Abstract
Regulatory authorities have indicated that new drugs to treat type 2 diabetes (T2D) should not be associated with an unacceptable increase in cardiovascular risk. Human genetics may be able to inform development of antidiabetic therapies by predicting cardiovascular and other health endpoints. We therefore investigated the association of variants in 6 genes that encode drug targets for obesity or T2D with a range of metabolic traits in up to 11,806 individuals by targeted exome sequencing, and follow-up in 39,979 individuals by targeted genotyping, with additional in silico follow up in consortia. We used these data to first compare associations of variants in genes encoding drug targets with the effects of pharmacological manipulation of those targets in clinical trials. We then tested the association those variants with disease outcomes, including coronary heart disease, to predict cardiovascular safety of these agents. A low-frequency missense variant (Ala316Thr;rs10305492) in the gene encoding glucagon-like peptide-1 receptor (GLP1R), the target of GLP1R agonists, was associated with lower fasting glucose and lower T2D risk, consistent with GLP1R agonist therapies. The minor allele was also associated with protection against heart disease, thus providing evidence that GLP1R agonists are not likely to be associated with an unacceptable increase in cardiovascular risk. Our results provide an encouraging signal that these agents may be associated with benefit, a question currently being addressed in randomised controlled trials. Genetic variants associated with metabolic traits and multiple disease outcomes can be used to validate therapeutic targets at an early stage in the drug development process.
Introduction
In 2008, the US Food and Drug Administration issued guidance for industry on new therapies to treat type 2 diabetes (T2D), recommending that sponsors should demonstrate that these treatments are “not associated with an unacceptable increase in cardiovascular risk” (1). This mandate challenges drug developers to prove safety during clinical trials, which is an expensive and late-phase strategy for the identification of such concerns. Instead, genetic approaches may aid in the identification of possible drug side-effects much earlier in the drug development process. Genetic variants can inform the treatment and prevention of human disease (2, 3), either reducing the prioritisation of potential targets (4, 5) or implicating new targets (6, 7). Functional exonic variants can be useful surrogates for drug effects, when, for example, a loss-of-function (LoF) variant may be a useful tool to understand the consequences of pharmacological inhibition of a particular target protein (7). Recent sequencing efforts have identified a large number of potentially functional low-frequency and rare exonic variants in human populations, even among genes under purifying selection (8–12). Although such variants may influence susceptibility to disease, the high cost of these sequencing approaches has previously meant that they have not been performed in the sample sizes required to allow routine investigation of their association with complex disease and related traits.
A recent targeted exome sequencing study of 202 genes encoding potential drug targets identified an abundance of potentially functional exonic variants (8). Among these 202 genes, six genes encoding drug targets licensed or in development by GlaxoSmithKline (GSK) for treatment of obesity and/or T2D were included. Recognizing that these data could be used to test for genetic variants mimicking pharmacological manipulation of the encoded protein (drug target), we investigated six genes encoding targets of relevance to obesity and T2D. These variants could then serve as tools to aid the broader evaluation of drug-related risk for adverse events mediated via on-target effects.
As a proof of concept for use of genetic variants to evaluate the cardiovascular safety of anti-diabetic agents, we evaluated the widely used glucose-lowering glucagon-like peptide-1 receptor (GLP1R) agonists (13). These agents are long-acting mimetics of the incretin hormone GLP1, which increases insulin secretion after oral consumption of glucose but not after glucose administered intravenously. There are uncertainties over the role of these agents in the aetiology of rare, adverse pancreatic events that have been reported following their usage (14). These therapies have been associated with weight loss (15) and reduced cardiovascular risk factors, and while a recent trial reported non-inferiority of GLP1R-agonists in cardiovascular safety(16), multiple trials evaluating cardiovascular safety have not yet been completed (17). We used a genetic variant in GLP1R that is associated with variation in fasting glucose levels and with T2D risk (18) to evaluate the cardiovascular safety of GLP1R agonists. The low-frequency variant protective for T2D was also protective for coronary heart disease (CHD). These findings support the notion that GLP1R agonists will not confer an increased cardiovascular risk in people. This study also demonstrates how genetic target validation approaches can be employed early in the drug development process to evaluate efficacy and safety.
Results
Association of genetic variants in genes encoding T2D and obesity drug targets
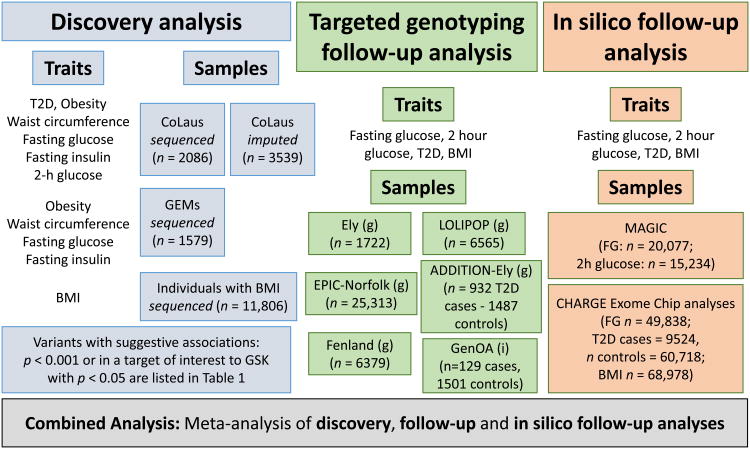
The study design consisted of initial discovery of variants with suggestive associations, to targeted genotyping and in silico follow-up analyses (Fig. 1). We investigated the association of 121 variants in six genes encoding therapeutic targets in use or in development for T2D or obesity (CNR2, DPP4, GLP1R, SLC5A1, HTR2C, MCHR1)—drawn from a recent targeted exome sequencing study of 202 genes encoding drug targets (8)—with variation in the following traits: T2D, obesity, body-mass index (BMI), waist circumference, fasting glucose, fasting insulin, and 2-h glucose (Fig. 1). In the “Discovery Analysis”, we identified seven variants potentially associated with T2D- or obesity-related traits (where p<0.001, or which were in a target of interest to GSK and p<0.05) (Table 1). For these seven variants, “Follow-Up Analysis” was performed by targeted genotyping in up to 39,979 additional individuals of European ancestry. Where possible, in silico follow-up analysis was performed for traits and variants available in large-scale genetic consortia data.
Figure 1. Overall study design, participating studies, and consortia.
Discovery analyses were performed using targeted exome sequencing of variation in six genes tested for association with seven traits. Variants were taken forward to follow-up by targeted genotyping. Additional in silico results were obtained using available association results. Combined results were obtained by fixed-effect meta-analysis of estimates from linear or logistic regression, as appropriate. Based on the 1331 statistical tests performed in discovery analyses, p<3.8×10-5 was used as the threshold for statistical significance. In targeted genotyping, (g) refers to studies that were directly genotyped for relevant markers, whereas (i) indicates those in which relevant variants were captured by imputation.
Table 1. Discovery, follow-up, and combined results for variants taken forward to follow-up.
Seven variants in six genes reached p<0.001 (or p<0.05 in target of interest to GSK) in sequence-based discovery analyses (Fig. 1) and were taken forward to follow-up in additional samples, by targeted genotyping and by in silico lookup from existing consortia. Data and P values are from fixed effect meta-analysis of linear regression for quantitative traits or logistic regression for binary disease status.
| Gene | Variant | Chr | Position (NCBI b37 genome alignment) |
Consequence | Trait | Effect allele |
Other allele |
MAF | Stage | Study |
n (case/control for binary trait) |
Beta (Odds ratio for binary trait) |
se [CI for OR] |
P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GLP1R | rs10305492 | 6 | 39046794 | A316T | Fasting glucose | A | G | 0.015 | Discovery | Sequenced CoLaus* | 1869 | -0.28 | 0.14 | 0.04 |
| Targeted follow-up | Additional CoLaus, ELY, Fenland, LOLIPOP, GEMS | 18,937 | -0.13 | 0.04 | 1.5×10-3 | |||||||||
| In silico follow-up | MAGIC (29) | 20,077 | -0.16 | 0.03 | 1.1×10-7- | |||||||||
| Combined | 40,883 | -0.15 | 0.02 | 2.6×10-10 | ||||||||||
|
| ||||||||||||||
| DPP4 | rs56179129 | 2 | 162890142 | V266I | Fasting glucose | T | C | 0.008 | Discovery | GEMS | 1416 | 0.61 | 0.21 | 3.6×10-3 |
| Targeted follow-up | CoLaus, ELY, LOLIPOP | 12934 | 0.00 | 0.07 | 0.95 | |||||||||
| In silico follow-up | CHARGE Exome chip (18) | 49838 | 0.00 | 0.03 | 0.16 | |||||||||
| Combined | 64188 | 0.01 | 0.03 | 0.71 | ||||||||||
|
| ||||||||||||||
| SLC5A1 | rs200410750 | 22 | 32439209 | 5′ UTR | Fasting Glucose | T | C | 0.001 | Discovery | Sequenced and imputed CoLaus | 5210 | 1.44 | 0.33 | 1.7×10-5 |
| Targeted follow-up | ELY, Fenland, LOLIPOP | 12707 | -0.16 | 0.27 | 0.56 | |||||||||
| In silico follow-up | n/a | NA | ||||||||||||
| Combined | 18059 | 0.51 | 0.19 | 0.01 | ||||||||||
|
| ||||||||||||||
| CNR2 | rs4649124 | 1 | 24201357 | Synonymous | 2 hour glucose | A | G | 0.420 | Discovery | Sequenced and imputed CoLaus | 505 | 0.18 | 0.06 | 0.01 |
| Targeted follow-up | ELY, Fenland | 6377 | 0.00 | 0.02 | 0.95 | |||||||||
| In silico follow-up | MAGIC (proxy: rs10917431)(49) | 15234 | -0.01 | 0.01 | 0.49 | |||||||||
| Combined | 22106 | 0.00 | 0.01 | 0.88 | ||||||||||
|
| ||||||||||||||
| CNR2 | rs2229579 | 1 | 24201162 | H316Y | T2D | T | C | 0.110 | Discovery | Sequenced and imputed CoLaus | 385/5241 | 0.73 | [0.55,0.97] | 0.03 |
| Targeted follow-up | ADDITION-Ely, NDS, LOLIPOP, GenOA | 7141/27096 | 1.06 | [0.99, 1.14] | 0.07 | |||||||||
| In silico follow-up | CHARGE Exome chip (18) | 9524/60718 | 0.96 | [0.90, 1.01] | 0.10 | |||||||||
| Combined | 17047/93225 | 0.99 | [0.95, 1.04] | 0.67 | ||||||||||
|
| ||||||||||||||
| HTR2C | rs56372597 | X | 113951968 | Intronic | BMI | A | G | 0.150 | Discovery | BMI | 10798 | 0.05 | 0.02 | 2.1×10-3 |
| Targeted follow-up | Additional CoLaus, ELY, EPIC, Fenland, LOLIPOP | 36983 | 0.00 | 0.01 | 0.92 | |||||||||
| In silico follow-up | n/a | NA | ||||||||||||
| Combined | 47781 | 0.01 | 0.01 | 0.13 | ||||||||||
|
| ||||||||||||||
| MCHR1 | rs117372135 | 22 | 41075523 | T25M | BMI | T | C | 0.002 | Discovery | BMI | 10952 | 0.62 | 0.15 | 4.5×10-5 |
| Targeted follow-up | Additional CoLaus, ELY, EPIC, Fenland, LOLIPOP | 37240 | 0.08 | 0.10 | 0.40 | |||||||||
| In silico follow-up | CHARGE adiposity Exome chip working group | 68978 | -0.04 | 0.07 | 0.59 | |||||||||
| Combined | 117170 | 0.08 | 0.05 | 0.13 | ||||||||||
Analyzed in sequenced CoLaus participants only owing to low imputation quality (R2 < 0.5) in additional CoLaus participants at the discovery stage. #Not analyzed in GEMS due to low number of carriers (< 5 minor alleles)
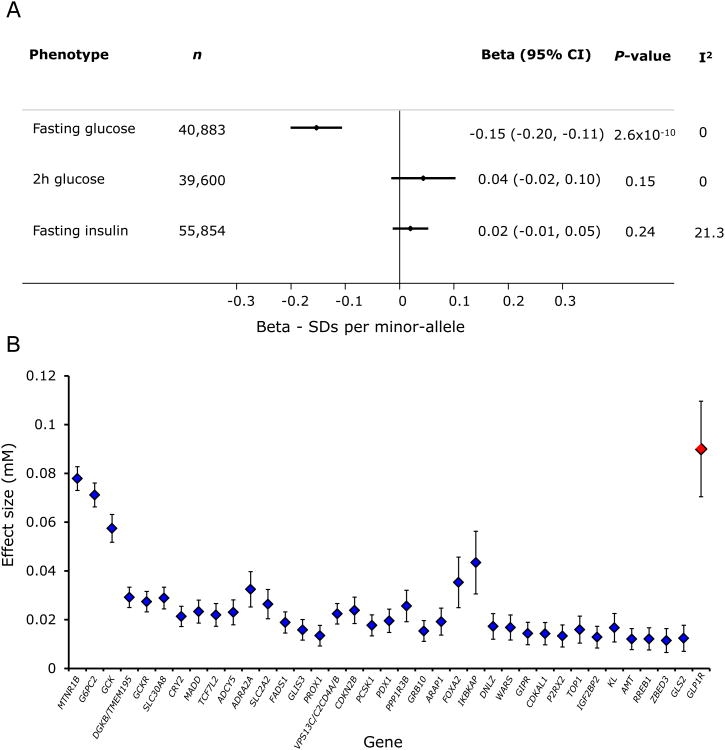
Initial discovery analyses included 1331 tests of association, with the threshold specified to reach significance in combined analyses being p<3.8×10-5. In a combined analysis of results from the different phases, we identified a low-frequency (∼1% minor allele frequency (MAF)) missense variant Ala316Thr; rs10305492 in the GLP1R gene to be associated with fasting glucose (Fig. 2A). The variant was in Hardy-Weinberg equilibrium in all genotyped samples (p > 0.2). The effect size (i.e. the difference per allele) of 0.09 mM was larger than most common variants previously reported for fasting glucose (Fig. 2B), and was recently found to be associated with fasting glucose in non-overlapping samples from large-scale analyses of coding variant associations with glycaemic traits (18). The combined analysis of the six other variants in Table 1 did not show evidence of association (p>3.8×10-5, by linear or logistic regression) with the suggestively associated trait in the discovery analysis (“Follow-up” p-values > 0.05; “Combined” p-values ≥ 0.005; Table 1).
Figure 2. Association of GLP1R variant (rs10305492) with glycaemic traits.
(A) Genetic variant association with glycaemic traits. Data are standard deviations per minor allele at rs10305492. Fasting glucose results are from the combined analysis (Table 1). Individual studies contributing to the associations for fasting insulin and 2-h glucose are in table S4. All results reflect point estimates and 95% confidence intervals (CI) from a fixed-effect meta-analysis of linear regression estimates. (B) Effect size of the GLP1R variant (in red) and loci previously reported to be associated with fasting glucose. Effect sizes are reported from discovery analyses of available MAGIC results (50), and from the combined estimate for the GLP1R variant in (A).
The GLP1R gene encodes the GLP1 receptor, the target for GLP-1, a hormone that mediates the augmented response to insulin secretion following oral glucose administration. This receptor is the target for the GLP1R-agonist class of T2D therapeutics and the association of this variant with fasting glucose mimicked a major effect of this class of agents. To further corroborate the utility of this variant as a surrogate indicator of pharmacological modulation of the receptor, we investigated its association with T2D and found that the minor allele was associated with lower risk of T2D [odds ratio (OR) = 0.83 [0.76, 0.91]; P = 9.4×10-5; in a fixed effect meta-analysis of log-odds ratios from studies and consortia listed in table S1 and in Supplementary Materials “Studies contributing to follow-up analyses of type-2 diabetes and obesity related traits”; ncases = 25,868, ncontrols = 122,393]. However, we saw no association of this GLP1R variant (Ala316Thr; rs10305492) with fasting insulin, nor with 2-h glucose (Fig. 2A).
Although there were no individuals carrying putative LoF variants in GLP1R in the targeted sequencing study, a single individual in the cohort-arm of the UK10K study had a LoF allele (W297*) but did not have an extreme glycaemic phenotype. This individual's fasting glucose and insulin concentrations were within the range of 95% of the values for this population. Nine high-confidence LoF variants in GLP1R were observed in the ExAC database (19). Eight were singletons and the most common had a frequency of less than 1/10,000, highlighting the difficulty in restricting analyses to individual LoF variants.
Association of GLP1R variant with quantitative traits and comparison with effects observed in clinical trials of GLP1R agonists
To further characterise the extent to which the GLP1R variant associations mirrored the effects of GLP1R-agonist therapy, we compared genetic associations to the metabolic effects observed in previously reported clinical trials (Fig. 3, table S2). GLP1R agonist therapy can result in lower fasting and post-challenge glucose, weight loss, a reduction in systolic blood pressure, reduced total- and LDL-cholesterol and an increase in resting heart rate. The effects of GLP1R-agonists on glycaemic measures (fasting glucose and 2-h glucose) were stronger than those on non-glycaemic factors (Fig. 3), which have been detectable only in some meta-analyses of clinical trials (20–23).
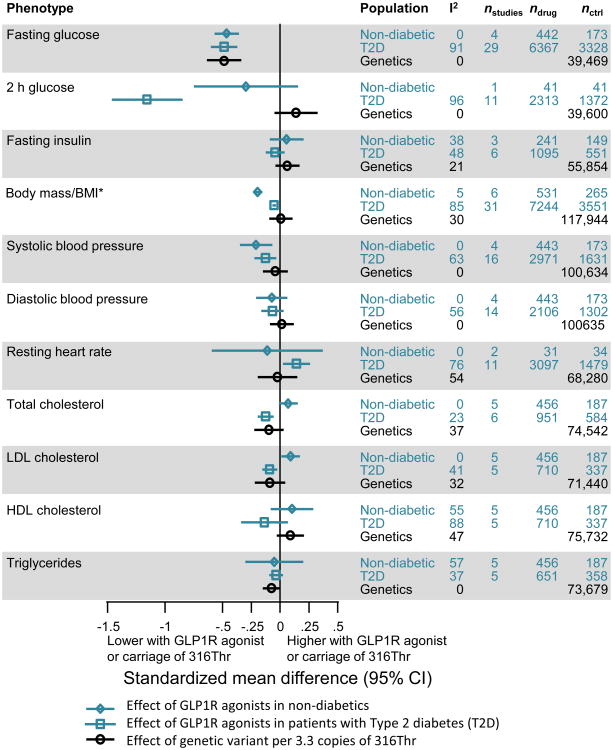
Figure 3. Comparison of GLP1R variant (rs10305492) associations with effects observed in clinical trials of GLP1R agonists in non-diabetic individuals and in individuals with T2D.
Genetic associations are all scaled to match the effects of GLP1R-agonists on fasting glucose (i.e. per 3.3 copies of the minor (A) allele). Genetic variant results are beta estimates and 95% confidence intervals from fixed effect meta-analysis of linear regression results Trial results are estimates from fixed-effect meta-analyses of standardised mean differences between treatment and comparison groups of the individual trials listed in table S3. *Trials reported effects on body mass, whereas genetic associations were only available for BMI.
Using fasting glucose as the benchmark, the per-allele association of the genetic variant with glucose (-0.15 SDs [-0.20,-0.11], from Fig. 2) was 3.3-fold weaker than the effect observed for GLP1R-agonist treatment (-0.49 [-0.60,-0.37], from Fig. 3). We therefore rescaled the genetic associations to account for this difference, by multiplying the magnitude of all observed genetic associations by 3.3 (Fig. 3), and demonstrated that there was little difference between the magnitude of association of the GLP1R variant and effects observed in clinical trials beyond that expected by chance (α=0.0025). An exception to this observation was the impact of GLP1R agonist therapy on weight in non-diabetic individuals when compared to the observed association between the variant and BMI (p=2.6×10-4, Cochrane's Q test) (table S2). The genetic variant was not associated with BMI (Fig. 3), whereas the agonist therapy caused a reduction in body mass in non-diabetic individuals but not in individuals with T2D (fig. S1, table S2). However, five of the six trials in non-diabetic individuals were performed in obese participants (table S3), whose higher starting weight may have enabled a greater weight loss.
GLP1R agonists appeared to have a greater effect on 2-h glucose than the magnitude of association observed for the variant (p=2.1×10-12, Cochrane's Q test) (Fig. 3; fig. S2; table S2). The difference was most pronounced in comparison to trials in individuals with T2D, among whom we observed heterogeneity in the effect of GLP1R agonists on 2-h glucose, even within drug class (I2=97%) (fig. S2B). There was no significant difference between the magnitude of genetic association and the impact of GLP1R agonist therapy on 2-h glucose in non-diabetic individuals (Fig. 3; table S2), although the number of people included in such trials was much smaller than in trials including individuals with T2D (table S3).
Association of GLP1R variant with disease outcomes
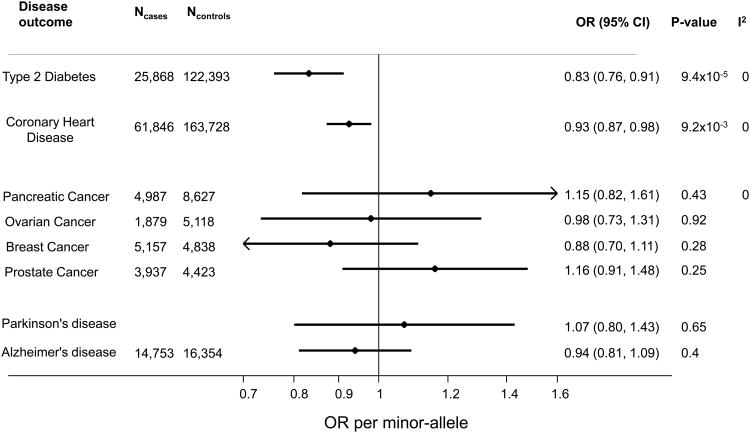
Our final aim was to describe the association of the GLP1R variant with CHD and other outcomes. In a large-scale international collaboration, we studied 61,846 individuals with CHD and 163,728 controls, and found that the fasting glucose–lowering allele of GLP1R was associated with protection against CHD (Fig. 4). The association with CHD is greater than the 1% reduction in risk that would be predicted based on the association of this variant with fasting glucose alone (24) (see Supplementary Methods on “Calculating the reduction in coronary heart disease risk attributable to lower fasting glucose levels”), suggesting that lowering of fasting glucose alone is unlikely to explain the observed association between the GLP1R variant and lower risk of CHD. Although not significant, carriage of the minor allele was associated with lower LDL cholesterol, triglycerides, systolic blood pressure, and higher HDL cholesterol.
Figure 4. Association of GLP1R variant (rs10305492) with disease outcomes.
Association with disease outcomes are reported per-minor allele at rs10305492. Data show odds ratios and 95% confidence intervals from logistic regression models.
Using data from international consortia, we found no evidence for association of the GLP1R variant with pancreatic cancer, although the confidence intervals were wide owing to the comparatively small sample size (4987 cases and 8627 controls) and low frequency of the allele (Fig. 4). There was no evidence of association with breast, ovarian, or prostate cancer risk. Given the interest in GLP1R agonist therapy for neurological disease, including Parkinson's (25) and Alzheimer's (26), we also investigated the association of the GLP1R variant with those diseases, but found no evidence of association (Fig. 4).
Discussion
Anticipating the side effects of drugs prior to phase III clinical trials could support drug discovery and development, reducing attrition rates and saving considerable time and money. The promise of human genetics in this endeavour (2, 3, 7, 27) depends on the availability of genetic variants that mimic pharmaceutical interventions. We undertook a systematic study to identify such genetic variants in the context of diabetes and obesity, and identified an association between fasting glucose and T2D with a missense variant in GLP1R, the gene encoding the GLP-1 receptor—the target of the GLP1R agonist class of T2D therapies. Regulatory authorities require evidence that therapies for T2D are not associated with unacceptable increases in cardiovascular risk. The reduced risk associated with the glucose-lowering genetic variant in GLP1R provides evidence that not only will GLP1R agonists meet this regulatory hurdle, but they may also reduce CHD events. Ongoing trials of GLP1R agonists are designed to resolve this uncertainty and will also augment the evidence on the broader validity of genetic approaches in drug-target validation.
A key consideration in assessing whether genetic variants can be used to understand therapeutic effects is how well the genetic variant mirrors the effects of pharmacological intervention at the same target. Genetic association data, here and reported previously (18), suggest that lifelong carriage of the minor GLP1R allele (at rs10305492) is associated with lower fasting glucose and lower risk of T2D, although not with 2-h glucose. Clinical trial data from individuals with T2D, who may have a diminished incretin effect, show that GLP1R agonists lower 2-h glucose considerably (28), whereas the effect on 2-h glucose is smaller in individuals without T2D (29), presumably because non-diabetic individuals are less likely to have an impaired incretin effect requiring therapeutic correction. Similarly, GLP1R agonists were associated with greater weight loss in obese individuals than in non-obese. Such a phenomenon has previously been suggested for the effects of GLP1R-agonism on blood pressure, where GLP1R-agonist therapy appears to lower blood pressure in individuals with high blood pressure but not in non-hypertensive individuals (30, 31). This highlights a limitation in the use of genetic variants in target validation: that the association of genetic variants is often tested in individuals of “normal” physiology, whereas clinical trials are generally performed in individuals with prevalent disease.
An important step in evaluating the utility of genomics in target validation is to understand the functional consequences of variants. For potential novel targets, whether the variant confers gain or loss of function informs the development of either an agonist or antagonist therapy. For example, LoF variants have been used to understand the consequences of antagonism of a novel drug target (7, 32). However, researchers have gained insights using variants validated as instruments when their phenotypic associations mirrored pharmacological action, even in the absence of strong functional insights into the mechanism of those variants (33). GLP1R-agonist therapy reduces fasting glucose in humans, as does administration of GLP1, regardless of the duration or severity of T2D (34). In mice, the loss of GLP1R leads to fasting hyperglycaemia (35, 36). Together, these findings in humans and in mice suggest that the glucose-lowering minor allele at rs10305492 confers gain of function. However, differences in basal activity of the human and murine GLP1R (37) limit our ability to extrapolate findings from GLP1R knockout mice to humans (15, 32). Previous attempts to characterise the effect of this variant in cellular models have been inconclusive (38, 39). The rarity of putative LoF alleles in the GLP1R impaired our ability to restrict analyses to such variants. Although the absence of definitive functional characterisation is a limitation of this study, our observation that the minor allele is strongly associated with lower fasting glucose levels and is protective against T2D supports the validity of the variant as a genetic instrument for GLP1R-agonist therapy. Future integration of large-scale human genetic data with functional characterisation in appropriate cell models will allow broader application of variants, other than those characterised as LoF, in target validation.
Although the GLP1R variant was not associated with any of the other non-glycaemic or quantitative cardiovascular parameters, there was insufficient evidence to suggest the genetic associations and pharmacological effects were different. Power calculations indicated that, to detect the expected association with systolic blood pressure or resting heart rate, a sample size of more than 250,000 individuals would be required. This is considerably larger than most current genetic consortia, although this limitation could soon be overcome as larger studies become available (40), further strengthening the promise of genomics in target validation. Although we did not observe overall evidence for association of variants other than the GLP1R variant, the discovery phase, from which we selected variants for follow-up, was relatively small in comparison to the overall sample and there remains a possibility of type II error in the discovery phase. As larger resources of genetic data become available, these potential concerns will also be reduced.
The detection of rare adverse effects of a drug remains a challenge. Pharmaco-epidemiological approaches using routine database analysis may identify rare adverse outcomes associated with treatment, but the approach is rarely conclusive because of confounding, particularly by indication. Our demonstration that the GLP1R variant is not associated with pancreatic, breast, prostate, or ovarian cancer, or with Parkinson's or Alzheimer's disease is limited by the upper bounds of the confidence intervals, which are too high to allow strong inference about the likely long-term safety of GLP1R agonists with regard to these outcomes. Although these data represent the largest resources available globally, the accumulation of studies with greater numbers of individuals with genetic data and robust disease outcome classification will considerably enhance the potential of this type of investigation. The comparisons of other traits and disease outcomes, beyond the primary indications, makes the assumption that pharmacological effects are mediated via “on-target” effects and not “off-target” effects (i.e. those mediated by effects of the agent on other non-specific targets). Although our results offer insight into the effects of GLP1R agonists, they do not necessarily apply to other agents targeting the incretin pathway through different mechanisms, such as by DPP-4 inhibition (41).
In conclusion, through a targeted exome sequencing approach, we identified that a low-frequency missense variant in GLP1R was associated with lower fasting glucose and risk of T2D, similar to the effects of GLP1R agonist therapy. This variant was also associated with lower risk of CHD, thus providing supportive evidence that these agents are not likely to be associated with an unacceptable increase in cardiovascular risk and may indeed be associated with benefit, a question currently being addressed in randomised controlled trials. We propose that future drug development and investment decisions could be informed by genomic data much earlier in the development process, providing insight into both efficacy and side-effects.
Methods
Study design
We studied six genes encoding therapeutic targets licensed or in development for obesity or T2D (CNR2, DPP4, GLP1R, SLC5A1, HTR2C, MCHR1), drawn from a recent targeted exome sequencing study of 202 genes encoding drug targets (8), which represented approximately 1% of the coding genome and 7% of all genes considered current or potential drug targets (8). In the “Discovery Analysis”, we investigated the association of common and rare variants in these six genes with seven T2D and obesity-related traits (Fig. 1). We analysed all variants which had i) MAF ≥ 0.5% or well imputed (R2 > 0.5) in CoLaus; ii) MAF ≥ 0.5% in GEMS; or iii) MAF ≥ 0.1% in BMI (given the larger sample size) in the CoLaus study (42), the GEMS study (43), or all individuals with BMI measurements. We examined 121 variants for association with six traits in the CoLaus study (6*121 = 726 tests); 4 traits in GEMS (4*121 = 484 tests); and 1 trait in the BMI study, comprising a total of 1331 tests of association. First, we analysed a subset of the population-based CoLaus study (n=2086) for T2D, obesity, waist circumference, fasting glucose, fasting insulin, and 2-h glucose traits. Second, in the GEMs dyslipidaemic case and normolipidaemic control study (ncases=787, ncontrols=792), we analysed obesity, waist circumference, fasting glucose, and fasting insulin traits. We performed discovery analyses in the CoLaus and GEMS studies separately due to the different study designs and traits analysed in attempt to maximise sensitivity to detect associations that might be masked by context-dependent associations. Third, BMI measures were available in a larger sample size from 11 studies (Fig. 1), and were analysed together. We provide the sample sizes for the discovery analyses in Figure 1 and trait-specific sample sizes in Table 1 (n = 505 – 11,806). We augmented the sequence data for the CoLaus study with imputed data in the remainder of the study (n=3539) where variants were imputable (R2>0.5) using a custom imputation process on individuals genotyped on the Affymetrix 500K chip but not included in the targeted sequencing experiment (Supplementary Materials).
Using results from the discovery analyses, we identified variants that were associated with T2D or obesity-related traits at the p<0.001 level or were located in genes encoding targets of strategic interest to GSK, including GLP1R, DPP4, CNR2, and HTR2C with a p value threshold of <0.05. To maximise sensitivity to detect associations in these genes of highest interest, we took forward to follow up those variants reaching p<0.05 in the discovery analyses. However, this did not affect the threshold for statistical significance or overall alpha value (3.8×10-5), for which we accounted for all association tests performed in the discovery analyses (N=1331). The principal reason for prioritising specific genes was to ensure a balance between sensitivity for targets of high priority to GSK and to maintain specificity: given that initial replication was performed by de novo large-scale targeted genotyping, we were practically unable to follow up vast numbers of variants. This does not bias the variants selected for follow-up, nor raise the risk of type I error. Indeed, the only variant we determined to be mimicking pharmacological manipulation was well beyond “genome-wide significance” even if all possible low-frequency and common variants in the genome had been tested.
We then genotyped seven variants in six genes in up to 39,979 follow-up participants of European ancestry drawn from multiple studies (Fig. 1): CoLaus (when GEMS was the discovery sample), GEMS (when CoLaus was the discovery set), Ely (44) (n=1722), EPIC-Norfolk (45) (n=25313), Fenland (46) (n=6379), and LOLIPOP (47) (n=6565) studies. The follow-up analysis of T2D included participants from the Norfolk Diabetes Study (Ncases=5587 and Ncontrols=19012), GenOA study (Ncases=129 and Ncontrols=1501), and individuals with T2D from the ADDITION study(48)(Ncases=816) who were combined with additional cases from the Ely study (Ncases=116) and compared to non-diabetic controls from the Ely study (Ncontrols=1487).
We also sought further in silico follow-up Analysis to further evaluate associations in collaborative studies utilizing results from the MAGIC and CHARGE consortia. Five of the seven variants were available for in silico analysis (table 1). Further details on each of the studies and consortia are provided in the Supplementary Materials and table S1 and S4.
Statistical analyses
We carried out genetic association analyses on variants identified via targeted sequencing using an additive genetic model by linear or logistic regression, adjusting for age and sex and other study-specific covariates. We combined study-specific estimates using fixed effect meta-analysis. We performed analyses on standardised variables (mean=0 and SD =1) and, as such, expressed effect sizes as standard deviations (SDs) for quantitative traits. In total, we analysed 121 single nucleotide variants (SNVs). Overall, we performed 1,331 tests of association in the discovery analyses and, as such, associations that were α<3.8×10-5 in the combined analysis were deemed to be statistically significant.
We performed targeted genotyping of selected variants from discovery analyses using Sequenom for the Ely, EPIC-Norfolk, Fenland, and ADDITION studies and KASPar for the LOLIPOP study. Imputed data were also available in the GenOA study using reference haplotypes from participants in the previous sequencing study (8). We carried out genetic association analyses in each study under an additive genetic model using linear or logistic regression, again adjusting for age, sex and study-specific covariates. We sought further in silico follow-up from summary association results from MAGIC and CHARGE consortia (table 1). We converted summary association result effect sizes to SDs using the SD of fasting glucose from the population-based Fenland study (SD = 0.65 mM) (46). We meta-analyzed results from the discovery analysis, follow-up analysis and in silico follow-up analysis using a fixed effect, inverse-variance weighted approach. The discovery analysis of the CoLaus study included association results from the sequence variants and imputed variants (table 1). In the entire CoLaus study, we later directly genotyped (KASPar technology) variants which had been imputed in the unsequenced CoLaus participants study as part of the original follow-up analysis. The combined analysis results in table 1 therefore represent those from the directly genotyped data.
For variants that showed statistically significant associations in the combined analysis (p<3.8×10-5), we investigated their association with a range of anthropometric, metabolic, and cardiovascular risk factors and disease outcomes in the studies described previously, as well as in additional studies described in table S1 and S4 and in Supplementary Materials. We also investigated the association of variants reaching statistical significance after follow up (α<3.8×10-5) with coronary heart disease (CHD) through targeted genotyping and collaboration with large-scale Exome chip consortia (table S1). For these variants, we also investigated association with a range of other disease outcomes (table S1), with a particular focus on diseases previously suggested as potential opportunities for repositioning (i.e. where existing drugs might be used for alternative indications). However, as the variant reaching statistical significance was not well covered on existing GWAS arrays or in HapMap, we were limited to those disease outcomes for which we could obtain association data. For genes that contained variants with p<3.8×10-5 in the combined analysis, we investigated the presence of putative LoF alleles in individuals in whom we had performed targeted sequencing (8) and in individuals with whole-genome sequencing from the UK10K study (www.uk10k.org).
Comparison of clinical trial effects and genetic associations
Randomized clinical trials of GLP1R-agonists were identified through previous systematic reviews and by performing a supplementary literature search, as detailed in the Supplementary Materials. Only trials with placebo or no-drug comparison groups (i.e. no trials with active comparison groups), with ≥ 4 weeks drug treatment (i.e. no single dose studies) and ≥ 10 participants per trial arm were included. Treatment effects were expressed in SDs before pooling across trials using random effects meta-analysis (see table S3 for details of clinical trials included). P-values derived from Cochrane's Q test were used as a guide to assess whether there were pairwise differences between the rescaled genetic and trial estimates.
Supplementary Material
Figure S1. Effects of GLP1R agonists on body weight.
Figure S2. Effects of GLP1R-agonists on 2h glucose.
Table S1. Study characteristics for disease traits.
Table S2. Comparison of heterogeneity between trial and rescaled genetic estimates.
Table S3. Details of randomized trials contributing to analyses of GLP1R-agonist effects included in Fig. 2
Table S4. Study characteristics for quantitative traits.
Acknowledgments
We are grateful for the contributions of the studies and consortia which provided summary results. For a complete list of Acknowledgements, see Supplementary Materials.
Funding: The work described in this manuscript was funded, in part, by GSK, and, in part, by the Medical Research Council (MC_UU_12015/1). CHARGE AD analyses were funded by R01 AG033193 (PI Seshadri). ADGC AD analyses were funded by UO1AG032984 (PI Schellenberg). Additional funding sources are outlined in the Supplementary Materials.
The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. Additional materials can be obtained by request to the corresponding authors and may be subject to MTA requirements.
Footnotes
Author contributions: RAS, MGE, NJW, DMW conceived and designed the study. RAS, DFF, LL, AYC, PS, RY, NG, AS, YC, TVV, HY, JAL, JHZ, SMW, JW, SW, NM, KM, AP, SJvdL, CG, AAAO, PA, LA, DA, IAO, BB, AB, IB, SBG, JCB, SB, MB, HB, EB, IBB, JBJ, SB, CC, MC, LAC, CC, JC, MdH, JAD, HME, GBE, EF, JF, TF, IF, NGF, FG, CG, SG, LH, JHJ, MEJ, JWJ, RK, FK, NDK, TJK, JK, ZKJ, ATK, KK, JK, AL, CL, GM, KLM, APM, KM, MMN, PBM, CN, SFN, PMN, BGN, CJP, DP, SP, GMP, MP, AP, CJP, JRQ, OR, CS, VS, MJS, NS, SJS, RS, NS, JAS, DJT, ST, RT, DLvdA, YTvdS, JV, MW, KW, JEA, LTA, JLA, ASB, JC, JD, DFE, RAE, JE, PWF, TMF, TH, JMMH, TJ, JK, ML, CL, MIM, JSP, OP, ER, JIR, DS, NJS, HS, PV, SOR, PD, JD, MOG, SK, JBM, MGE, NJW, DMW participated in the acquisition and/or analysis of data. LL, PS, JAL, JHZ, SJS and JMMH performed analyses and/or provided statistical guidance. IB, SB, MB, HB, EB, IBB, LAC, KLM, PBM, SJS, MW, LTA, JD, DFE, ML, CL, MIM, JSP, PV, SOR, PD, JD, MOG, SK, JBM, MGE, NJW, DMW supervised analyses. RAS, DFF, LL, MGE, NJW, DMW wrote the manuscript, with input from all authors.
Competing interests: J.L. A, M.G. E, L. L, and D.M. W are GSK stockholders. All named authors are solely responsible for the content of the manuscript. The current manuscript was a collaborative effort between academic and GSK scientists and analyses focused on a subset of genes included in the original sequencing paper (8), which focused on target genes of interest to GlaxoSmithKline. Since October 2015, D.F. has been a full time employee of Bayer AG, Germany.
Data and materials availability: Data on glycemic traits were contributed by MAGIC investigators and were downloaded from www.magicinvestigators.org. Associations with T2D in a GWAS of Europeans were obtained from The DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) investigators (http://diagram-consortium.org/downloads.html).
References and notes
- 1.US FDA. Guidance for Industry Diabetes Mellitus — Evaluating Cardiovascular Risk in New Antidiabetic Therapies to Treat Type 2 Diabetes. 2008 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm071627.pdf.
- 2.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 3.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 4.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart AFR, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett MS, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki ML, Perola M, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PIW, Klungel OH, Maitland-van der Zee AH, Peters BJM, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VHM, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WMM, a Boer JM, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, König IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox Ka, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schäfer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O'Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacho J, Tybjaerg-Hansen A, Jensen JS, Grande P, Sillesen H, Nordestgaard BG. Genetically elevated C-reactive protein and ischemic vascular disease. N Engl J Med. 2008;359:1897–1908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 7.Myocardial Infarction Genetics Consortium Investigators. Inactivating Mutations in NPC1L1 and Protection from Coronary Heart Disease. N Engl J Med. 2014 doi: 10.1056/NEJMoa1405386. 141112140016008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zöllner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–3. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, Mc Gee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson Da, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, Wheeler Da, Sabo A, Lusk C, Weiss KG, Akbar H, Cree A, Hawes AC, Newsham I, Varghese RT, Villasana D, Gross S, Joshi V, Santibanez J, Morgan M, Chang K, Iv WH, Templeton AR, Boerwinkle E, Gibbs R, Sing CF. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson Da, Bamshad MJ, Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovshin JA, Drucker DJ. Incretin-based therapies for type 2 diabetes mellitus. Nat Rev Endocrinol. 2009;5:262–9. doi: 10.1038/nrendo.2009.48. [DOI] [PubMed] [Google Scholar]
- 14.Cure P, Pileggi A, Alejandro R. Exenatide and rare adverse events. N Engl J Med. 2008;358:1969–70. doi: 10.1056/NEJMc0707137. discussion 1971–2. [DOI] [PubMed] [Google Scholar]
- 15.Astrup A, Rössner S, Van Gaal L, Rissanen A, Niskanen L, Al Hakim M, Madsen J, Rasmussen MF, Lean MEJ NN8022-1807 Study Group. Effects of liraglutide in the treatment of obesity: a randomised, double-blind, placebo-controlled study. Lancet (London, England) 2009;374:1606–16. doi: 10.1016/S0140-6736(09)61375-1. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer MA, Claggett B, Diaz R, Dickstein K, Gerstein HC, Køber LV, Lawson FC, Ping L, Wei X, Lewis EF, Maggioni AP, McMurray JJV, Probstfield JL, Riddle MC, Solomon SD, Tardif JC. Lixisenatide in Patients with Type 2 Diabetes and Acute Coronary Syndrome. N Engl J Med. 2015;373:2247–2257. doi: 10.1056/NEJMoa1509225. [DOI] [PubMed] [Google Scholar]
- 17.Sivertsen J, Rosenmeier J, Holst JJ, Vilsbøll T. The effect of glucagon-like peptide 1 on cardiovascular risk. Nat Rev Cardiol. 2012;9:209–22. doi: 10.1038/nrcardio.2011.211. [DOI] [PubMed] [Google Scholar]
- 18.Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich L, Hidalgo B, Fox K, Huffman JE, An P, Lu Y, Rasmussen-Torvik LJ, Grarup N, Ehm MG, Li L, Baldridge AS, Stančáková A, Abrol R, Besse C, Boland A, Bork-Jensen J, Fornage M, Freitag DF, Garcia ME, Guo X, Hara K, Isaacs A, Jakobsdottir J, Lange LA, Layton JC, Li M, Hua Zhao J, Meidtner K, Morrison AC, Nalls MA, Peters MJ, Sabater-Lleal M, Schurmann C, Silveira A, Smith AV, Southam L, Stoiber MH, Strawbridge RJ, Taylor KD, Varga TV, Allin KH, Amin N, Aponte JL, Aung T, Barbieri C, Bihlmeyer NA, Boehnke M, Bombieri C, Bowden DW, Burns SM, Chen Y, Chen YD, Cheng CY, Correa A, Czajkowski J, Dehghan A, Ehret GB, Eiriksdottir G, Escher SA, Farmaki AE, Frånberg M, Gambaro G, Giulianini F, Goddard WA, Goel A, Gottesman O, Grove ML, Gustafsson S, Hai Y, Hallmans G, Heo J, Hoffmann P, Ikram MK, Jensen RA, Jørgensen ME, Jørgensen T, Karaleftheri M, Khor CC, Kirkpatrick A, Kraja AT, Kuusisto J, Lange EM, Lee IT, Lee WJ, Leong A, Liao J, Liu C, Liu Y, Lindgren CM, Linneberg A, Malerba G, Mamakou V, Marouli E, Maruthur NM, Matchan A, Mc Kean-Cowdin R, Mc Leod O, Metcalf GA, Mohlke KL, Muzny DM, Ntalla I, Palmer ND, Pasko D, Peter A, Rayner NW, Renström F, Rice K, Sala CF, Sennblad B, Serafetinidis I, Smith JA, Soranzo N, Speliotes EK, Stahl EA, Stirrups K, Tentolouris N, Thanopoulou A, Torres M, Traglia M, Tsafantakis E, Javad S, Yanek LR, Zengini E, Becker DM, Bis JC, Brown JB, Adrienne Cupples L, Hansen T, Ingelsson E, Karter AJ, Lorenzo C, Mathias RA, Norris JM, Peloso GM, Sheu WHH, Toniolo D, Vaidya D, Varma R, Wagenknecht LE, Boeing H, Bottinger EP, Dedoussis G, Deloukas P, Ferrannini E, Franco OH, Franks PW, Gibbs RA, Gudnason V, Hamsten A, Harris TB, Hattersley AT, Hayward C, Hofman A, Jansson JH, Langenberg C, Launer LJ, Levy D, Oostra BA, O'Donnell CJ, O'Rahilly S, Padmanabhan S, Pankow JS, Polasek O, Province MA, Rich SS, Ridker PM, Rudan I, Schulze MB, Smith BH, Uitterlinden AG, Walker M, Watkins H, Wong TY, Zeggini E, Laakso M, Borecki IB, Chasman DI, Pedersen O, Psaty BM, Shyong Tai E, van Duijn CM, Wareham NJ, Waterworth DM, Boerwinkle E, Linda Kao WH, Florez JC, Loos RJF, Wilson JG, Frayling TM, Siscovick DS, Dupuis J, Rotter JI, Meigs JB, Scott RA, Goodarzi MO. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Exome Aggregation Consortium. Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O'Donnell-Luria A, Ware J, Hill A, Cummings B, Tukiainen T, Birnbaum D, Kosmicki J, Duncan L, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Cooper D, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M, Moonshine AL, Natarajan P, Orozco L, Peloso G, Poplin R, Rivas M, Ruano-Rubio V, Ruderfer D, Shakir K, Stenson P, Stevens C, Thomas B, Tiao G, Tusie-Luna M, Weisburd B, Won HH, Yu D, Altshuler D, Ardissino D, Boehnke M, Danesh J, Roberto E, Florez J, Gabriel S, Getz G, Hultman C, Kathiresan S, Laakso M, McCarroll S, McCarthy M, McGovern D, McPherson R, Neale B, Palotie A, Purcell S, Saleheen D, Scharf J, Sklar P, Patrick S, Tuomilehto J, Watkins H, Wilson J, Daly M, MacArthur D. Analysis of protein-coding genetic variation in 60,706 humans. 2015 doi: 10.1101/030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno JL, Willett KC, Desilets AR. Exenatide as a novel weight loss modality in patients without diabetes. Ann Pharmacother. 2012;46:1700–6. doi: 10.1345/aph.1R372. [DOI] [PubMed] [Google Scholar]
- 21.Vilsbøll T, Christensen M, Junker AE, Knop FK, Gluud LL. Effects of glucagon-like peptide-1 receptor agonists on weight loss: systematic review and meta-analyses of randomised controlled trials. BMJ. 2012;344:d7771. doi: 10.1136/bmj.d7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson LE, Holt TA, Rees K, Randeva HS, O'Hare JP. Effects of exenatide and liraglutide on heart rate, blood pressure and body weight: systematic review and meta-analysis. BMJ Open. 2013;3 doi: 10.1136/bmjopen-2012-001986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt LJ, Habacher W, Augustin T, Krahulec E, Semlitsch T. A systematic review and meta-analysis of the efficacy of lixisenatide in the treatment of patients with type 2 diabetes. Diabetes Obes Metab. 2014 doi: 10.1111/dom.12269. [DOI] [PubMed] [Google Scholar]
- 24.The Emerging Risk Factors Collaboration, Diabetes mellitus, fasting glucose, and risk of cause-specific death. N Engl J Med. 2011;364:829–41. doi: 10.1056/NEJMoa1008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harkavyi A, Abuirmeileh A, Lever R, Kingsbury AE, Biggs CS, Whitton PS. Glucagon-like peptide 1 receptor stimulation reverses key deficits in distinct rodent models of Parkinson's disease. J Neuroinflammation. 2008;5:19. doi: 10.1186/1742-2094-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Duffy KB, Ottinger MA, Ray B, Bailey Ja, Holloway HW, Tweedie D, Perry T, Mattson MP, Kapogiannis D, Sambamurti K, Lahiri DK, Greig NH. GLP-1 receptor stimulation reduces amyloid-beta peptide accumulation and cytotoxicity in cellular and animal models of Alzheimer's disease. J Alzheimers Dis. 2010;19:1205–19. doi: 10.3233/JAD-2010-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory AP, Dendrou Ca, Attfield KE, Haghikia A, Xifara DK, Butter F, Poschmann G, Kaur G, Lambert L, Leach Oa, Prömel S, Punwani D, Felce JH, Davis SJ, Gold R, Nielsen FC, Siegel RM, Mann M, Bell JI, Mc Vean G, Fugger L. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–11. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjems LL, Holst JJ, Vølund A, Madsbad S. The influence of GLP-1 on glucose-stimulated insulin secretion: effects on beta-cell sensitivity in type 2 and nondiabetic subjects. Diabetes. 2003;52:380–386. doi: 10.2337/diabetes.52.2.380. [DOI] [PubMed] [Google Scholar]
- 29.Dushay J, Gao C, Gopalakrishnan GS, Crawley M, Mitten EK, Wilker E, Mullington J, Maratos-Flier E. Short-term exenatide treatment leads to significant weight loss in a subset of obese women without diabetes. Diabetes Care. 2012;35:4–11. doi: 10.2337/dc11-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ussher JR, Drucker DJ. Cardiovascular actions of incretin-based therapies. Circ Res. 2014;114:1788–803. doi: 10.1161/CIRCRESAHA.114.301958. [DOI] [PubMed] [Google Scholar]
- 31.Grimm M, Han J, Weaver C, Griffin P, Schulteis CT, Dong H, Malloy J. Efficacy, safety and tolerability of exenatide once weekly in patients with type 2 diabetes mellitus: an integrated analysis of the DURATION trials. Postgrad Med. 2013;125:47–57. doi: 10.3810/pgm.2013.05.2660. [DOI] [PubMed] [Google Scholar]
- 32.Flannick J, Thorleifsson G, Beer NL, Jacobs SBR, Grarup N, Burtt NP, Mahajan A, Fuchsberger C, Atzmon G, Benediktsson R, Blangero J, Bowden DW, Brandslund I, Brosnan J, Burslem F, Chambers J, Cho YS, Christensen C, Douglas DA, Duggirala R, Dymek Z, Farjoun Y, Fennell T, Fontanillas P, Forsén T, Gabriel S, Glaser B, Gudbjartsson DF, Hanis C, Hansen T, Hreidarsson AB, Hveem K, Ingelsson E, Isomaa B, Johansson S, Jørgensen T, Jørgensen ME, Kathiresan S, Kong A, Kooner J, Kravic J, Laakso M, Lee JY, Lind L, Lindgren CM, Linneberg A, Masson G, Meitinger T, Mohlke KL, Molven A, Morris AP, Potluri S, Rauramaa R, Ribel-Madsen R, Richard A, Rolph T, Salomaa V, Segrè AV, Skärstrand H, Steinthorsdottir V, Stringham HM, Sulem P, Tai ES, Teo YY, Teslovich T, Thorsteinsdottir U, Trimmer JK, Tuomi T, Tuomilehto J, Vaziri-Sani F, Voight BF, Wilson JG, Boehnke M, McCarthy MI, Njølstad PR, Pedersen O, Groop L, Cox DR, Stefansson K, Altshuler D. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–63. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JEL, Shah T, Sofat R, Stender S, Johnson PCD, Scott Ra, Leusink M, Verweij N, Sharp SJ, Guo Y, Giambartolomei C, Chung C, Peasey A, Amuzu A, Li K, Palmen J, Howard P, Cooper Ja, Drenos F, Li YR, Lowe G, Gallacher J, Stewart MCW, Tzoulaki I, Buxbaum SG, van der DL, Forouhi ANG, Onland-Moret NC, van der Schouw YT, Schnabel RB, Hubacek Ja, Kubinova R, Baceviciene M, Tamosiunas A, Pajak A, Topor-Madry R, Stepaniak U, Malyutina S, Baldassarre D, Sennblad B, Tremoli E, de Faire U, Veglia F, Ford I, Jukema JW, Westendorp RGJ, de Borst GJ, de Jong Pa, Algra A, Spiering W, der Zee AHM, Klungel OH, de Boer A, Doevendans Pa, Eaton CB, Robinson JG, Duggan D, Kjekshus J, Downs JR, Gotto AM, Keech AC, Marchioli R, Tognoni G, Sever PS, Poulter NR, Waters DD, Pedersen TR, Amarenco P, Nakamura H, McMurray JJV, Lewsey JD, Chasman DI, Ridker PM, Maggioni AP, Tavazzi L, Ray KK, Seshasai SRK, Manson JE, Price JF, Whincup PH, Morris RW, Lawlor Da, Smith GD, Ben-Shlomo Y, Schreiner PJ, Fornage M, Siscovick DS, Cushman M, Kumari M, Wareham NJ, Verschuren WMM, Redline S, Patel SR, Whittaker JC, Hamsten A, Delaney Ja, Dale C, Gaunt TR, Wong A, Kuh D, Hardy R, Kathiresan S, Castillo Ba, van der Harst P, Brunner EJ, Tybjaerg-Hansen A, Marmot MG, Krauss RM, Tsai M, Coresh J, Hoogeveen RC, Psaty BM, Lange La, Hakonarson H, Dudbridge F, Humphries SE, Talmud PJ, Kivimäki M, Timpson NJ, Langenberg C, Asselbergs FW, Voevoda M, Bobak M, Pikhart H, Wilson JG, Reiner AP, Keating BJ, Hingorani AD, Sattar N. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2014:1–11. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toft-Nielsen MB, Madsbad S, Holst JJ. Determinants of the effectiveness of glucagon-like peptide-1 in type 2 diabetes. J Clin Endocrinol Metab. 2001;86:3853–3860. doi: 10.1210/jcem.86.8.7743. [DOI] [PubMed] [Google Scholar]
- 35.Scrocchi LA, Brown TJ, Ma Clusky N, Brubaker PL, Auerbach AB, Joyner AL, Drucker DJ. Glucose intolerance but normal satiety in mice with a null mutation in the glucagon-like peptide 1 receptor gene. Nat Med. 1996;2:1254–8. doi: 10.1038/nm1196-1254. [DOI] [PubMed] [Google Scholar]
- 36.Ayala JE, Bracy DP, James FD, Burmeister Ma, Wasserman DH, Drucker DJ. Glucagon-like peptide-1 receptor knockout mice are protected from high-fat diet-induced insulin resistance. Endocrinology. 2010;151:4678–87. doi: 10.1210/en.2010-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert R, Wenzel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 38.Fortin J, Schroeder JC, Zhu Y, Beinborn M, Kopin AS. Pharmacological characterization of human incretin receptor missense variants. J Pharmacol Exp Ther. 2010;332:274–80. doi: 10.1124/jpet.109.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koole C, Wootten D, Simms J, Valant C, Miller LJ, Christopoulos A, Sexton PM. Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol Pharmacol. 2011;80:486–97. doi: 10.1124/mol.111.072884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–4. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 41.Drucker DJ, Nauck Ma. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 42.Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, Hayoz D, Paccaud F, Preisig M, Song KS, Yuan X, Danoff TM, Stirnadel Ha, Waterworth D, Mooser V, Waeber G, Vollenweider P. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyszynski DF, Waterworth DM, Barter PJ, Cohen J, Kesäniemi YA, Mahley RW, Mc Pherson R, Waeber G, Bersot TP, Sharma SS, Nolan V, Middleton LT, Sundseth SS, Farrer La, Mooser V, Grundy SM. Relation between atherogenic dyslipidemia and the Adult Treatment Program-III definition of metabolic syndrome (Genetic Epidemiology of Metabolic Syndrome Project) Am J Cardiol. 2005;95:194–8. doi: 10.1016/j.amjcard.2004.08.091. [DOI] [PubMed] [Google Scholar]
- 44.Forouhi NG, Luan J, Hennings S, Wareham NJ. Incidence of Type 2 diabetes in England and its association with baseline impaired fasting glucose: the Ely study 1990-2000. Diabet Med. 2007;24:200–7. doi: 10.1111/j.1464-5491.2007.02068.x. [DOI] [PubMed] [Google Scholar]
- 45.Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, Wareham N. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer. 1999;80(1):95–103. [PubMed] [Google Scholar]
- 46.Rolfe EDL, Loos RJF, Druet C, Stolk RP, Ekelund U, Griffin SJ, Forouhi NG, Wareham NJ, Ong KK. Association between birth weight and visceral fat in adults. Am J Clin Nutr. 2010;92:347–52. doi: 10.3945/ajcn.2010.29247. [DOI] [PubMed] [Google Scholar]
- 47.Kooner JS, Chambers JC, Aguilar-Salinas Ca, Hinds DA, Hyde CL, Warnes GR, Gómez Pérez FJ, Frazer Ka, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–51. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 48.Echouffo-Tcheugui JB, Simmons RK, Williams KM, Barling RS, Prevost aT, Kinmonth AL, Wareham NJ, Griffin SJ. The ADDITION-Cambridge trial protocol: a cluster -- randomised controlled trial of screening for type 2 diabetes and intensive treatment for screen-detected patients. BMC Public Health. 2009;9:136. doi: 10.1186/1471-2458-9-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU, Kao WHL, Li M, Glazer NL, Manning AK, Luan J, Stringham HM, Prokopenko I, Johnson T, Grarup N, Boesgaard TW, Lecoeur C, Shrader P, O'Connell J, Ingelsson E, Couper DJ, Rice K, Song K, Andreasen CH, Dina C, Köttgen A, Le Bacquer O, Pattou F, Taneera J, Steinthorsdottir V, Rybin D, Ardlie K, Sampson M, Qi L, van Hoek M, Weedon MN, Aulchenko YS, Voight BF, Grallert H, Balkau B, Bergman RN, Bielinski SJ, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Buchanan Ta, Bumpstead SJ, Cavalcanti-Proença C, Charpentier G, Chen YDI, Chines PS, Collins FS, Cornelis M, Crawford GJ, Delplanque J, Doney A, Egan JM, Erdos MR, Firmann M, Forouhi NG, Fox CS, Goodarzi MO, Graessler J, Hingorani A, Isomaa B, Jørgensen T, Kivimaki M, Kovacs P, Krohn K, Kumari M, Lauritzen T, Lévy-Marchal C, Mayor V, Mc Ateer JB, Meyre D, Mitchell BD, Mohlke KL, Morken Ma, Narisu N, Palmer CNa, Pakyz R, Pascoe L, Payne F, Pearson D, Rathmann W, Sandbaek A, Sayer AA, Scott LJ, Sharp SJ, Sijbrands E, Singleton A, Siscovick DS, Smith NL, Sparsø T, Swift AJ, Syddall H, Thorleifsson G, Tönjes A, Tuomi T, Tuomilehto J, Valle TT, Waeber G, Walley A, Waterworth DM, Zeggini E, Zhao JH, Illig T, Wichmann HE, Wilson JF, van Duijn C, Hu FB, Morris AD, Frayling TM, Hattersley AT, Thorsteinsdottir U, Stefansson K, Nilsson P, Syvänen AC, Shuldiner AR, Walker M, Bornstein SR, Schwarz P, Williams GH, Nathan DM, Kuusisto J, Laakso M, Cooper C, Marmot M, Ferrucci L, Mooser V, Stumvoll M, Loos RJF, Altshuler D, Psaty BM, Rotter JI, Boerwinkle E, Hansen T, Pedersen O, Florez JC, Mc Carthy MI, Boehnke M, Barroso I, Sladek R, Froguel P, Meigs JB, Groop L, Wareham NJ, Watanabe RM. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–8. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Mägi R, Strawbridge RJ, Rehnberg E, Gustafsson S, Kanoni S, Rasmussen-Torvik LJ, Yengo L, Lecoeur C, Shungin D, Sanna S, Sidore C, Johnson PCD, Jukema JW, Johnson T, Mahajan A, Verweij N, Thorleifsson G, Hottenga JJ, Shah S, Smith AV, Sennblad B, Gieger C, Salo P, Perola M, Timpson NJ, Evans DM, Pourcain BS, Wu Y, Andrews JS, Hui J, Bielak LF, Zhao W, Horikoshi M, Navarro P, Isaacs A, O'Connell JR, Stirrups K, Vitart V, Hayward C, Esko T, Mihailov E, Fraser RM, Fall T, Voight BF, Raychaudhuri S, Chen H, Lindgren CM, Morris AP, Rayner NW, Robertson N, Rybin D, Liu CT, Beckmann JS, Willems SM, Chines PS, Jackson AU, Kang HM, Stringham HM, Song K, Tanaka T, Peden JF, Goel A, Hicks Aa, An P, Müller-Nurasyid M, Franco-Cereceda A, Folkersen L, Marullo L, Jansen H, Oldehinkel AJ, Bruinenberg M, Pankow JS, North KE, Forouhi NG, Loos RJF, Edkins S, Varga TV, Hallmans G, Oksa H, Antonella M, Nagaraja R, Trompet S, Ford I, Bakker SJL, Kong A, Kumari M, Gigante B, Herder C, Munroe PB, Caulfield M, Antti J, Mangino M, Small K, Miljkovic I, Liu Y, Atalay M, Kiess W, James AL, Rivadeneira F, Uitterlinden AG, Palmer CNa, Doney ASF, Willemsen G, Smit JH, Campbell S, Polasek O, Bonnycastle LL, Hercberg S, Dimitriou M, Bolton JL, Fowkes GR, Kovacs P, Lindström J, Zemunik T, Bandinelli S, Wild SH, Basart HV, Rathmann W, Grallert H, Maerz W, Kleber ME, Boehm BO, Peters A, Pramstaller PP, Province Ma, Borecki IB, Hastie ND, Rudan I, Campbell H, Watkins H, Farrall M, Stumvoll M, Ferrucci L, Waterworth DM, Bergman RN, Collins FS, Tuomilehto J, Watanabe RM, de Geus EJC, Penninx BW, Hofman A, Oostra Ba, Psaty BM, Vollenweider P, Wilson JF, Wright AF, Hovingh GK, Metspalu A, Uusitupa M, Magnusson PKE, Kyvik KO, Kaprio J, Price JF, Dedoussis GV, Deloukas P, Meneton P, Lind L, Boehnke M, Shuldiner AR, van Duijn CM, Morris AD, Toenjes A, Peyser Pa, Beilby JP, Körner A, Kuusisto J, Laakso M, Bornstein SR, Schwarz PEH, Lakka Ta, Rauramaa R, Adair LS, Smith GD, Spector TD, Illig T, de Faire U, Hamsten A, Gudnason V, Kivimaki M, Hingorani A, Keinanen-Kiukaanniemi SM, Saaristo TE, Boomsma DI, Stefansson K, van der Harst P, Dupuis J, Pedersen NL, Sattar N, Harris TB, Cucca F, Ripatti S, Salomaa V, Mohlke KL, Balkau B, Froguel P, Pouta A, Jarvelin MR, Wareham NJ, Bouatia-Naji N, McCarthy MI, Franks PW, Meigs JB, Teslovich TM, Florez JC, Langenberg C, Ingelsson E, Prokopenko I, Barroso I. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JRB, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Böttcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbatón-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks Aa, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin MR, Jhun Ma, Johnson PCD, Jukema JW, Jula A, Kao WH, Kaprio J, Kardia SLR, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik KO, Laakso M, Lakka T, Lannfelt L, Lathrop GM, Launer LJ, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos RJF, Luan J, Lyssenko V, Mägi R, Magnusson PKE, Marmot M, Meneton P, Mohlke KL, Mooser V, Morken Ma, Miljkovic I, Narisu N, O'Connell J, Ong KK, Oostra Ba, Palmer LJ, Palotie A, Pankow JS, Peden JF, Pedersen NL, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser Pa, Polasek O, Pramstaller PP, Province Ma, Räikkönen K, Rauramaa R, Rehnberg E, Rice K, Rotter JI, Rudan I, Ruokonen A, Saaristo T, Sabater-Lleal M, Salomaa V, Savage DB, Saxena R, Schwarz P, Seedorf U, Sennblad B, Serrano-Rios M, Shuldiner AR, Sijbrands EJG, Siscovick DS, Smit JH, Small KS, Smith NL, Smith AV, Stančáková A, Stirrups K, Stumvoll M, Sun YV, Swift AJ, Tönjes; A, Tuomilehto J, Trompet S, Uitterlinden AG, Uusitupa M, Vikström M, Vitart V, Vohl MC, Voight BF, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Wheeler E, Widen E, Wild SH, Willems SM, Willemsen G, Wilson JF, Witteman JCM, Wright AF, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham NJ, Mc Carthy MI, Barroso I, Watanabe RM, Florez JC, Dupuis J, Meigs JB, Langenberg C. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O'Brien ET, Poulter NR, Sever P, Shields DC, Thom S, Wannamethee SG, Whincup PH, Brown MJ, Connell JM, Dobson RJ, Howard PJ, Mein Ca, Onipinla A, Shaw-Hawkins S, Zhang Y, Davey Smith G, Day INM, Lawlor Da, Goodall AH, Fowkes FG, Abecasis GR, Elliott P, Gateva V, Braund PS, Burton PR, Nelson CP, Tobin MD, van der Harst P, Glorioso N, Neuvrith H, Salvi E, Staessen Ja, Stucchi A, Devos N, Jeunemaitre X, Plouin PF, Tichet J, Juhanson P, Org E, Putku M, Sõber S, Veldre G, Viigimaa M, Levinsson A, Rosengren A, Thelle DS, Hastie CE, Hedner T, Lee WK, Melander O, Wahlstrand B, Hardy R, Wong A, Cooper Ja, Palmen J, Chen L, Stewart AFR, Wells Ga, Westra HJ, Wolfs MGM, Clarke R, Franzosi MG, Goel A, Hamsten A, Lathrop M, Peden JF, Seedorf U, Watkins H, Ouwehand WH, Sambrook J, Stephens J, Casas JP, Drenos F, Holmes MV, Kivimaki M, Shah S, Shah T, Talmud PJ, Whittaker J, Wallace C, Delles C, Laan M, Kuh D, Humphries SE, Nyberg F, Cusi D, Roberts R, Newton-Cheh C, Franke L, Stanton AV, Dominiczak AF, Farrall M, Hingorani AD, Samani NJ, Caulfield MJ, Munroe PB. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.den Hoed M, Eijgelsheim M, Esko T, Brundel BJJM, Peal DS, Evans DM, Nolte IM, Segrè AV, Holm H, Handsaker RE, Westra HJ, Johnson T, Isaacs A, Yang J, Lundby A, Zhao JH, Kim YJ, Go MJ, Almgren P, Bochud M, Boucher G, Cornelis MC, Gudbjartsson D, Hadley D, van der Harst P, Hayward C, den Heijer M, Igl W, Jackson AU, Kutalik Z, Luan J, Kemp JP, Kristiansson K, Ladenvall C, Lorentzon M, Montasser ME, Njajou OT, O'Reilly PF, Padmanabhan S, St Pourcain B, Rankinen T, Salo P, Tanaka T, Timpson NJ, Vitart V, Waite L, Wheeler W, Zhang W, Draisma HHM, Feitosa MF, Kerr KF, Lind Pa, Mihailov E, Onland-Moret NC, Song C, Weedon MN, Xie W, Yengo L, Absher D, Albert CM, Alonso A, Arking DE, de Bakker PIW, Balkau B, Barlassina C, Benaglio P, Bis JC, Bouatia-Naji N, Brage S, Chanock SJ, Chines PS, Chung M, Darbar D, Dina C, Dörr M, Elliott P, Felix SB, Fischer K, Fuchsberger C, de Geus EJC, Goyette P, Gudnason V, Harris TB, Hartikainen AL, Havulinna AS, Heckbert SR, Hicks Aa, Hofman A, Holewijn S, Hoogstra-Berends F, Hottenga JJ, Jensen MK, Johansson A, Junttila J, Kääb S, Kanon B, Ketkar S, Khaw KT, Knowles JW, Kooner AS, Kors Ja, Kumari M, Milani L, Laiho P, Lakatta EG, Langenberg C, Leusink M, Liu Y, Luben RN, Lunetta KL, Lynch SN, Markus MRP, Marques-Vidal P, Mateo Leach I, McArdle WL, McCarroll Sa, Medland SE, Miller Ka, Montgomery GW, Morrison AC, Müller-Nurasyid M, Navarro P, Nelis M, O'Connell JR, O'Donnell CJ, Ong KK, Newman AB, Peters A, Polasek O, Pouta A, Pramstaller PP, Psaty BM, Rao DC, Ring SM, Rossin EJ, Rudan D, Sanna S, Scott Ra, Sehmi JS, Sharp S, Shin JT, Singleton AB, Smith AV, Soranzo N, Spector TD, Stewart C, Stringham HM, Tarasov KV, Uitterlinden AG, Vandenput L, Hwang SJ, Whitfield JB, Wijmenga C, Wild SH, Willemsen G, Wilson JF, Witteman JCM, Wong A, Wong Q, Jamshidi Y, Zitting P, Boer JMa, Boomsma DI, Borecki IB, van Duijn CM, Ekelund U, Forouhi NG, Froguel P, Hingorani A, Ingelsson E, Kivimaki M, Kronmal Ra, Kuh D, Lind L, Martin NG, Oostra Ba, Pedersen NL, Quertermous T, Rotter JI, van der Schouw YT, Verschuren WMM, Walker M, Albanes D, Arnar DO, Assimes TL, Bandinelli S, Boehnke M, de Boer Ra, Bouchard C, Caulfield WLM, Chambers JC, Curhan G, Cusi D, Eriksson J, Ferrucci L, van Gilst WH, Glorioso N, de Graaf J, Groop L, Gyllensten U, Hsueh WC, Hu FB, Huikuri HV, Hunter DJ, Iribarren C, Isomaa B, Jarvelin MR, Jula A, Kähönen M, Kiemeney La, van der Klauw MM, Kooner JS, Kraft P, Iacoviello L, Lehtimäki T, Lokki MLL, Mitchell BD, Navis G, Nieminen MS, Ohlsson C, Poulter NR, Qi L, Raitakari OT, Rimm EB, Rioux JD, Rizzi F, Rudan I, Salomaa V, Sever PS, Shields DC, Shuldiner AR, Sinisalo J, Stanton AV, Stolk RP, Strachan DP, Tardif JC, Thorsteinsdottir U, Tuomilehto J, van Veldhuisen DJ, Virtamo J, Viikari J, Vollenweider P, Waeber G, Widen E, Cho YS, Olsen JV, Visscher PM, Willer C, Franke L, Erdmann J, Thompson JR, Pfeufer A, Sotoodehnia N, Newton-Cheh C, Ellinor PT, Stricker BHC, Metspalu A, Perola M, Beckmann JS, Smith GD, Stefansson K, Wareham NJ, Munroe PB, Sibon OCM, Milan DJ, Snieder H, Samani NJ, Loos RJF. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–31. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan Aa, Beane-Freeman L, Bracci PM, Buring J, Canzian F, Duell EJ, Gallinger S, Giles GG, Goodman GE, Goodman PJ, Jacobs EJ, Kamineni A, Klein AP, Kolonel LN, Kulke MH, Li D, Malats N, Olson SH, Risch Ha, Sesso HD, Visvanathan K, White E, Zheng W, Abnet CC, Albanes D, Andreotti G, Austin Ma, Barfield R, Basso D, Berndt SI, Boutron-Ruault MC, Brotzman M, Büchler MW, Bueno-de-Mesquita HB, Bugert P, Burdette L, Campa D, Caporaso NE, Capurso G, Chung C, Cotterchio M, Costello E, Elena J, Funel N, Gaziano JM, Giese Na, Giovannucci EL, Goggins M, Gorman MJ, Gross M, Haiman Ca, Hassan M, Helzlsouer KJ, Henderson BE, Holly Ea, Hu N, Hunter DJ, Innocenti F, Jenab M, Kaaks R, Key TJ, Khaw KT, Klein Ea, Kogevinas M, Krogh V, Kupcinskas J, Kurtz RC, La Croix A, Landi MT, Landi S, Le Marchand L, Mambrini A, Mannisto S, Milne RL, Nakamura Y, Oberg AL, Owzar K, Patel AV, Peeters PHM, Peters U, Pezzilli R, Piepoli A, Porta M, Real FX, Riboli E, Rothman N, Scarpa A, Shu XO, Silverman DT, Soucek P, Sund M, Talar-Wojnarowska R, Taylor PR, Theodoropoulos GE, Thornquist M, Tjønneland A, Tobias GS, Trichopoulos D, Vodicka P, Wactawski-Wende J, Wentzensen N, Wu C, Yu H, Yu K, Zeleniuch-Jacquotte A, Hoover R, Hartge P, Fuchs C, Chanock SJ, Stolzenberg-Solomon RS, Amundadottir LT. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46:994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R, Ikram MA, Ioannidis JPA, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;056:1–7. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Kesteloot H. Anthropometric, lifestyle and metabolic determinants of resting heart rate. A population study. Eur Heart J. 1999;20:103–10. doi: 10.1053/euhj.1999.1230. [DOI] [PubMed] [Google Scholar]
- 58.Gauderman J, Ph D, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006:35–50. http://hydra.usc.edu/gxe.
- 59.Sarwar N, Gao P, Seshasai SRK, Gobin R, Kaptoge S, Di Angelantonio E, Ingelsson E, Lawlor Da, Selvin E, Stampfer M, Stehouwer CDa, Lewington S, Pennells L, Thompson a, Sattar N, White IR, Ray KK, Danesh J. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375:2215–22. doi: 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forouhi NG, Ye Z, Rickard AP, Khaw KT, Luben R, Langenberg C, Wareham NJ. Circulating 25-hydroxyvitamin D concentration and the risk of type 2 diabetes: results from the European Prospective Investigation into Cancer (EPIC)-Norfolk cohort and updated meta-analysis of prospective studies. Diabetologia. 2012;55:2173–82. doi: 10.1007/s00125-012-2544-y. [DOI] [PubMed] [Google Scholar]
- 61.Swarbrick MM, Waldenmaier B, Pennacchio La, Lind DL, Cavazos MM, Geller F, Merriman R, Ustaszewska A, Malloy M, Scherag A, Hsueh WC, Rief W, Mauvais-Jarvis F, Pullinger CR, Kane JP, Dent R, Mc Pherson R, Kwok PY, Hinney A, Hebebrand J, Vaisse C. Lack of support for the association between GAD2 polymorphisms and severe human obesity. PLoS Biol. 2005;3:e315. doi: 10.1371/journal.pbio.0030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langenberg C, Sharp S, Forouhi NG, Franks PW, Schulze MB, Kerrison N, Ekelund U, Barroso I, Panico S, Tormo MJ, Spranger J, Griffin S, van der Schouw YT, Amiano P, Ardanaz E, Arriola L, Balkau B, Barricarte a, Beulens JWJ, Boeing H, Bueno-de-Mesquita HB, Buijsse B, Chirlaque Lopez MD, Clavel-Chapelon F, Crowe FL, de Lauzon-Guillan B, Deloukas P, Dorronsoro M, Drogan D, Froguel P, Gonzalez C, Grioni S, Groop L, Groves C, Hainaut P, Halkjaer J, Hallmans G, Hansen T, Huerta Castaño JM, Kaaks R, Key TJ, Khaw KT, Koulman a, Mattiello a, Navarro C, Nilsson P, Norat T, Overvad K, Palla L, Palli D, Pedersen O, Peeters PH, Quirós JR, Ramachandran a, Rodriguez-Suarez L, Rolandsson O, Romaguera D, Romieu I, Sacerdote C, Sánchez MJ, Sandbaek a, Slimani N, Sluijs I, Spijkerman aMW, Teucher B, Tjonneland a, Tumino R, van der DL, Verschuren AWMM, Tuomilehto J, Feskens E, McCarthy M, Riboli E, Wareham NJ. Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia. 2011;54:2272–82. doi: 10.1007/s00125-011-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondière UR, Hémon B, Casagrande C, Vignat J, Overvad K, Tjønneland a, Clavel-Chapelon F, Thiébaut a, Wahrendorf J, Boeing H, Trichopoulos D, Trichopoulou a, Vineis P, Palli D, Bueno-De-Mesquita HB, Peeters PHM, Lund E, Engeset D, González; Ca, Barricarte a, Berglund G, Hallmans G, Day NE, Key TJ, Kaaks R, Saracci R. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–24. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 64.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 65.Danesh J, Saracci R, Berglund G, Feskens E, Overvad K, Panico S, Thompson S, Fournier A, Clavel-Chapelon F, Canonico M, Kaaks R, Linseisen J, Boeing H, Pischon T, Weikert C, Olsen A, Tjønneland A, Johnsen SP, Jensen MK, Quirós JR, Svatetz CAG, Pérez MJS, Larrañaga N, Sanchez CN, Iribas CM, Bingham S, Khaw KT, Wareham N, Key T, Roddam A, Trichopoulou A, Benetou V, Trichopoulos D, Masala G, Sieri S, Tumino R, Sacerdote C, Mattiello A, Verschuren WMM, Bueno-de-Mesquita HB, Grobbee DE, van der Schouw YT, Melander O, Hallmans G, Wennberg P, Lund E, Kumle M, Skeie G, Ferrari P, Slimani N, Norat T, Riboli E. EPIC-Heart: the cardiovascular component of a prospective study of nutritional, lifestyle and biological factors in 520,000 middle-aged participants from 10 European countries. Eur J Epidemiol. 2007;22:129–41. doi: 10.1007/s10654-006-9096-8. [DOI] [PubMed] [Google Scholar]
- 66.Bingham S, Riboli E. Diet and cancer--the European Prospective Investigation into Cancer and Nutrition. Nat Rev Cancer. 2004;4:206–15. doi: 10.1038/nrc1298. [DOI] [PubMed] [Google Scholar]
- 67.Shepherd J, Cobbe SM, Ford I, Isles CG, Lorimer AR, MacFarlane PW, McKillop JH, Packard CJ. Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. West of Scotland Coronary Prevention Study Group. N Engl J Med. 1995;333:1301–7. doi: 10.1056/NEJM199511163332001. [DOI] [PubMed] [Google Scholar]
- 68.Shepherd J, Blauw GJ, Murphy MB, Bollen ELEM, Buckley BM, Cobbe SM, Ford I, Gaw A, Hyland M, Jukema JW, Kamper AM, Macfarlane PW, Meinders AE, Norrie J, Packard CJ, Perry IJ, Stott DJ, Sweeney BJ, Twomey C, Westendorp RGJ. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–30. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 69.Evans A, Salomaa V, Kulathinal S, Asplund K, Cambien F, Ferrario M, Perola M, Peltonen L, Shields D, Tunstall-Pedoe H, Kuulasmaa K. MORGAM (an international pooling of cardiovascular cohorts) Int J Epidemiol. 2005;34:21–7. doi: 10.1093/ije/dyh327. [DOI] [PubMed] [Google Scholar]
- 70.Kulathinal S, Karvanen J, Saarela O, Kuulasmaa K. Case-cohort design in practice - experiences from the MORGAM Project. Epidemiol Perspect Innov. 2007;4:15. doi: 10.1186/1742-5573-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 72.Assimes TL, Hólm H, Kathiresan S, Reilly MP, Thorleifsson G, Voight BF, Erdmann J, Willenborg C, Vaidya D, Xie C, Patterson CC, Morgan TM, Burnett MS, Li M, Hlatky MA, Knowles JW, Thompson JR, Absher D, Iribarren C, Go A, Fortmann SP, Sidney S, Risch N, Tang H, Myers RM, Berger K, Stoll M, Shah SH, Thorgeirsson G, Andersen K, Havulinna AS, Herrera JE, Faraday N, Kim Y, Kral BG, Mathias RA, Ruczinski I, Suktitipat B, Wilson AF, Yanek LR, Becker LC, Linsel-Nitschke P, Lieb W, König IR, Hengstenberg C, Fischer M, Stark K, Reinhard W, Winogradow J, Grassl M, Grosshennig A, Preuss M, Schreiber S, Wichmann HE, Meisinger C, Yee J, Friedlander Y, Do R, Meigs JB, Williams G, Nathan DM, MacRae CA, Qu L, Wilensky RL, Matthai WH, Qasim AN, Hakonarson H, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser VE, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Martinelli N, Olivieri O, Trabetti E, Malerba G, Pignatti PF, Guiducci C, Mirel D, Parkin M, Hirschhorn JN, Asselta R, Duga S, Musunuru K, Daly MJ, Purcell S, Eifert S, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Ouwehand WH, Deloukas P, Scholz M, Cambien F, Huge A, Scheffold T, Salomaa V, Girelli D, Granger CB, Peltonen L, McKeown PP, Altshuler D, Melander O, Devaney JM, Epstein SE, Rader DJ, Elosua R, Engert JC, Anand SS, Hall AS, Ziegler A, O'Donnell CJ, Spertus JA, Siscovick D, Schwartz SM, Becker D, Thorsteinsdottir U, Stefansson K, Schunkert H, Samani NJ, Quertermous T. Lack of association between the Trp719Arg polymorphism in kinesin-like protein-6 and coronary artery disease in 19 case-control studies. J Am Coll Cardiol. 2010;56:1552–63. doi: 10.1016/j.jacc.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hallmans G, Agren A, Johansson G, Johansson A, Stegmayr B, Jansson JH, Lindahl B, Rolandsson O, Söderberg S, Nilsson M, Johansson I, Weinehall L. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand J Public Health Suppl. 2003;61:18–24. doi: 10.1080/14034950310001432. [DOI] [PubMed] [Google Scholar]
- 74.Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J. Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 75.Hills SA, Balkau B, Coppack SW, Dekker JM, Mari A, Natali A, Walker M, Ferrannini E. The EGIR-RISC STUDY (The European group for the study of insulin resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives. Diabetologia. 2004;47:566–70. doi: 10.1007/s00125-004-1335-5. [DOI] [PubMed] [Google Scholar]
- 76.Lean MEJ, Carraro R, Finer N, Hartvig H, Lindegaard ML, Rössner S, Van Gaal L, Astrup A NN8022-1807 Investigators. Tolerability of nausea and vomiting and associations with weight loss in a randomized trial of liraglutide in obese, non-diabetic adults. Int J Obes (Lond) 2014;38:689–97. doi: 10.1038/ijo.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelly AS, Rudser KD, Nathan BM, Fox CK, Metzig AM, Coombes BJ, Fitch AK, Bomberg EM, Abuzzahab MJ. The effect of glucagon-like peptide-1 receptor agonist therapy on body mass index in adolescents with severe obesity: a randomized, placebo-controlled, clinical trial. JAMA Pediatr. 2013;167:355–60. doi: 10.1001/jamapediatrics.2013.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenstock J, Klaff LJ, Schwartz S, Northrup J, Holcombe JH, Wilhelm K, Trautmann M. Effects of exenatide and lifestyle modification on body weight and glucose tolerance in obese subjects with and without pre-diabetes. Diabetes Care. 2010;33:1173–5. doi: 10.2337/dc09-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Elkind-Hirsch K, Marrioneaux O, Bhushan M, Vernor D, Bhushan R. Comparison of single and combined treatment with exenatide and metformin on menstrual cyclicity in overweight women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:2670–8. doi: 10.1210/jc.2008-0115. [DOI] [PubMed] [Google Scholar]
- 80.Aviles-Olmos I, Dickson J, Kefalopoulou Z, Djamshidian A, Ell P, Soderlund T, Whitton P, Wyse R, Isaacs T, Lees A, Limousin P, Foltynie T. Exenatide and the treatment of patients with Parkinson's disease. J Clin Invest. 2013;123:2730–6. doi: 10.1172/JCI68295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madsbad S, Schmitz O, Ranstam J, Jakobsen G, Matthews DR NN2211-1310 International Study Group. Improved glycemic control with no weight increase in patients with type 2 diabetes after once-daily treatment with the long-acting glucagon-like peptide 1 analog liraglutide (NN2211): a 12-week, double-blind, randomized, controlled trial. Diabetes Care. 2004;27:1335–42. doi: 10.2337/diacare.27.6.1335. [DOI] [PubMed] [Google Scholar]
- 82.Seino Y, Rasmussen MF, Zdravkovic M, Kaku K. Dose-dependent improvement in glycemia with once-daily liraglutide without hypoglycemia or weight gain: A double-blind, randomized, controlled trial in Japanese patients with type 2 diabetes. Diabetes Res Clin Pract. 2008;81:161–8. doi: 10.1016/j.diabres.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 83.Marre M, Shaw J, Brändle M, Bebakar WMW, Kamaruddin NA, Strand J, Zdravkovic M, Le Thi TD, Colagiuri S LEAD-1 SU study group. Liraglutide, a once-daily human GLP-1 analogue, added to a sulphonylurea over 26 weeks produces greater improvements in glycaemic and weight control compared with adding rosiglitazone or placebo in subjects with Type 2 diabetes (LEAD-1 SU) Diabet Med. 2009;26:268–78. doi: 10.1111/j.1464-5491.2009.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nauck M, Frid A, Hermansen K, Shah NS, Tankova T, Mitha IH, Zdravkovic M, Düring M, Matthews DR LEAD-2 Study Group. Efficacy and safety comparison of liraglutide, glimepiride, and placebo, all in combination with metformin, in type 2 diabetes: the LEAD (liraglutide effect and action in diabetes)-2 study. Diabetes Care. 2009;32:84–90. doi: 10.2337/dc08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forst T, Michelson G, Ratter F, Weber MM, Anders S, Mitry M, Wilhelm B, Pfützner A. Addition of liraglutide in patients with Type 2 diabetes well controlled on metformin monotherapy improves several markers of vascular function. Diabet Med. 2012;29:1115–8. doi: 10.1111/j.1464-5491.2012.03589.x. [DOI] [PubMed] [Google Scholar]
- 86.Zinman B, Gerich J, Buse JB, Lewin A, Schwartz S, Raskin P, Hale PM, Zdravkovic M, Blonde L LEAD-4 Study Investigators. Efficacy and safety of the human glucagon-like peptide-1 analog liraglutide in combination with metformin and thiazolidinedione in patients with type 2 diabetes (LEAD-4 Met+TZD) Diabetes Care. 2009;32:1224–30. doi: 10.2337/dc08-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Russell-Jones D, Vaag A, Schmitz O, Sethi BK, Lalic N, Antic S, Zdravkovic M, Ravn GM, Simó R Liraglutide Effect and Action in Diabetes 5 (LEAD-5) met+SU Study Group. Liraglutide vs insulin glargine and placebo in combination with metformin and sulfonylurea therapy in type 2 diabetes mellitus (LEAD-5 met+SU): a randomised controlled trial. Diabetologia. 2009;52:2046–55. doi: 10.1007/s00125-009-1472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim D, MacConell L, Zhuang D, Kothare PA, Trautmann M, Fineman M, Taylor K. Effects of once-weekly dosing of a long-acting release formulation of exenatide on glucose control and body weight in subjects with type 2 diabetes. Diabetes Care. 2007;30:1487–93. doi: 10.2337/dc06-2375. [DOI] [PubMed] [Google Scholar]
- 89.Kadowaki T, Namba M, Yamamura A, Sowa H, Wolka AM, Brodows RG. Exenatide exhibits dose-dependent effects on glycemic control over 12 weeks in Japanese patients with suboptimally controlled type 2 diabetes. Endocr J. 2009;56:415–24. doi: 10.1507/endocrj.k08e-296. [DOI] [PubMed] [Google Scholar]
- 90.Moretto TJ, Milton DR, Ridge TD, Macconell LA, Okerson T, Wolka AM, Brodows RG. Efficacy and tolerability of exenatide monotherapy over 24 weeks in antidiabetic drug-naive patients with type 2 diabetes: a randomized, double-blind, placebo-controlled, parallel-group study. Clin Ther. 2008;30:1448–60. doi: 10.1016/j.clinthera.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 91.Kendall DM, Riddle MC, Rosenstock J, Zhuang D, Kim DD, Fineman MS, Baron AD. Effects of exenatide (exendin-4) on glycemic control over 30 weeks in patients with type 2 diabetes treated with metformin and a sulfonylurea. Diabetes Care. 2005;28:1083–91. doi: 10.2337/diacare.28.5.1083. [DOI] [PubMed] [Google Scholar]
- 92.DeFronzo RA, Ratner RE, Han J, Kim DD, Fineman MS, Baron AD. Effects of exenatide (exendin-4) on glycemic control and weight over 30 weeks in metformin-treated patients with type 2 diabetes. Diabetes Care. 2005;28:1092–100. doi: 10.2337/diacare.28.5.1092. [DOI] [PubMed] [Google Scholar]
- 93.Buse JB, Henry RR, Han J, Kim DD, Fineman MS, Baron AD Exenatide-113 Clinical Study Group. Effects of exenatide (exendin-4) on glycemic control over 30 weeks in sulfonylurea-treated patients with type 2 diabetes. Diabetes Care. 2004;27:2628–35. doi: 10.2337/diacare.27.11.2628. [DOI] [PubMed] [Google Scholar]
- 94.Apovian CM, Bergenstal RM, Cuddihy RM, Qu Y, Lenox S, Lewis MS, Glass LC. Effects of exenatide combined with lifestyle modification in patients with type 2 diabetes. Am J Med. 2010;123:468.e9–17. doi: 10.1016/j.amjmed.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 95.Buse JB, Bergenstal RM, Glass LC, Heilmann CR, Lewis MS, Kwan AYM, Hoogwerf BJ, Rosenstock J. Use of twice-daily exenatide in Basal insulin-treated patients with type 2 diabetes: a randomized, controlled trial. Ann Intern Med. 2011;154:103–12. doi: 10.7326/0003-4819-154-2-201101180-00300. [DOI] [PubMed] [Google Scholar]
- 96.Gill A, Hoogwerf BJ, Burger J, Bruce S, Macconell L, Yan P, Braun D, Giaconia J, Malone J. Effect of exenatide on heart rate and blood pressure in subjects with type 2 diabetes mellitus: a double-blind, placebo-controlled, randomized pilot study. Cardiovasc Diabetol. 2010;9:6. doi: 10.1186/1475-2840-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liutkus J, Rosas Guzman J, Norwood P, Pop L, Northrup J, Cao D, Trautmann M. A placebo-controlled trial of exenatide twice-daily added to thiazolidinediones alone or in combination with metformin. Diabetes Obes Metab. 2010;12:1058–65. doi: 10.1111/j.1463-1326.2010.01251.x. [DOI] [PubMed] [Google Scholar]
- 98.Zinman B, Hoogwerf BJ, Durán García S, Milton DR, Giaconia JM, Kim DD, Trautmann ME, Brodows RG. The effect of adding exenatide to a thiazolidinedione in suboptimally controlled type 2 diabetes: a randomized trial. Ann Intern Med. 2007;146:477–85. doi: 10.7326/0003-4819-146-7-200704030-00003. [DOI] [PubMed] [Google Scholar]
- 99.DeFronzo RA, Triplitt C, Qu Y, Lewis MS, Maggs D, Glass LC. Effects of exenatide plus rosiglitazone on beta-cell function and insulin sensitivity in subjects with type 2 diabetes on metformin. Diabetes Care. 2010;33:951–7. doi: 10.2337/dc09-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Derosa G, Franzetti IG, Querci F, Carbone A, Ciccarelli L, Piccinni MN, Fogari E, Maffioli P. Exenatide plus metformin compared with metformin alone on β-cell function in patients with Type 2 diabetes. Diabet Med. 2012;29:1515–23. doi: 10.1111/j.1464-5491.2012.03699.x. [DOI] [PubMed] [Google Scholar]
- 101.Derosa G, Franzetti IG, Querci F, Carbone A, Ciccarelli L, Piccinni MN, Fogari E, Maffioli P. Variation in inflammatory markers and glycemic parameters after 12 months of exenatide plus metformin treatment compared with metformin alone: a randomized placebo-controlled trial. Pharmacotherapy. 2013;33:817–26. doi: 10.1002/phar.1301. [DOI] [PubMed] [Google Scholar]
- 102.Gao Y, Yoon KH, Chuang LM, Mohan V, Ning G, Shah S, Jang HC, Wu TJ, Johns D, Northrup J, Brodows R. Efficacy and safety of exenatide in patients of Asian descent with type 2 diabetes inadequately controlled with metformin or metformin and a sulphonylurea. Diabetes Res Clin Pract. 2009;83:69–76. doi: 10.1016/j.diabres.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 103.Seino Y, Takami A, Boka G, Niemoeller E, Raccah D PDY6797 investigators. Pharmacodynamics of the glucagon-like peptide-1 receptor agonist lixisenatide in Japanese and Caucasian patients with type 2 diabetes mellitus poorly controlled on sulphonylureas with/without metformin. Diabetes Obes Metab. 2014;16:739–47. doi: 10.1111/dom.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ratner RE, Rosenstock J, Boka G DRI6012 Study Investigators. Dose-dependent effects of the once-daily GLP-1 receptor agonist lixisenatide in patients with Type 2 diabetes inadequately controlled with metformin: a randomized, double-blind, placebo-controlled trial. Diabet Med. 2010;27:1024–32. doi: 10.1111/j.1464-5491.2010.03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bolli GB, Munteanu M, Dotsenko S, Niemoeller E, Boka G, Wu Y, Hanefeld M. Efficacy and safety of lixisenatide once daily vs. placebo in people with Type 2 diabetes insufficiently controlled on metformin (GetGoal-F1) Diabet Med. 2014;31:176–84. doi: 10.1111/dme.12328. [DOI] [PubMed] [Google Scholar]
- 106.Pinget M, Goldenberg R, Niemoeller E, Muehlen-Bartmer I, Guo H, Aronson R. Efficacy and safety of lixisenatide once daily versus placebo in type 2 diabetes insufficiently controlled on pioglitazone (GetGoal-P) Diabetes Obes Metab. 2013;15:1000–7. doi: 10.1111/dom.12121. [DOI] [PubMed] [Google Scholar]
- 107.Riddle MC, Aronson R, Home P, Marre M, Niemoeller E, Miossec P, Ping L, Ye J, Rosenstock J. Adding once-daily lixisenatide for type 2 diabetes inadequately controlled by established basal insulin: a 24-week, randomized, placebo-controlled comparison (GetGoal-L) Diabetes Care. 2013;36:2489–96. doi: 10.2337/dc12-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Riddle MC, Forst T, Aronson R, Sauque-Reyna L, Souhami E, Silvestre L, Ping L, Rosenstock J. Adding once-daily lixisenatide for type 2 diabetes inadequately controlled with newly initiated and continuously titrated basal insulin glargine: a 24-week, randomized, placebo-controlled study (GetGoal-Duo 1) Diabetes Care. 2013;36:2497–503. doi: 10.2337/dc12-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ahrén B, Leguizamo Dimas A, Miossec P, Saubadu S, Aronson R. Efficacy and safety of lixisenatide once-daily morning or evening injections in type 2 diabetes inadequately controlled on metformin (GetGoal-M) Diabetes Care. 2013;36:2543–50. doi: 10.2337/dc12-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rosenstock J, Hanefeld M, Shamanna P, Min KW, Boka G, Miossec P, Zhou T, Muehlen-Bartmer I, Ratner RE. Beneficial effects of once-daily lixisenatide on overall and postprandial glycemic levels without significant excess of hypoglycemia in type 2 diabetes inadequately controlled on a sulfonylurea with or without metformin (GetGoal-S) J Diabetes Complications. 28:386–92. doi: 10.1016/j.jdiacomp.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 111.Seino Y, Min KW, Niemoeller E, Takami A EFC10887 GETGOAL-L Asia Study Investigators. Randomized, double-blind, placebo-controlled trial of the once-daily GLP-1 receptor agonist lixisenatide in Asian patients with type 2 diabetes insufficiently controlled on basal insulin with or without a sulfonylurea (GetGoal-L-Asia) Diabetes Obes Metab. 2012;14:910–7. doi: 10.1111/j.1463-1326.2012.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu Pan C, Han P, Liu X, Yan S, Feng P, Zhou Z, Lv X, Tian H, Jin Kui Y, Su B, Shang S, Niemoeller E. Lixisenatide treatment improves glycaemic control in Asian patients with type 2 diabetes mellitus inadequately controlled on metformin with or without sulfonylurea: a randomized, double-blind, placebo-controlled, 24-week trial (GetGoal-M-Asia) Diabetes Metab Res Rev. 2014;30:726–35. doi: 10.1002/dmrr.2541. [DOI] [PubMed] [Google Scholar]
- 113.Fonseca VA, Alvarado-Ruiz R, Raccah D, Boka G, Miossec P, Gerich JE EFC6018 GetGoal-Mono Study Investigators. Efficacy and safety of the once-daily GLP-1 receptor agonist lixisenatide in monotherapy: a randomized, double-blind, placebo-controlled trial in patients with type 2 diabetes (GetGoal-Mono) Diabetes Care. 2012;35:1225–31. doi: 10.2337/dc11-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJF, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WHL, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JRB, Platou CGP, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney ASF, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurðsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen AC, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CNa, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka Ta, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel KHJ, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, Mc Carthy MI. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lorenz M, Pfeiffer C, Steinsträsser A, Becker RHA, Rütten H, Ruus P, Horowitz M. Effects of lixisenatide once daily on gastric emptying in type 2 diabetes--relationship to postprandial glycemia. Regul Pept. 2013;185:1–8. doi: 10.1016/j.regpep.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effects of GLP1R agonists on body weight.
Figure S2. Effects of GLP1R-agonists on 2h glucose.
Table S1. Study characteristics for disease traits.
Table S2. Comparison of heterogeneity between trial and rescaled genetic estimates.
Table S3. Details of randomized trials contributing to analyses of GLP1R-agonist effects included in Fig. 2
Table S4. Study characteristics for quantitative traits.