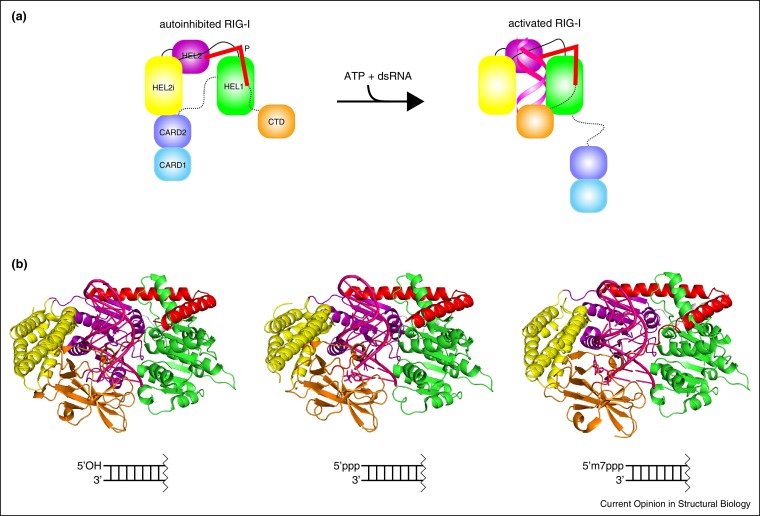
Figure 2.
dsRNA recognition mechanism for RLRs. (a) Domain architecture of RIG-I and model for structural rearrangements upon dsRNA recognition. CARD1 (cyan), CARD2 (blue), helicase HEL1 (green), helicase insertion domain HEL2i (yellow), helicase HEL2 (purple), the regulatory pincer motif P (red), and C-terminal domain CTD (orange). LGP2 lacks the N-terminal CARDs. In the autoinhibited conformation, the N-terminal CARDs are sequestered from signaling and the pincer maintains RIG-I in an autoinhibited state (PDB: 4A2W). Binding of dsRNA and ATP to the CTD brings HEL2i in contact with dsRNA (PDB: 2YKG). The change in conformation upon dsRNA and ATP binding presumably releases the CARD domains for signaling. (b) Specificity of 5′ recognition revealed through the recent structures with 5′OH (left, PDB: 5F9F), 5′ppp (center, PDB: 5F9H), and 5′m7Gppp (right, PDB: 5F98). Figures generated by PyMOL [67].
In the context of RLR signaling, MDA-5 and RIG-I can activate effector molecule MAVS [10]. CARD-CARD interactions between RLRs and MAVS lead to activation of interferon kinases, such as Tank binding kinase-1 (TBK-1) and interferon kB kinase ɛ (IKKɛ), that can phosphorylate interferon regulator factors 3 (IRF3) and 7 (IRF7) [1]. Phosphorylation and nuclear localization of IRF3/7, as well as nuclear factor κB (NFκB), result in type I interferon (IFN) production (Figure 1) [1]. IFN-α/β produced as a result of these signaling events can function in an autocrine and paracrine manner, leading to the induction of a large number of antiviral molecules [1, 6, 11] (Figure 1). Signaling initiated by type I IFNs result in the production of IFN stimulated genes (ISGs), which include numerous host factors that limit virus replication. Among the ISGs, IFIT (Interferon Induced proteins with tetratricopeptide repeats (TPR)) family proteins have been identified as important contributors to antiviral activity through a single-stranded RNA (ssRNA)-dependent mechanism that is incompletely understood at present [12, 13]. IFIT proteins can be expressed in an IFN independent and dependent manner. While all IFIT proteins share the TPR motifs, different isoforms have varied tissue specific and temporal expression profiles [12, 13]. Moreover, IFIT can multimerize and the different multimerization patterns are likely to have different functional outcomes. Here we will review recent advances in our understanding based on structural and biochemical studies of RIG-I-RNA interactions with 5′ modified RNA and discuss how dsRNA and ssRNA recognition are different using studies of IFIT proteins. Additionally, we will discuss how viruses may target such recognition as well as remaining gaps in our understanding of this important early immune response to viral infection.
RIG-I and MDA-5 are multidomain autoinhibited proteins
A series of structures of autoinhibited RIG-I as well as dsRNA-bound forms of RIG-I provided key snap shots, which provide insight into RIG-I regulation, including RNA dependent structural rearrangements and signaling [14••, 15••, 16••, 17••] (Figure 2). In the autoinhibited form, the two N-terminal CARD domains of RIG-I form a head to tail interaction, where the N-terminus of the CARD2 head interacts directly with the C-terminal region of CARD1 and also interact with Heli domain [17••]. These CARD-helicase interactions may also prevent access to MAVS directly or by blocking ubiquitination by TRIM25 [18] and interactions with unanchored polyubiquitin chains [19], which are important for persistent downstream signaling.
RNA recognition and signaling in RLRs require multiple domains.
In RIG-I, interaction with dsRNA involves multiple domains. The CTD, which can directly bind to dsRNA [20•, 21•] and the ATPase core are both required for dsRNA recognition and signaling [22••, 23••]. In addition, the pincer motif (also called the bridging domain or regulatory element) is critical for RIG-I activation as its interactions with the ATPase core functions as an allosteric coupler that facilitates RNA recognition and signaling [24••]. Consistent with these findings that multiple domains within the RLR architecture is important for signaling, the helicase domain alone binds dsRNA with low affinity, and the addition of the CTD markedly improves dsRNA binding [25••, 26••].
Many characteristics of RNA are important for RLR activation. Double strandedness [7•, 27, 28, 29] as well as the ‘blunt’ ends of dsRNA are important elements of RNA recognition by RLRs and are required to activate RLRs [5, 30]. However, selection of self RNA from non-self RNA and therefore persistent signaling require additional characteristics. These include 5′ triphosphate (5′ppp) [21•, 31, 32, 33], 5′ diphosphate (5′ pp) [34•, 35•], and the panhandle structure formed by the 5′ and 3′ untranslated regions (UTRs) [35•, 36]. Yoneyama et al. [7•] suggested that dsRNA binding and ATPase activity are both required for RLR signaling and this proposal was recently demonstrated in two independent studies, where ATP binding, but not ATP hydrolysis was shown to be important for signaling [22••, 26••, 37, 38]. Based on these studies a model emerges where critical interactions of dsRNA with the CTD and subsequently with the helicase result in the reorientation of the pincer domain leading to ATP hydrolysis and release of the N-terminal CARDs for signal transduction. Recent reports also point to ATP hydrolysis as a potential mechanism to discriminate among some RNA lacking PAMP-like characteristics where signaling is aborted. If PAMP characteristics in the bound RNA is observed, then RLRs can engage in more sustained signaling, potentially leading to higher burst activity and limited viral infections. RIG-I and MDA-5 can bind short and long dsRNA, with shorter dsRNA acting as better activators for RIG-I [22••, 39] and longer RNA are better activators of MDA-5 [9, 40, 41•, 42•]. Additional studies point to the stability of the dsRNA stem region suggesting stable double stranded RNA is necessary for persistent signaling during viral infections [43, 44], but not sufficient as a dumbbell RNA lacking blunt ends can bind RIG-I yet display attenuated signaling [22••]. A recent study used mass spectrometry to assess how RIG-I domains change conformation upon RNA ligand and ATP binding [45••]. This study, which used RNA ligands from the Rawling et al. study [22••], revealed that differences in hydrogen-deuterium exchange (HDX) patterns were consistent with major conformational changes upon RNA and ATP binding, as well as with end recognition and binding to double stranded stem regions. These studies collectively show that multiple interactions within the RLRs and multiple properties of RNA are important to confer specificity in order to discriminate between self vs non-self RNA.
Can RLRs discriminate among the 5′ cap structures?
During viral infections, a major source of cytosolic RNA appears to be from replicating viruses. While data is limited in terms of sequence and structural compositions of these RNAs, emerging studies point to the 5′ cap structure as a major element that hosts use to discriminate between self vs non-self patterns. The eukaryotic cap structure contains a triphosphorylated linked 5′-5′ with a N7 methylated guanosine (Figure 1b). Methylation status of this structure, including N7 and 2′O positions result in various permutations of the cap, cap0, cap1, and cap2 designations (see Figure 1b). While the potential role of the cap structure in RLR recognition has been previous speculated, the first evidence of the specificity of recognition was recently defined by Schuberth-Wagner et al., where the significance of the synthetic modified 5′ppp-RNA and its influence on innate immune signaling were assessed. These studies revealed that various 5′ modifications can drive specificity of binding to RIG-I [46••]. For example, the cap (no methylations) like 5′ppp is a strong activator of IFN signaling whereas cap0 is able to partially stimulate IFNs, consistent with its lower than 5′ppp binding affinity for RIG-I [46••]. Previous studies of RIG-I bound to dsRNA that included either 5′ppp or 5′pp suggested that the binding pocket may be able to accommodate the cap structure [21•, 33], but studies with cap0 revealed lower binding than that observed for 5′ppp. Additional modifications at the 2′O position of +1 or +2 nucleotides in the presence or absence of N7 methylation resulted in suboptimal or completely abolished signaling. Of note, the ability of RIG-I to bind RNA is consistent with IFN signaling in cell-based studies [22••, 23••, 34•, 38]; however, there are some deviations that suggest the potential for additional layers of regulation.
The structural basis for 5′ cap recognition was recently provided by Davarkar et al. [26••], where chemically synthesized RNA was used to assess the conformational tolerance to the various 5′ RNA ends (Figure 2b–d) [26••]. In comparison to the structures of 5′ OH and 5′ PPP bound to RIG-I, the G nucleotide base is located within the binding site of the cap0 bound to RIG-I structure but pointing away from the basic patch within RIG-I CTD. The disordering of motif IVa accommodates the positioning and prevents additional steric hindrance [26••]. This orientation, which was previously predicted, is the most favorable since addition of the m7G moiety may also provide electrostatic repulsion with respect to the basic charge within the binding pockets. Thus, a combination of steric and electrostatic forces are likely controlling the interactions within the binding pocket.
Previous studies of RIG-I bound to 5′ppp RNA [14••, 21•, 25••, 33, 39] as well as recent studies [26••, 46••] identified a specific contact between the 2′O and the sidechain of H830, which is a highly conserved residue. Interestingly, both studies revealed that cap1 structures neither bound RIG-I with significant affinity nor stimulated signaling and IFN induction. However, a H830A mutation, which relieves the steric hindrance, allowed the positioning of the 2′O methyl group within the tight binding site.
Collectively, these studies suggest that the m7G cap, which is important for mRNA stability and protein translation, is also a significant determinant of self vs non-self tolerance. Of note, these studies have been performed with RIG-I. Whether MDA-5 also has similar specificity has yet to be determined. Importantly, since MDA-5 is thought to recognize long dsRNA and rely less on the specific nature of the 5′ RNA ends, the impact of the 5′ composition remains an open question.
IFIT proteins differentiate the 5′ end of ssRNA RNA
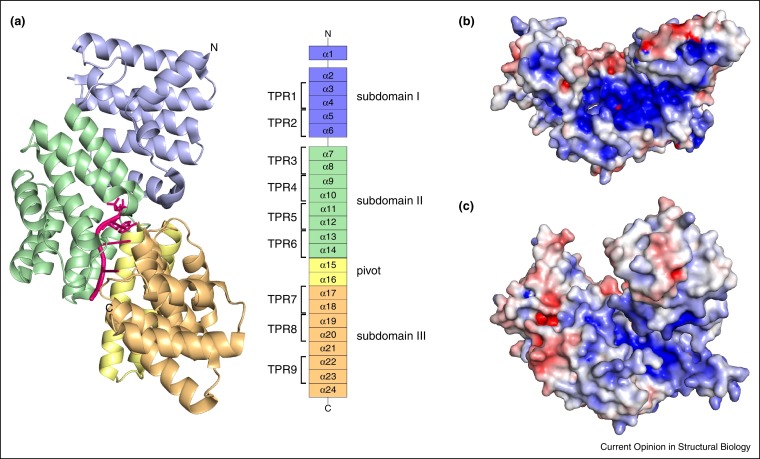
Unlike the RLRs which recognize dsRNA, the IFIT family of proteins target ssRNA and discriminate RNA on the basis of the 5′ modification [12, 13, 47•]. IFIT proteins, which are produced in large amounts in response to IFNs, have been shown to have antiviral functions. Tetratricopeptide repeats (TPRs) are helix turn helix motifs present in proteins that engage in protein–protein interactions. All IFIT proteins contain between 8 and 12 TPR motifs with additional intervening alpha helices. The relative orientation of the TPRs can facilitate conformational changes, which are important for IFIT function. IFITs are part of IFN induced response element signaling, which promotes the production of a large number of antiviral genes, such as protein kinase R (PKR) that inhibits cellular protein production [12, 13]. IFIT proteins are among the most highly expressed in response to IFNs. In humans, IFIT1, IFIT2, IFIT3, and IFIT5 as well as IFIT1B and IFIT1P1, a pseudogene, are expressed, while mouse expresses Ifit1, Ifit2, and Ifit3 along with Ifit1b, Ifit1c and Ifit3b, with the latter three genes largely uncharacterized [13]. Structures of a truncated form of IFIT1 [48••], full length IFIT2 [49, 50], IFIT5 [48••, 51], and IFIT5 bound to 5′ppp RNA [48••] have been solved (Figure 3 ), which highlight some of the key properties that allow IFIT proteins to engage ssRNA in a 5′ end dependent manner [48••, 50]. Unlike RLRs, where the RNA recognition is carried out by multiple domains, different TPRs in IFIT proteins form the binding pocket. Importantly, all IFIT structures to date show a significant basic patch (Figure 3b and c), which can accommodate ssRNA. Specificity of the 5′ ssRNA has been explored biochemically, which support the notion that 2′O methylation is an important property for binding, recognition, and signaling by IFIT family of proteins [36, 52•]. Biochemical studies on the basis of the available structural data and molecular modeling led to the dissection of the RNA binding site [52•] and identified nucleotide preferences for some IFIT family proteins such as the AU preference for IFIT2 [49].
Figure 3.
Recognition of 5′ modified ssRNA by IFIT family of proteins. (a) Structure of human IFIT5 bound to 5′ppp ssRNA model (oligo A; PDB: 4HOT [48••]; left). Domain organization of IFIT5 (right). Electrostatic potential shown on the surface of (b) IFIT2 (PDB: 4G1T [50]) and (c) IFIT5 (PDB: 4HOQ [48••]). Surface potential is indicated by red, white, and blue colors representing negative, neutral, and positive electrostatic potential, respectively (−5 to +5 kBT e−1). Figures generated by PyMOL [67] with the Adaptive Poisson-Boltzmann Solver (APBS) Plugin implemented within PyMOL.
Multiple viral mechanisms to evade 5′ end recognition
Viral immune evasion mechanisms to inhibit innate immune responses are observed for nearly all RNA viruses. In particular, many RNA viruses are detected by PRRs, such as RIG-I and MDA-5. For example, Rhabdoviruses (Vesicular Stomatitis virus and Rabies virus), Paramyxoviruses (Sendai virus, Respiratory syncytial virus, and Newcastle disease virus), Orthomyxoviruses (influenza A and B), and Filoviruses (Ebola virus (EBOV) and Marburg virus (MARV)) are primarily detected by RIG-I, whereas for Picornaviruses (EMCV, Coronavirus, and murine hepatitis virus, and murine norovirus-1 type I), MDA-5 likely is the primary PRR [53, 54]. RIG-I and MDA-5 are thought to be important for detection of positive sense RNA viruses, such as Flaviviruses (Dengue virus and West Nile viruses) as well as double stranded RNA viruses from the Reovirus family (Rotavirus) [53, 54, 55, 56, 57]. Whether or not these differences are due to specific recognition by RLRs remains to be defined.
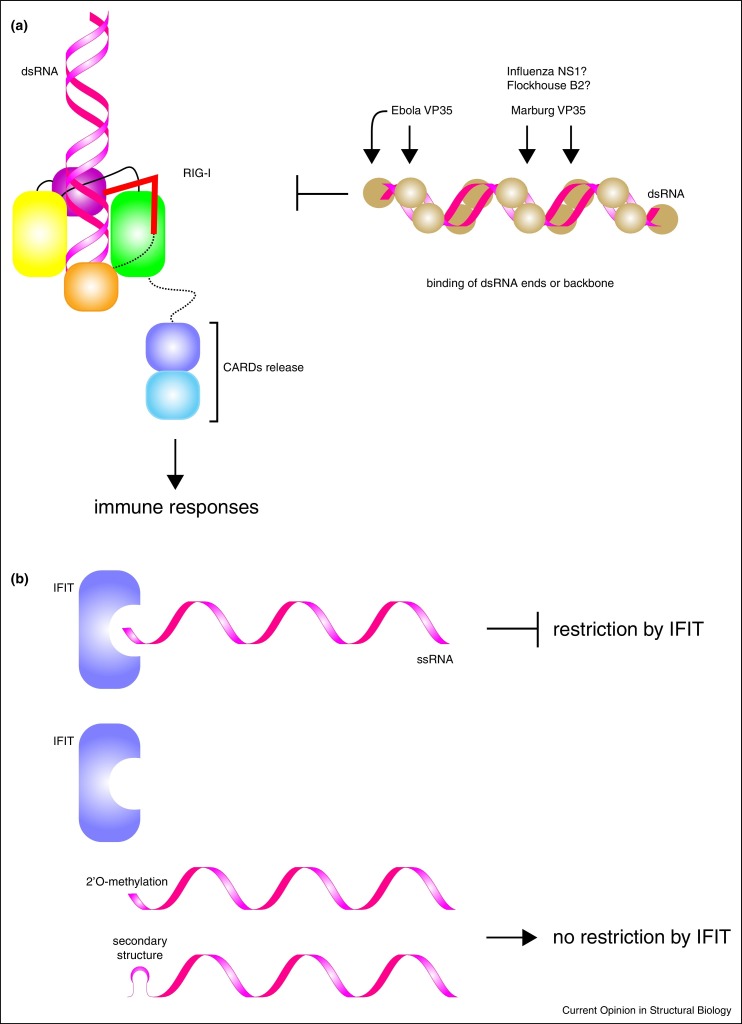
Among the mechanisms that antagonize RLRs, filoviruses (EBOVs and MARVs) as well as influenza virus utilize specific virally encoded antagonists that target RNA PAMPs and these viral antagonists do not directly bind RLRs. Therefore, it is important to discuss the potential implications for the role of 5′ RNA moieties in the context of RLRs or more specifically RIG-I activation. Previous structural studies have shown that influenza A virus (IAV) non-structural protein 1 (NS1) and EBOV/MARV viral protein 35 (VP35) IFN inhibitory domain (IID) can directly bind to short [58•, 59•, 60•] and long double stranded RNA (Figure 4a) [61•, 62•]. Specific contacts between the dsRNA and the viral proteins are consistent with a model where the viral proteins ‘hide’ RNA PAMPs. Despite structural similarities, EBOV and MARV VP35 proteins show significant differences in their ability to recognize blunt ends of RNA. EBOV VP35 IID shows significant interactions with the blunt ends [58•], whereas MARV VP35 does not associate with RNA blunt ends and correspondingly loses its ability to antagonize RIG-I when activated by short dsRNA.
Figure 4.
Viruses use a variety of mechanisms to ‘hide’ RNA, including 5′ RNA recognition by RLRs and IFIT proteins. (a) Examples of dsRNA ‘hiding’ by coating dsRNA by Ebola and Marburg viral VP35 proteins, influenza NS1, and flock house virus B2. Of these proteins, Ebola virus VP35 is also known to bind 5′ dsRNA blunt ends. (b) 5′ ssRNA detection by IFIT proteins is evaded by many viruses through 2′O methylation by viral/host methyltransferases or by utilizing a highly stable RNA secondary structure.
Similar to RLRs, viruses also antagonize ssRNA recognition by IFIT proteins, with the most predominant mechanism being the acquisition of the 2′O methylated cap structure (5′ cap1 or 5′ cap2) by a viral specific methyltransferase or by stealing the cap structure from cellular RNA [63]. In West Nile virus (WNV) [64••] or Japanese encephalitis virus (JEV) [46••, 65••], viruses generated with mutations within the methyltransferase domain results in IFIT sensitivity, where the mutant viruses, but not wildtype viruses were subjected to restriction by Ifit1. Encephalomyocarditis virus (EMCV) is a picornavirus, which uses a 5′ viral peptide to evade detection, while members of the Togaviridae family uses a highly stable 5′ RNA secondary structure [66•] (Figure 4b). In each situation, the absence of the 2′O methylation is compensated in order to avoid detection by IFIT. High resolution structural information for each of these systems will likely provide additional insights into the specific molecular mechanisms that underlie these immune evasion mechanisms.
Concluding remarks
Modification of the 5′ RNA cap allows safe passage into the cytosol for protein translation by providing increased stability and protection from 5′ exonucleases. Additionally, the 5′ cap structure of RNA is thought synergistically enhance protein translation. Recent studies indicate that in addition to facilitating protein translation, the 5′ cap structures promote self vs non-self-discrimination. Given the ability to detect and respond to viral infections due to the immune responses triggered by 5′ RNA recognition, it is not surprising that viruses have developed mechanisms to antagonize proteins that detect cap structures. We have seen remarkable progress in our understanding of host recognition of viral RNA. Future work to address how RNA cap recognition leads to sustained downstream immune signaling will enhance our understanding of this rapidly developing area at a key host-pathogen interface. Such studies are also expected to provide new opportunities to develop therapeutics by targeting cap-dependent signaling.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Conflict of interest statement
None declared.
Acknowledgments
Work in our laboratory is supported by in part by National Institute of Health (NIH) grants (R01AI107056 (Leung), R01AI123926 (Amarasinghe), R01AI114654 (Basler), U191099565 (Ting), U19AI109945 (Basler), U19AI109664 (Basler), and by Department of the Defense, Defense Threat Reduction Agency grants HDTRA1-14-0013 (Basler) and HDTRA1-12-1-0051 (Basler). The content of the information does not necessarily reflect the position or the policy of the federal government and no official endorsement should be inferred.
References
- 1.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang M.A., Kim E.K., Now H., Nguyen N.T., Kim W.J., Yoo J.Y., Lee J., Jeong Y.M., Kim C.H., Kim O.H. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funabiki M., Kato H., Miyachi Y., Toki H., Motegi H., Inoue M., Minowa O., Yoshida A., Deguchi K., Sato H. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald M.E., Rawling D.C., Vela A., Pyle A.M. An evolving arsenal: viral RNA detection by RIG-I-like receptors. Curr Opin Microbiol. 2014;20:76–81. doi: 10.1016/j.mib.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]; Initially described autorepression of RIG-I and laid the foundation for studies on the dsRNA-mediated immunomodulatory mechanisms of RLRs.
- 8.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Berke I.C., Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu B., Hur S. How RIG-I like receptors activate MAVS. Curr Opin Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 12.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fensterl V., Sen G.C. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. [14••, 15••, 16••, 17••] reported the first multidomain RLR structures. These included autoinhibited and activated conformations as well as RIG-I bound to dsRNA. Together, these studies provided important insight into how multiple domains within RLRs modulate activity upon RNA binding.
- 15••.Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S., Carell T., Hopfner K.P. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [14••].
- 16••.Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Jr., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [14••].
- 17••.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]; See annotation to Ref. [14••].
- 18.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 19.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]; Refs. [20•, 21•] provided the structural basis for dsRNA binding by isolated C-terminal domains, including insight into the blunt end recognition.
- 21•.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [20•].
- 22••.Rawling D.C., Fitzgerald M.E., Pyle A.M. Establishing the role of ATP for the function of the RIG-I innate immune sensor. Elife. 2015:4. doi: 10.7554/eLife.09391. [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. [22••, 23••, 24••, 25••] established the relationship between RIG-I RNA detection and conformational changes, including those that respond to ATP binding. Importantly, these manuscripts highlight a critical role for ATP binding.
- 23••.Ramanathan A., Devarkar S.C., Jiang F., Miller M.T., Khan A.G., Marcotrigiano J., Patel S.S. The autoinhibitory CARD2-Hel2i interface of RIG-I governs RNA selection. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1299. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [22••].
- 24••.Rawling D.C., Kohlway A.S., Luo D., Ding S.C., Pyle A.M. The RIG-I ATPase core has evolved a functional requirement for allosteric stabilization by the Pincer domain. Nucleic Acids Res. 2014;42:11601–11611. doi: 10.1093/nar/gku817. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [22••].
- 25••.Vela A., Fedorova O., Ding S.C., Pyle A.M. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J Biol Chem. 2012;287:42564–42573. doi: 10.1074/jbc.M112.385146. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [22••].
- 26••.Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provided the structural basis for m7G cap and 2′O methylation recognition. Also see Ref. [46••].
- 27.Hausmann S., Marq J.B., Tapparel C., Kolakofsky D., Garcin D. RIG-I and dsRNA-induced IFNbeta activation. PLoS ONE. 2008;3:e3965. doi: 10.1371/journal.pone.0003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt A., Schwerd T., Hamm W., Hellmuth J.C., Cui S., Wenzel M., Hoffmann F.S., Michallet M.C., Besch R., Hopfner K.P. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawling D.C., Pyle A.M. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 32.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. [34•, 35•] established 5-diphosphates as important agonists of RIG-I.
- 35•.Luo D., Kohlway A., Vela A., Pyle A.M. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure. 2012;20:1983–1988. doi: 10.1016/j.str.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [34•].
- 36.Habjan M., Andersson I., Klingstrom J., Schumann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Muhlberger E. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louber J., Brunel J., Uchikawa E., Cusack S., Gerlier D. Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5. BMC Biol. 2015;13:54. doi: 10.1186/s12915-015-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassig C., Matheisl S., Sparrer K.M., de Oliveira Mann C.C., Moldt M., Patel J.R., Goldeck M., Hartmann G., Garcia-Sastre A., Hornung V. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife. 2015:4. doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T., Hohng S., Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci U S A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T., Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. [41•, 42•] provided insight into MDA5 mediated signaling via protein–protein interactions of the CARDs.
- 42•.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]; See annotation to Ref. [41•].
- 43.Anchisi S., Guerra J., Garcin D. RIG-I ATPase activity and discrimination of self-RNA versus non-self-RNA. MBio. 2015;6:e02349. doi: 10.1128/mBio.02349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anchisi S., Guerra J., Mottet-Osman G., Garcin D. Mismatches in the influenza A virus RNA panhandle prevent retinoic acid-inducible gene I (RIG-I) sensing by impairing RNA/RIG-I complex formation. J Virol. 2015;90:586–590. doi: 10.1128/JVI.01671-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Zheng J., Yong H.Y., Panutdaporn N., Liu C., Tang K., Luo D. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015;43:1216–1230. doi: 10.1093/nar/gku1329. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided an important experimental tool to evaluate various structural snapshots in the context of solution state conformational changes associated with ligand binding by RLRs.
- 46••.Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reported how m7G cap and 2′O methylation recognition allow for self vs non-self differentiation. Also see Ref. [26••].
- 47•.Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R., Burckstummer T., Stefanovic A., Krieger S., Bennett K.L. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]; Identified IFIT proteins for their ability to distinguish ssRNA 5′ ends.
- 48••.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]; Defined the structural basis for 5′ RNA specificity of IFIT5 and identified the structural features that allow IFIT proteins to distinguish ssRNA from dsRNA.
- 49.Feng F., Yuan L., Wang Y.E., Crowley C., Lv Z., Li J., Liu Y., Cheng G., Zeng S., Liang H. Crystal structure and nucleotide selectivity of human IFIT5/ISG58. Cell Res. 2013;23:1055–1058. doi: 10.1038/cr.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z., Liang H., Zhou Q., Li Y., Chen H., Ye W., Chen D., Fleming J., Shu H., Liu Y. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katibah G.E., Lee H.J., Huizar J.P., Vogan J.M., Alber T., Collins K. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49:743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp-mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evaluated the molecular basis of RNA selectivity by IFIT proteins through structure-based biochemical analyses.
- 53.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 54.Ramos H.J., Gale M., Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., Garcia-Sastre A., Katze M.G. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broquet A.H., Hirata Y., McAllister C.S., Kagnoff M.F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 57.Leung D.W., Basler C.F., Amarasinghe G.K. RIG-I like receptors. Trends Microbiol. 2012 doi: 10.1016/j.tim.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Leung D.W., Prins K.C., Borek D.M., Farahbakhsh M., Tufariello J.M., Ramanan P., Nix J.C., Helgeson L.A., Otwinowski Z., Honzatko R.B. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]; Defined blunt end recognition as a basis for RLR inhibition by Ebola proteins. Together with Refs. [59•, 60•, 61•, 62•], established the basis for RLR inhibition by filoviruses.
- 59•.Leung D.W., Shabman R.S., Farahbakhsh M., Prins K.C., Borek D.M., Wang T., Muhlberger E., Basler C.F., Amarasinghe G.K. Structural and functional characterization of Reston Ebola VP35 Interferon Inhibitory Domain. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [58•].
- 60•.Bale S., Julien J.P., Bornholdt Z.A., Krois A.S., Wilson I.A., Saphire E.O. Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. J Virol. 2013;87:10385–10388. doi: 10.1128/JVI.01452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [58•].
- 61•.Ramanan P., Edwards M.R., Shabman R.S., Leung D.W., Endlich-Frazier A.C., Borek D.M., Otwinowski Z., Liu G., Huh J., Basler C.F. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [58•].
- 62•.Bale S., Julien J.P., Bornholdt Z.A., Kimberlin C.R., Halfmann P., Zandonatti M.A., Kunert J., Kroon G.J., Kawaoka Y., Macrae I.J. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS Pathog. 2012;8:e1002916. doi: 10.1371/journal.ppat.1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [58•].
- 63.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrated that viral encoded methyltransferases provide a key immune evasion of mechanisms against IFIT proteins.
- 65••.Kimura T., Katoh H., Kayama H., Saiga H., Okuyama M., Okamoto T., Umemoto E., Matsuura Y., Yamamoto M., Takeda K. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol. 2013;87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [64••].
- 66•.Hyde J.L., Gardner C.L., Kimura T., White J.P., Liu G., Trobaugh D.W., Huang C., Tonelli M., Paessler S., Takeda K. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014 doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]; Described how RNA structural stability facilitates evasion of IFIT antiviral activity.
- 67.DeLano W.L. San Carlos, CA, USA; DeLano Scientific: 2002. The PyMOL Molecular Graphics System. [Google Scholar]