Abstract
Ketolides are the latest derivatives developed from the macrolide erythromycin to improve antimicrobial activity. All macrolides and ketolides bind to the 50S ribosomal subunit, where they come into contact with adenosine 2058 (A2058) within domain V of the 23S rRNA and block protein synthesis. An additional interaction at nucleotide A752 in the rRNA domain II is made via the synthetic carbamate-alkyl-aryl substituent in the ketolides HMR3647 (telithromycin) and HMR3004, and this interaction contributes to their improved activities. Only a few macrolides, including tylosin, come into contact with domain II of the rRNA and do so via interactions with nucleotides G748 and A752. We have disrupted these macrolide-ketolide interaction sites in the rRNA to assess their relative importance for binding. Base substitutions at A752 were shown to confer low levels of resistance to telithromycin but not to HMR3004, while deletion of A752 confers low levels of resistance to both ketolides. Mutations at position 748 confer no resistance. Substitution of guanine at A2058 gives rise to the MLSB (macrolide, lincosamide, and streptogramin B) phenotype, which confers resistance to all the drugs. However, resistance to ketolides was abolished when the mutation at position 2058 was combined with a mutation in domain II of the same rRNA. In contrast, the same dual mutations in rRNAs conferred enhanced resistance to tylosin. Our results show that the domain II interactions of telithromycin and HMR3004 differ from each other and from those of tylosin. The data provide no indication that mutations within domain II, either alone or in combination with an A2058 mutation, can confer significant levels of telithromycin resistance.
Many groups of clinically useful antibiotics inhibit pathogen growth by preventing the synthesis of new proteins on the bacterial ribosome (13, 48). Generally, inhibition occurs when a functionally important site on the ribosome is blocked by the binding of a single antibiotic molecule. One such site is dedicated to peptide bond formation (the peptidyltransferase center) and is situated at the mouth of the peptide exit tunnel on the 50S ribosomal subunit (32). This region is adjacent to the binding site for macrolide, lincosamide, and streptogramin B (MLSB) antibiotics (19, 41). Thus, the primary inhibitory effect of lincosamides and streptogramin B drugs is to block the formation of peptide bonds, and this reaction is also inhibited to various degrees by the 16-membered-lactone-ring macrolides. However, the main inhibitory effect of macrolides is to block the passage of the newly synthesized peptide chain though the exit tunnel; and in the case of the smaller 14-membered-ring macrolides, whose structures do not reach into the peptidyltransferase center, this is probably their sole mode of action on the assembled ribosome (37, 47).
To exert an effective blockade against the growing peptide chain, it is essential that macrolide antibiotics attach securely to their binding sites within the tunnel. The primary attachment site of all macrolide antibiotics is at adenosine 2058 (A2058) of 23S rRNA (19, 41). Close by are several other accessible rRNA nucleotides that are also used as contact sites if they fall within the reach of a drug's chemical substituents (3, 19, 41). In the study described in this article, we investigated how the macrolide tylosin and the ketolide derivatives of the macrolide erythromycin come into contact with nucleotides in domain II of 23S rRNA.
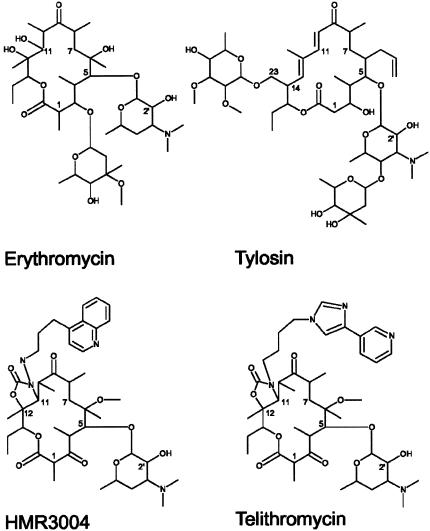
Erythromycin is a naturally occurring 14-membered-ring macrolide that has been derivatized in numerous ways to improve its pharmaceutical properties (5). The latest derivatives, the ketolides, are equipped with 3-keto and 6-methoxy groups that improve acid stability, and in the case of HMR3647 (telithromycin) and HMR3004 (Fig. 1), an alkyl-aryl chain extends from a carbamate at lactone ring positions C-11 and C-12 (4). The C-11 and C-12 extension interacts with nucleotide A752 within domain II of 23S rRNA, and derivatives of telithromycin and HMR3004 lacking the C-11 and C-12 extension have ribosome binding affinities that are reduced more than 1,000-fold (20, 53). Erythromycin does not make direct contact with nucleotide A752, as this compound is too small to span the distance (approximately 15 Å) from its primary contact site at A2058 (Fig. 2), on the opposite face of the exit tunnel (41). The interaction between MLSB antibiotics and this region of the rRNA domain II is not unprecedented, however, and contact between the streptogramin B drug vernamycin and nucleotide A752 has previously been documented (31); furthermore, binding of the 16-membered-ring macrolide tylosin (Fig. 1) involves interaction with the neighboring nucleotide, G748 (19, 30).
FIG. 1.
Structures of the macrolide and ketolide compounds used in this study. The chemical groups that contact domain II of 23S rRNA are the mycinose sugar at C-23 of tylosin and the alkyl-aryl substituents extending from C-11 to C-12 of telithromycin and HMR3004.
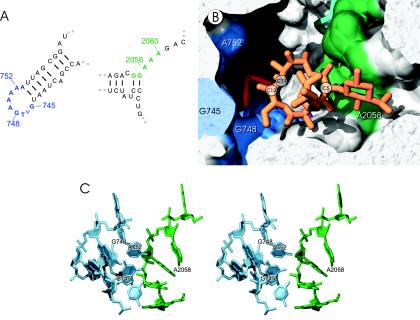
FIG. 2.
Macrolide and ketolide target site in 23S rRNA. (A) Secondary structures of the nucleotides that make up the target site, with the relevant portions of domain II in blue on the left and those of domain V in green on the right. (B) Cross-section of the ribosomal tunnel drawn from the crystallographic coordinates (41), showing erythromycin (orange) bound within the macrolide and ketolide binding site. Positions C-5, C-11, and C-12 of the lactone ring are labeled to show the orientation of erythromycin (and the other macrolides and ketolides) within the site; rRNA nucleotides on the inner surface of the ribosomal tunnel are color coded as described for panel A. The path of the tunnel is from the peptidyltransferase center (on the far right and slightly above the plane of the picture) and past the drug binding site, where it turns down into the plane of the picture (black area). The tips of the ribosomal proteins L4 (turquoise), L22 (red), and L35 (magenta, behind the C-2 lactone ring) protrude into the tunnel. (C) Stereo view showing the perspective of the domain II and V nucleotides that make up the macrolide and ketolide binding site color coded as described for panel A; ribosomal proteins and antibiotics are omitted.
Resistance to MLSB antibiotics is most commonly caused by alterations within the ribosome binding site that perturb the drug-rRNA interaction. The best-documented examples of this mechanism involve nucleotide A2058, where base methylation (44, 51) or mutation (49) confers resistance to MLSB antibiotics. Similarly, methylation of the tylosin contact site at G748 confers specific resistance to a subgroup of macrolides that are structurally related to tylosin (30). Telithromycin is the first ketolide to be approved for clinical use, and it has been demonstrated to be an effective antimicrobial agent (11, 24-27). It is yet too soon to evaluate fully the extent to which resistance to telithromycin might develop in clinical isolates and whether this will involve mutation in domain II of the rRNA. However, given the many cases of rRNA mutations that confer macrolide resistance (14, 46, 49), plus a recent report of telithromycin resistance in a laboratory isolate of Streptococcus pneumoniae from which nucleotide A752 had been deleted (7), we believed that it was important to evaluate the potential for resistance mechanisms of this type.
We have introduced mutations at nucleotides G748 and A752 within domain II of 23S rRNA to disrupt key contact sites for the ketolides telithromycin and HMR3004 and for the macrolide tylosin. The main interaction site of the drugs at A2058 was also mutagenized and combined with the domain II mutations. The effects of the single and dual rRNA mutations on the potencies of the antimicrobials enabled us to assess the likelihood that these types of resistance mechanisms will occur in clinical strains.
MATERIALS AND METHODS
Escherichia coli strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli strain DH1 (40) was used for the initial isolation of plasmids containing mutant rRNA alleles. Two strains, AS19rlmAI and TA531, were used to evaluate the phenotypes conferred by the mutant rRNAs. Strain AS19rlmAI is a derivative of AS19 that has lost its ability to methylate 23S rRNA nucleotide G745; both AS19 and AS19rlmAI are sensitive to macrolide drugs, probably due to a defective tolC gene. In strain TA531, none of the seven chromosomally encoded rRNA (rrn) operons are functional, and the strain is dependent on a plasmid, pHK-rrnC+, for rRNA production.
TABLE 1.
Bacterial strains, plasmids, and oligodeoxynucleotides used in the study
| Strain, plasmid, and primer | Description | Reference(s) |
|---|---|---|
| Strains | ||
| DH1 | F−endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 | 40 |
| AS19a | Highly permeable to macrolide and ketolide drugs | 42 |
| AS19rlmAI | rlmAI knockout strain of AS19; lacks methylation at 23S rRNA G745 | 28 |
| TA531 | Δ(rrnE rrnB rrnH rrnA) rrnG::lacZ+rrnD::cat+rrnC::cat+ ΔrecA/pTRNA66/pHK-rrnC+ | 1 |
| MC250 | Δ(rrnE rrnB rrnH rrnA) rrnG::lacZ+rrnD::cat+rrnC::cat+ ΔrecA, rpsL121/pTRNA66/prrnS12 (rrnB+rpsL+) | 34 |
| Plasmids | ||
| pFK5 | AmprrrnB+, A2058G in 23S rRNA gene | 8 |
| pNK | AmprrrnB+, wild-type rRNA genes | 52 |
| pUC19 | Ampr, polycloning site | 33, 54 |
| pTRNA66 | Spcr, tRNA genes | 1 |
| pHK-rrnC+ | KanrrrnC+, wild-type rRNA genes | 1 |
| Primers | ||
| −21 | Binds after the 3′ end of the pUC19 polycloning site | |
| −40 | Binds in front of the 5′ end of the pUC19 polycloning site | |
| SD2 | For sequencing of 23S rRNA domain V at A2058 | |
| SD23 | For sequencing of 23S RNA domain II around A752 | |
| 5′ mutagenesis primer | 5′-CGACTAATGTTGAAAAATTAGCGG-3′b | |
| 3′ mutagenesis primer | 3′-GCTGATTACAACTTTTTAATCGCC-5′b |
In terms, the highly permeable phenotypes of this strain and strain AS19rlmAI are probably caused by a defective efflux mechanism due to a tolC mutation.
The positions underlined in bold correspond to nucleotides 748 and 752, respectively, where base substitutions or nucleotide deletions were introduced into the rrnB 23S rRNA gene. A single change was made in each of the 5′ mutagenesis primers and was used with the complementary change in the 3′ mutagenesis primer.
Plasmid pHK-rrnC+ is equipped with a kanamycin resistance gene; plasmids pUC19, pNK, and pFK5 have a β-lactamase gene and were maintained with ampicillin (at 100 μg/ml, unless stated otherwise) in the growth medium. The E. coli strains were grown at 37°C in Luria broth or on agar plates with the same nutrients (40). For determination of the growth rates of the TA531 strains, 1,000-fold dilutions of overnight cultures were grown in Luria broth with and without ampicillin, and cell density was measured be determination of the optical density at 600 nm every 30 min to calculate the cell doubling times.
Site-directed mutagenesis.
Nucleotides 748 and 752 in the plasmid-encoded 23S rRNA gene were mutagenized by PCR with two sets of oligonucleotides (22, 23). Briefly, complementary oligonucleotides with the desired mutations were designed (Table 1), and each was used in separate PCRs with a primer overlapping the SacI or the SphI site within domain II of the E. coli rrnB 23S rRNA gene. The products from the first two reactions were mixed in a second PCR mixture to generate a 900-bp SacI-SphI fragment with the desired mutation. Each SacI-SphI fragment was cloned into plasmid pUC19, and its structure was checked by sequencing before it was inserted in its original context into the 23S rRNA genes of plasmids pNK and pFK5. Plasmid pNK contains a wild-type rrnB operon, and plasmid pFK5 is essentially the same as pNK but has a A-to-G substitution at 23S rRNA position 2058 (Table 1). Mutant plasmids were used to transform E. coli strain DH1, and the plasmid structures were verified by restriction digestion and sequencing (CEQ 2000; Beckman) before they were used to transform the other E. coli strains.
Expression of mutant 23S rRNAs.
The pNK and pFK5 plasmid derivatives were moved into E. coli strains AS19rlmAI and TA531. In TA531, displacement of the resident plasmid pHK-rrnC+ is required to obtain homozygous strains with a mutated rrnB operon. After transformation of TA531 with pNK or pFK5 derivatives, the cells were grown for at least 20 generations in Luria broth medium containing ampicillin, before they were diluted 105-fold and plated onto agar with ampicillin. Individual colonies were screened for the loss of kanamycin resistance, indicating replacement of the original pHK-rrnC+ plasmid by pNK or pFK5. The structures of the pNK and pFK5 derivatives were verified as described above for each strain.
MIC measurements.
Overnight cultures of AS19rlmAI and TA531 cells containing pNK and pFK5 derivatives were diluted 105-fold and were plated onto agar containing 25 μg of ampicillin per ml, the macrolide antibiotics erythromycin and tylosin (Sigma), and the ketolides telithromycin (HMR3647) and HMR3004 (Aventis Pharma). For TA531 cells, the macrolide and ketolide concentrations were increased in 1-μg/ml steps up to 5 μg/ml, 5-μg/ml steps up to 50 μg/ml, and 50-μg/ml steps at higher concentrations; for AS19rlmAI cells, the drug concentrations started at 0.25 μg/ml and were increased in twofold steps to 1,024 μg/ml. The agar plates were incubated at 37°C, and after 20 h the MICs were scored as the lowest concentration at which no growth was observed.
Ribosome purification and rRNA sequencing.
Ribosomes were prepared from strain TA531 by sonication and centrifugation, and rRNA was extracted as described previously (9). The relevant regions in domains II and V of the 23S rRNAs were sequenced by primer extension with reverse transcriptase (45).
RESULTS
Choice of E. coli strains.
The ribosomes from E. coli are those that have been the best studied by biochemical and genetics means, and this remains the only bacterium in which rRNA nucleotide modifications have been comprehensively mapped. There are, however, three obstacles inherent in using E. coli to study the effects of rRNA mutations on drug resistance. First, the bacterium has a gram-negative wall and outer membrane that act as a barrier to most MLSB drugs. This problem was overcome by using permeant E. coli strain AS19 (Table 1), which can absorb a range of MLSB drugs (28). Second, E. coli 23S rRNA is methylated at nucleotide G745 (17), which is situated at the MLSB drug binding site, where it forms a noncanonical base pair with A752 (2, 21). This methylation is absent in gram-positive bacteria (29), and as it was uncertain whether it would affect the binding of MLSB drugs (especially in combination with rRNA mutations), we inactivated the gene encoding the G745-specific methyltransferase, rlmAI, on the AS19 chromosome (Table 1).
The third drawback with E. coli is that, for technical reasons, mutagenized 23S rRNA genes are expressed from a plasmid-encoded rRNA (rrn) operon. E. coli has seven chromosome-encoded (wild-type) rrn operons, and thus, strains with mutant rrn plasmids come to possess a heterologous population of ribosomes (43). For the 2058G mutation expressed from the type of plasmid used here, about 55% of ribosomes contain the mutant 23S rRNA (9), leaving a residual 45% of wild-type ribosomes that can cloud the evaluation of mutant rRNA phenotypes. This concern has been removed by inactivating all the chromosomal rrn operons in strain TA531 and ensuring that rRNA is produced only from the plasmid-encoded rrn operon (1).
These changes in the E. coli strains do have their biological costs in terms of cell growth rates, and the virtues of strains AS19rlmAI and TA531 have not yet been combined into a single strain. Thus, we have studied separate aspects of macrolide and ketolide binding using strains AS19rlmAI and TA531.
Nucleotides mutagenized within domain II of 23S rRNA.
Two nucleotides, G748 and A752, in domain II of 23S rRNA have been associated with the interaction with macrolides and ketolides and were thus obvious candidates for mutagenesis. The three possible single-base substitutions were made at each position, in addition to deletion of G748 and A752 and insertion of an extra adenosine next to A752.
Mutations at the main macrolide and ketolide contact site, A2058, have already been well studied (43, 49, 50). However, the studies with the mutation at position 2058 have often been done with heterogeneous ribosome populations, and to our knowledge, no studies have ever been done with this mutation in combination with domain II mutations. In the present study, rRNAs with single-site mutations were expressed first in TA531 to observe the phenotypes of the mutants with homogeneous mutations in the ribosomes (Table 2). Next, the mutated rRNAs were expressed in AS19rlmAI cells, enabling us to investigate less permeant drugs such as tylosin, as well as the phenotypes of strains with dual mutations that proved to be lethal unless they were expressed in a heterozygous strain.
TABLE 2.
MICs of erythromycin and ketolides for rRNA mutant strains of E. coli TA531
| Mutationa | MIC (μg/ml) for TA531 cellsb
|
Doubling time (min)c | ||
|---|---|---|---|---|
| Erythromycin | Telithromycin | HMR3004 | ||
| None (wild type) | 45 | 5 | 3 | 57 ± 2.9 |
| 752G | 40 | 15 | 3 | 57 |
| 752U | 30 | 15 | 3 | 57 |
| 752C | 45 | 20 | 3 | 57 |
| 752A-ins | 45 | 20 | 3 | 57 |
| 752Δ | 30 | 25 | 20 | 60 ± 2.5 |
| 748A, U, C, and Δ | 45 | 5 | 3 | 57 |
| 2058G | 400d | 200 | 200 | 57 ± 3.2e |
The bases A, C, G, and U indicate the substitutions made at the given nucleotide positions in 23S rRNA. A-ins, an adenosine was inserted at this position; Δ, the nucleotide was deleted. Substitutions of A, C, and U were also introduced at position 745, and none of these changes affected growth in the presence of the drugs tested here (data not shown).
Strain TA531 has no active chromosomal copies of rrn, and all rRNAs are derived from plasmids expressing rrnB. Strains are thus homozygous for the 23S rRNA mutations shown. We were unsuccessful in attempts to create strains that were homozygous for 23S rRNA containing a domain II mutation (at position 748 or 752) combined with the 2058G domain V mutation (see text).
Growth rates were initially estimated by comparison of colony sizes on agar plates (a minimum of five platings). Mutants that grew at rates different from those for the wild-type cells were cultured in liquid broth in the absence of MLSB antibiotics to estimate their doubling times. The doubling times shown here are the means ± standard deviations from three or more growth experiments (with ampicillin in the broth). The doubling time of strain DH1 was 40 ± 1.2 min when it was grown under the same conditions as strain TA531; no significant difference in growth was seen for DH1 cells expressing the various mutations.
MICs were generally 30 to 50% lower than the values shown here when these mutations were heterozygously expressed in E. coli strain DH1; significant exceptions are the ketolide MICs for the mutant with the 2058G mutation, for which the erythromycin, telithromycin, and HMR3004 MICs were 200, 15, and 10 μg/ml, respectively (9).
Values here are for the 2058G mutation expressed from rrnB in plasmid pFK5. TA531 cells containing an independently constructed rrnB plasmid with 2058G (50) were found to have growth rates that were indistinguishable from those of cells with wild-type or pFK5 plasmids (data not shown).
Homozygous expression of rRNA mutations.
TA531 cells with the rRNAs with mutations at single sites (Table 2) were all kanamycin sensitive, indicating that these cells had been cured of the pHK-rrnC+ plasmid with the wild-type rrnC operon. Furthermore, direct sequencing of the rRNA verified that the ribosome populations in these cells were homogeneous and contained only mutant 23S rRNA encoded by the pNK- and pFK5-encoded rrnB operon. Expression of all the rRNAs with mutations at single sites in TA531 made it possible to assess resistant phenotypes and whether the mutations carry a biological cost, indicated by slower growth rates.
With the exception of the nucleotide 752 deletion, which slowed the growth rate by approximately 5% (Table 2), none of the single-site mutations in domain II or the A2058G substitution detectibly altered the rate of cell growth. Each of the single substitutions at A752, as well as deletion of this nucleotide, conferred mild resistance to telithromycin. Of these mutations, the A752 deletion conferred the most obvious telithromycin-resistant phenotype and was the only domain II mutation to give HMR3004 resistance. None of the changes at position 752 conferred resistance to erythromycin; and no resistance to erythromycin, telithromycin, or HMR3004 resulted from the changes at nucleotide 745 or 748. Predictably, the 2058G substitution conferred high levels of resistance to all three drugs (Table 2).
Plasmids were constructed in which each of the mutations at position 752 (and 748U) was combined with 2058G. None of the plasmids with the dual mutations was capable of displacing resident plasmid pHK-rrnC+ from TA531 cells. The use of a modified version of this cell-plasmid system (34) showed that forcing the loss of a plasmid-encoding wild-type rrn with any of the pFK5 derivatives with double mutations was lethal to the cell (data not shown). Thus, cells harboring the dual mutations are viable only in the heterozygous state with functional wild-type rrn operons.
Mutant rRNA expression in AS19rlmAI cells.
Markedly lower drug concentrations were required to inhibit the permeable strain AS19rlmAI (Table 3) compared to those required to inhibit the TA531 strain; in particular, study of the macrolide tylosin was made possible by using the permeable strain (the MIC of tylosin is >500 μg/ml for TA531). In AS19rlmAI, the rRNAs with single-site mutations are expressed together with the chromosome-encoded wild-type rRNAs, and this mixture confers ketolide resistance phenotypes that are diluted and less evident than those of TA531 cells. No detectable change in the growth rate of AS19rlmAI was caused by any of the rRNA mutations (data not shown).
TABLE 3.
MICs of the macrolide and the ketolide antibiotics for rRNA mutant strains of E. coli AS19rlmAI
| Mutationa | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Erythromycin | Tylosin | Telithromycin | HMR3004 | |
| None (wild type) | 0.5 | 1 | 0.5 | 0.5 |
| 752G | 0.5 | 1 | 1 | 0.5 |
| 752U | 0.5 | 1 | 1 | 0.5 |
| 752C | 0.5 | 1 | 1 | 0.5 |
| 752A-ins | 1 | 2 | 1 | 1 |
| 752Δ | 0.5 | 2 | 1 | 1 |
| 748U | 0.5 | 1 | 0.5 | 0.5 |
| 2058G | 1,024 | 16 | 64 | 64 |
| 2058G + 752G | 32 | 64 | 1 | 1 |
| 2058G + 752U | 32 | 128 | 2 | 1 |
| 2058G + 752C | 32 | 64 | 2 | 1 |
| 2058G + 752Δ | 2 | 4 | 1 | 1 |
| 2058G + 752A-ins | 32 | 64 | 2 | 1 |
| 2058G + 748U | 64 | 32 | 1 | 2 |
All strains expressed 23S rRNA both from a plasmid-encoded (mutagenized) copy of rrnB and from the chromosomally encoded (wild-type) rrn operons and are, thus, with the exception of the nonmutagenized control, heterozygous with respect to 23S rRNA. Plasmids encoding single mutations in 23S rRNA domain II (at nucleotides 748 and 752) are pNK derivatives; plasmids encoding the nucleotide 2058 mutant and mutant combinations are pFK5 derivatives (Table 1). See footnote a of Table 2 for definition of the abbreviations and designations.
AS19rlmAI cells expressing the rRNA with the 2058G mutation exhibited exceptionally strong resistance to erythromycin and distinct but lower levels of resistance to the ketolides telithromycin and HMR3004. This is consistent with previous observations with strain DH1, in which a heterologous population of ribosomes with 2058G conferred high levels of erythromycin resistance and significantly lower levels of ketolide resistance (Table 2, footnote d). The disproportionately low level of ketolide resistance in 2058G heterozygotes could be linked to the bactericidal mode of action of telithromycin (35, 36) against cells with sensitive (wild-type) ribosomes. The 2058G mutation causes a modest increase in the tylosin MIC.
Combination of the substitutions at position 752 with 2058G partially (for erythromycin) or completely (for ketolides) neutralized the resistance phenotype conferred by 2058G (Table 3). The rRNAs with dual mutations were expressed to the same extent as the rRNAs with single mutations, they were assembled into ribosomes (established by direct sequencing of the rRNA), and they caused no detectible change in the rate of growth of AS19rlmAI cells. Thus, despite the loss of erythromycin and ketolide resistance, the rRNAs with dual mutations do appear to be functional, as they are present in translating ribosomes. Moreover, several combinations of these mutations confer improved resistance to tylosin.
DISCUSSION
We have assessed the importance of key nucleotides in domain II of 23S rRNA for macrolide and ketolide binding. Base substitutions at nucleotide 752 confer modest resistance to telithromycin when cells contain a homogeneous population of mutant ribosomes (Table 2). Thus, the antimicrobial activity of telithromycin is most effective when there is an adenine at position 752. This indicates that telithromycin interacts specifically with A752, possibly via hydrogen bonds between one of the heterocyclic rings on the C-11 to C-12 extension (Fig. 1) and chemical groups of the adenine. However, substitutions at A752 confer no resistance to the ketolide HMR3004, even though both HMR3004 and telithromycin protect the base at A752 against chemical modification (20, 53). These observations suggest that HMR3004 interacts with nucleotide 752 in a less discriminatory manner than telithromycin, possibly by stacking of the aryl group of the HMR3004 C-11 to C-12 extension onto the base at position 752. Deletion of nucleotide 752 would disrupt both types of interactions, and accordingly, this mutation confers resistance to both ketolides (Table 2). None of the changes at nucleotide 752 confer erythromycin resistance, which fits with the lack of direct contact between erythromycin and this nucleotide (41). Similarly, the MIC data show no evidence of an interaction between HMR3004, telithromycin, or erythromycin and the neighboring nucleotide, G748 (Table 2); and this nucleotide is outside the range of drug interaction in the crystal structures (3, 41).
The highest levels of resistance in these studies was conferred by the A-to-G substitution at nucleotide 2058. The 2′-hydroxyl of the desosamine sugar on erythromycin, telithromycin, and, presumably, HMR3004 makes a hydrogen bond to the N-6 position of A2058 (3, 41); and disruption of this interaction by mutation (49) or methylation of A2058 (51) confers the well-documented MLSB resistance phenotype.
Gram-negative bacteria are generally impermeable to tylosin, and the membrane defects in strain AS19rlmAI made it possible to study the effects of this macrolide. Tylosin is one of the few naturally occurring macrolides to contact domain II of the 23S rRNA (38), interacting via its mycinose sugar with nucleotide G748 (19, 30). While the single-site domain II mutations (Table 3) have no appreciable effect on tylosin binding and the 2058G mutation confers only mild resistance, tylosin resistance is significantly enhanced by the combination of these mutations. The 752U-2058G combination gave the clearest tylosin-resistant phenotype, and this is consistent with the proximity of these two nucleotides to tylosin shown in the crystal structure and by chemical protection data (19, 38). The 748U-2058G combination conferred slightly higher levels of resistance than either mutation on its own, although there was no evidence of the synergistic effect that was seen with methylation of the bases at these two positions (30).
The level of erythromycin resistance conferred by 2058G was markedly reduced when the 2058G mutation was combined with any of the domain II mutations, and the ketolide-resistant phenotypes were abolished (Table 3). The rRNAs with the dual mutations are clearly functional because, in addition to the improved resistance to tylosin that they confer, these rRNAs were shown to be present in translationally active ribosomes. However, the lack of success at creating strains with a homogeneous population of ribosomes with the dual mutations suggests that the rRNAs are in some way defective and require the presence of wild-type ribosomes to sustain their growth. Although we do not understand this phenomenon, it is not without precedent, as examples of rRNA mutations that are phenotypically silent on their own but that become deleterious in combination have been documented (39).
The function of the rRNAs has remained highly preserved in different organisms throughout evolution (15), and this is reflected in the almost perfect conservation of the rRNA secondary structure (6, 18) tertiary and quaternary structures (2, 21, 55). Consequently, observations made for the rRNA of one species of bacterium can invariably be extrapolated to other species. The results reported here show that single-site mutations in domain II of E. coli 23S rRNA confer only minor levels of ketolide resistance and that the combination of these mutations with the 2058G mutation abolishes the ketolide-resistant phenotype conferred by 2058G. From the present set of data, there is no indication that mutations in domain II of 23S rRNA will come to pose a clinical problem by conferring telithromycin resistance to bacterial pathogens. However, as bacteria display tremendous resourcefulness in their ability to alter the rRNA conformation and attain macrolide resistance (10, 12, 16), it would be prudent to monitor pathogens continually for the emergence of new resistance mechanisms.
Acknowledgments
We thank Birte Vester for discussions and for generously providing plasmids with the 2058G mutation.
We gratefully acknowledge support from the Danish Biotechnology Instrument Centre, the Danish Natural Sciences Research Council, the European Commission's Fifth Framework Program (grant QLK2-CT2000-00935), and the Nucleic Acid Centre of the Danish Grundforskningsfond.
REFERENCES
- 1.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 3.Berisio, R., J. Harms, F. Schluenzen, R. Zarivach, H. A. Hansen, P. Fucini, and A. Yonath. 2003. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryskier, A. 2000. Ketolides—telithromycin, an example of a new class of antibacterial agents. Clin. Microbiol. Infect. 6:661-669. [DOI] [PubMed] [Google Scholar]
- 5.Bryskier, A. J., J. P. Butzler, H. C. Neu, and P. M. Tulkens. 1993. Macrolides: chemistry, pharmacology and clinical uses. Arnette Blackwell, Paris, France.
- 6.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douthwaite, S., and C. Aagaard. 1993. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23S rRNA peptidyl transferase loop. J. Mol. Biol. 232:725-731. [DOI] [PubMed] [Google Scholar]
- 9.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, D. J., S. Douthwaite, I. Morrissey, S. Bakker, J. Poehlsgaard, L. Jakobsen, and D. Felmingham. 2003. Macrolide resistance by ribosomal mutation in clinical isolates of Streptococcus pneumoniae from the PROTEKT 1999-2000 study. Antimicrob. Agents Chemother. 47:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, D. J., I. Morrissey, S. Bakker, and D. Felmingham. 2002. Molecular characterization of macrolide resistance mechanisms among Streptococcus pneumoniae and Streptococcus pyogenes isolated from the PROTEKT 1999-2000 study. J. Antimicrob. Chemother. 50(Suppl. S1):39-47. [DOI] [PubMed] [Google Scholar]
- 12.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8:181-188. [DOI] [PubMed] [Google Scholar]
- 13.Gale, E. F., E. Cundliffe, P. E. Reynolds, M. H. Richmond, and M. J. Waring. 1981. The molecular basis of antibiotic action. John Wiley & Sons, London, United Kingdom.
- 14.Garza-Ramos, G., L. Xiong, P. Zhong, and A. Mankin. 2001. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183:6898-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66:679-716. [DOI] [PubMed] [Google Scholar]
- 16.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 17.Gustafsson, C., and B. C. Persson. 1998. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutell, R. R., N. Larsen, and C. R. Woese. 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 58:10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623-631. [DOI] [PubMed] [Google Scholar]
- 21.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 24.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibilities to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jalava, J., J. Kataja, H. Seppala, and P. Huovinen. 2001. In vitro activities of the novel ketolide telithromycin (HMR 3647) against erythromycin-resistant Streptococcus species. Antimicrob. Agents Chemother. 45:789-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlov, R. S., T. M. Bogdanovitch, P. C. Appelbaum, L. Ednie, L. S. Stratchounski, M. R. Jacobs, and B. Bozdogan. 2002. Antistreptococcal activity of telithromycin compared with seven other drugs in relation to macrolide resistance mechanisms in Russia. Antimicrob. Agents Chemother. 46:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclercq, R. 2001. Overcoming antimicrobial resistance: profile of a new ketolide antibacterial, telithromycin. J. Antimicrob. Chemother. 48(Suppl. B):9-23. [DOI] [PubMed] [Google Scholar]
- 28.Liu, M., and S. Douthwaite. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, M., and S. Douthwaite. 2002. Methylation at nucleotide G745 or G748 in 23S rRNA distinguishes gram-negative from gram-positive bacteria. Mol. Microbiol. 44:195-204. [DOI] [PubMed] [Google Scholar]
- 30.Liu, M., and S. Douthwaite. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl. Acad. Sci. USA 99:14658-14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 32.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 33.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 34.O'Connor, M., W. M. Lee, A. Mankad, C. L. Squires, and A. E. Dahlberg. 2001. Mutagenesis of the peptidyltransferase center of 23S rRNA: the invariant U2449 is dispensable. Nucleic Acids Res. 29:710-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odenholt, I., E. Lowdin, and O. Cars. 2001. Pharmacodynamics of telithromycin in vitro against respiratory tract pathogens. Antimicrob. Agents Chemother. 45:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pankuch, G. A., M. A. Visalli, M. R. Jacobs, and P. C. Appelbaum. 1998. Susceptibilities of penicillin- and erythromycin-susceptible and -resistant pneumococci to HMR 3647 (RU 66647), a new ketolide, compared with susceptibilities to 17 other agents. Antimicrob. Agents Chemother. 42:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poehlsgaard, J., and S. Douthwaite. 2003. Macrolide antibiotic interaction and resistance on the bacterial ribosome. Curr. Opin. Investig. Drugs 4:140-148. [PubMed] [Google Scholar]
- 38.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Correa, D., and A. E. Dahlberg. 2004. Genetic evidence against the 16S ribosomal RNA helix 27 conformational switch model. RNA 10:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 42.Sekiguchi, M., and S. Iida. 1967. Mutants of Escherichia coli permeable to actinomycin. Proc. Natl. Acad. Sci. USA 58:2315-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigmund, C. D., M. Ettayebi, A. Borden, and E. A. Morgan. 1988. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 164:673-690. [DOI] [PubMed] [Google Scholar]
- 44.Skinner, R., E. Cundliffe, and F. J. Schmidt. 1983. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 258:12702-12706. [PubMed] [Google Scholar]
- 45.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 46.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenson, T., M. Lovmar, and M. Ehrenberg. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005-1014. [DOI] [PubMed] [Google Scholar]
- 48.Vázquez, D. 1979. Inhibitors of protein biosynthesis. Springer-Verlag, Berlin, Germany.
- 49.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vester, B., and R. A. Garrett. 1987. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie 69:891-900. [DOI] [PubMed] [Google Scholar]
- 51.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]