Summary
Liver cancer genome sequencing revealed target pathways for liver cancer prevention/therapy and the reason for genomic instability in liver cancer.
Abstract
The incidence of liver cancer has increased in recent years. Worldwide, liver cancer is common: more than 600000 related deaths are estimated each year. In the USA, about 27170 deaths due to liver cancer are estimated for 2016. Liver cancer is highly resistant to conventional chemotherapy and radiotherapy. For all stages combined, the 5-year survival rate is 15–17%, leaving much to be desired for liver cancer prevention and therapy. Heterogeneity, which can originate from genomic instability, is one reason for poor outcome. About 80–90% of liver cancers are hepatocellular carcinoma (HCC), and recent cancer genome sequencing studies have revealed frequently mutated genes in HCC. In this review, we discuss the cause of the tumor heterogeneity based on the functions of genes that are frequently mutated in HCC. We overview the functions of the genes that are most frequently mutated (e.g. TP53, CTNNB1, AXIN1, ARID1A and WWP1) that portray major pathways leading to HCC and identify the roles of these genes in preventing genomic instability. Notably, the pathway analysis suggested that oxidative stress management may be critical to prevent accumulation of DNA damage and further mutations. We propose that both chromosome instability (CIN) and microsatellite instability (MIN) are integral to the hepatic carcinogenesis process leading to heterogeneity in HCC and that the pathways leading to heterogeneity may be targeted for prognosis, prevention and treatment.
Introduction: liver cancer
Liver cancer is common worldwide, especially in Southeast Asia and sub-Saharan Africa, where hepatitis virus infection is endemic. More than 600000 deaths from liver cancer are estimated worldwide each year. In contrast to an overall decreasing trend in cancer deaths, the incidence of and deaths by liver cancer in the USA have increased in recent years. Approximately 27170 deaths are estimated for 2016 (1). About 90% of liver cancer is hepatocellular carcinoma (HCC); the remaining 10% is cholangiocarcinoma (bile duct cancer). Cirrhosis and nonalcoholic fatty liver disease are prominent risk factors in the USA and are often associated with alcohol abuse and obesity, respectively.
In the USA, the overall 5-year survival rate for patients with liver cancer is 15–17% (1). Surgery is performed at early stages, when the 5-year survival is 31%. However, less than half of all patients with liver cancer are diagnosed at an early stage. Later stage liver cancers are quite resistant to current chemo- and radiotherapies. For patients with later stage cancer, survival rates drop to 11% (regional) and 3% (metastatic). Clinical trials with newer immunotherapies have shown some signs of promise (2), but more time is needed to assess the results on a larger scale. Thus, better therapies to treat liver cancer, along with diagnostic, prognostic and preventive measures for high-risk groups, such as those with hepatitis virus, cirrhosis or nonalcoholic fatty liver disease, and those who have been exposed to dietary aflatoxin, are desperately needed.
Heterogeneity in HCC
Liver cancer is highly heterogeneous in terms of morphology, genome composition and mutated genes (3,4). Not only is HCC heterogeneous among patients, multiple HCCs that occurred in a patient also showed significant heterogeneity (5) and signs of tumor evolution (5–8). The heterogeneity of HCCs in a patient may result in difficulty in clinic for designing customized targeted approach for HCCs in such patients.
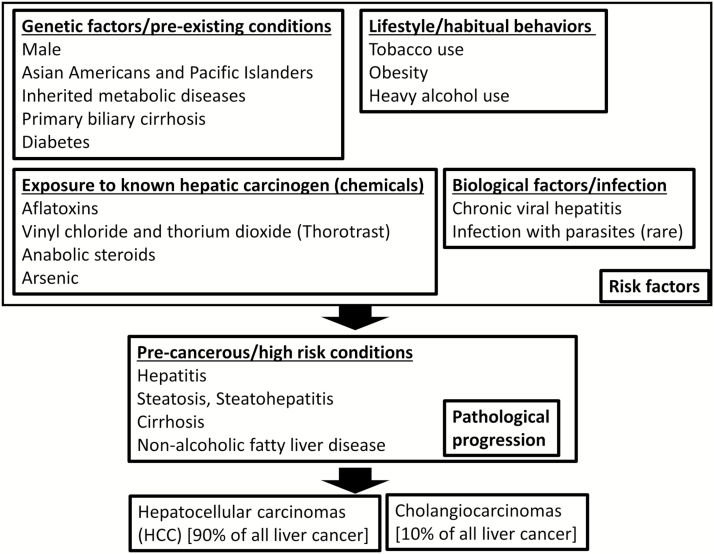
Since 90% of liver cancer is HCC and most studies have been conducted with HCC, in this review, we primarily discuss the results from HCC studies. Risk factors and known causes for HCC include genetic factors (e.g. male gender, metabolic syndrome and diabetes), carcinogens (e.g. aflatoxin), lifestyle/habitual behaviors (e.g. tobacco smoking, alcohol abuse and obesity) and biological factors/infection (e.g. hepatitis virus) (1,3,4). Exposure to the risk factors depends on environment. Thus, there is much variation among geological regions and time frame in liver cancer etiology.
In Africa and Southeast Asia, hepatitis virus and aflatoxin exposure are estimated to be accountable for 70–90% of HCC (9). Hepatitis virus infection is a leading risk factor for HCC in the USA as well. Among 1500 patients at United States Veterans Administration hospitals who developed HCC from 2005 through 2010, the annual proportion of nonalcoholic steatohepatitis-related HCC (7.5–12.0%) and HCC cases associated with hepatitis B (1.4–3.5%) remained relatively stable. The proportion of HCC cases associated with HCV increased from 61.0% in 2005 to 74.9% in 2010, whereas HCC cases associated with alcohol abuse alone decreased from 21.9% in 2005 to 15.7% in 2010 (Figure 1) (10,11). Recent studies utilizing cancer deep sequencing suggest that particular risk factors, such as hepatitis virus infection, aflatoxin exposure or alcohol abuse, may have stronger links to specific sets of pathways and types of HCC; from the standpoint of affected pathways, there may be several HCC subtypes (12–22).
Figure 1.
Risk factors and paths to liver cancer. HCC count as 90% of all liver cancers, whereas remaining 10% as cholangiocarcinoma (cancer in bile duct). Major known risk factors can be categorized as genetic factors, carcinogens, lifestyle/habitual behaviors and biological factors/infection. Although significant numbers of HCC are related to lifestyle, HBV/HCV-positive HCC remain most common (55% of HCC) in the USA [based on Mittal et al. (10)]. Human liver cancer genome sequencing studies have begun to uncover relationship between risk factor/probable causes and mutated genes. Profiling of gene mutations according to the risk factors should follow.
These findings from several groups and HCCs in different regions (especially in USA, Japan and China) indicated complicating factors for genomics approaches. Statistical results from conventional HCC tumor sequencing may be influenced by the composition of the tumor sources. For example, deep sequencing results of HCCs from regions where hepatitis virus infection is endemic may portray a mutation and pathway profile that is different from sequencing results from HCCs resulting from nonalcoholic steatohepatitis/nonalcoholic fatty liver disease. Hence, HCC heterogeneity must be investigated with a view toward identifying both etiological cause and affected pathways in the resulting HCC. This is important because, in the clinic, high heterogeneity means lack of consistency in therapeutic outcome. Targeted therapy, like sorafenib, may prove ineffective due to its limited range of applicability. Therefore, molecular tumor typing should be refined and guidelines for personalized cancer therapy approach should be established with molecular markers to improve therapy and prevention.
Major types of genomic instability: (CIN and MIN)
Many liver cancers show high degrees of genomic instability. Genomic instability is roughly categorized as mitotic error-mediated chromosome instability (CIN) and DNA metabolism defect-mediated microsatellite instability (MIN). Although CIN and MIN can coexist, in most cancers, one form is dominant. In HCC, CIN is prevalent (23,24). CIN is generally linked to poorer prognosis than MIN. One reason for this poor prognosis is that genomic instability can serve as a mutator, increasing the rate of mutation that may permit the cancer to adapt to challenges from therapies (25). CIN in particular can cause large-scale genomic alterations, such as chromothripsis or copy number variations, and is hazardous to genomic integrity (26).
Mouse models for CIN and cancer, including HCC
As stated in the previous section, clinical samples can be affected by etiological fluctuations. As a complementary approach, genetically defined animal models for genomic instability can provide more conclusive proof in terms of biology. In the past years, carcinogenesis studies with CIN mouse models emerged (27–31). Tumor developments were observed in the CIN models, and the models were investigated with more interest in certain organs that proved to be prone to cancer (i.e. lung and liver) or whose cancer carries high CIN (i.e. colon) thus far. With these models, several new findings about CIN on carcinogenesis and on cancer have emerged. In an experiment with a CIN model, mitotic checkpoint component Mad2-overexpressing mice, lung tumor recurrence was enhanced with Mad2 overexpression, indicating that the presence of high CIN can increase recurrence and, hence, poor prognosis (32). In an experiment with another CIN mouse model, regulator of chromosome cohesion and centrosome integrity Sgo1 haploinsufficient mice, spontaneous HCC were observed, and with hepatic and colonic carcinogen Azoxymethane treatments, HCC developed in 7 of 10 Sgo1 mice, whereas of the 9 control mice none developed HCC, showing a significant increase in proneness to HCC (P < 0.05) (33), also supporting the notion of pro-carcinogenic effect of CIN. However, another important notion that emerged is that CIN can also work against carcinogenesis, by increasing cell death (34–36). In an experiment with aged cenpe−/+ mice, spontaneous HCC development rate was lower, which was interpreted as a result of increased cell death in the liver (facilitated by CIN and genomic damage) in the model (34). It is theorized that carcinogenesis or tumor suppression with CIN is a result of a balance between CIN-mediated DNA damage/mutation (with low-grade aneuploidy) and CIN-facilitated cell death (with high-grade aneuploidy) (34–36).
In addition, CIN may influence tumor outcome with other routes than genomic instability and countering cell death. In the Sgo1 mice, colonic transcriptome was altered in pro-carcinogenic manners, including activation of oncogenic signaling (e.g. Wnt, PPAR and insulin), reduced oxidative stress response and reduced immune function (37). Transcriptomic changes were observed in other organs, including the lung and liver, where cancer proneness in the model was found, and the transcriptome alterations showed similar characteristics to the colon to some extent (lung: 38, liver: C.V. Rao et al., in preparation). The transcriptome results suggest that introducing genomic instability in the tissue may create an environment that is pro-carcinogenic and pro-cancer survival (e.g. activation of oncogenic pathway(s), reduced oxidative stress response and reduced immune function) (37,38).
The results from the CIN mouse models showed some similarities to those in human HCC. Wang et al. identified human Sgo1 protein accumulation in human HCC and proposed Sgo1 as a potential target for therapeutic intervention (39). The HCC developed in Sgo1−/+ mouse model also showed Sgo1 accumulation. Moreover, the mouse model study indicated that DNA-damaging reagents induced accumulation of Sgo1 in the nucleus, suggesting that the Sgo1 protein may be accumulated in nucleus in HCC with a possible functional role of DNA damage response or repair (33).
Other paths from genomic instability to carcinogenesis
Further, genomic instability can generate aneuploid cells. Aneuploidy affects the transcriptome and proteome, leading to proteotoxic stress and activation of the endoplasmic reticulum stress response. Therefore, aneuploidy can modulate characteristics of the cells and the microenvironment (40). Cells with genomic damage can go senescent, which may contribute to the deterioration of surrounding tissue functions through senescence-associated secretory phenotype (SASP) (41). SASP can occur in human liver. The hepatocyte SASP included characteristic factors such as interleukin (IL)-8 and IL-6, as well as novel components such as SAA4, IL-32 and fibrinogen (42). The switch in secretome is regulated by Notch 1 in mice hepatocytes. Notch signaling seems to modulate SASP composition in senescent hepatocytes, controlling the immune reaction in the liver and thereby negatively regulating the elimination of senescent hepatocytes, at least in part through suppressing T-lymphocyte recruitment to the liver (43). SASP is involved not only in aging but in HCC therapeutic interventions such as chemotherapy and radiotherapy as well. For example, SASP is shown to be induced by radiation treatments on rat liver in vivo (44). Chemotherapy drug treatment can cause SASP in liver, and attenuation of SASP can promote HCC progression, thus SASP can play a suppressive role on HCC. A variant of histone H2A, macroH2A1, is a marker of senescence-associated heterochromatic foci. Chemotherapeutic and DNA-demethylating agent 5-aza-deoxycytidine (5-aza-dC) induced senescence through epigenetic regulation by macroH2A1 and DNA methylation, and depletion of macroH2A1 amplified the antiproliferative effects of 5-aza-deoxycytidine in HCC cells (45). Thus, genomic instability may lead to tissue dysfunction and carcinogenesis through many emerging paths.
HBV or HCV infection can cause genomic instability in the liver
Genomic instability has a variety of causes, including mutation or epigenetic misregulation in genes that maintain genomic integrity, for example, CIN- or MIN-related genes, virus or retrotransposon, genotoxic agents and radiation or thermal stress. Reduced removal of cells with genomic instability due to immune dysfunction, cell death defect or issues with tissue homeostasis can also increase genomic instability (46). The most relevant genomic instability-inducing agents in the liver are hepatitis virus B (HBV) and hepatitis virus C (HCV) (9). HBV and HCV can induce genomic instability in at least three ways: (i) by interfering with mitotic regulator proteins directly, (ii) by integrating the viral genome into various sites in the host genome and (iii) by causing lingering inflammation in the liver. For (i), HBV viral protein HBX can bind directly to mitotic spindle checkpoint protein BubR1 (47). Overexpressed HCV viral protein NS5A can disturb mitotic processes and cause genomic instability (48). Interfering with these mitotic regulator proteins can cause CIN-type genomic instability. For (ii), HBV encodes the regulatory HBx protein, the primary role of which is to promote transcription of the viral genome, and the viral genome then persists as an extrachromosomal DNA circle in infected cells (49). Viral genome integration occurs at various sites in the genome, but when integration occurs at a locus where a carcinogenesis-relevant gene lies and interferes with the gene’s function, the effect may later be detected. For (iii), genomic instability can result in cell death. Perhaps due to an increase in dying cells, inflammation markers and inflammatory cytokines like IL6 tend to be high in tissues with genomic instability. In Sgo1 CIN model mice, hepatic IL6 expression and serum HCC marker alpha-fetoprotein amount increased, consistent with the notion that genomic instability alone can lead to sterile tissue inflammation in liver and other organs (33).
Are the genes and pathways that are frequently mutated in HCC involved in genomic instability?
Earlier, we examined the functions of genes that are frequently mutated in colon cancer in order to elucidate why CIN is so prevalent in colon cancer (80–90% of occurrence) and identified the progressive nature of CIN in colon cancer. That is, many of the frequently mutated genes in colon cancer (e.g. APC, TP53 and FBXW7) maintain mitotic fidelity, thereby preventing CIN. As mutations accumulate, degree of CIN progressively increases (31). Because HCC also exhibits a high degree of heterogeneity and genomic instability that may or may not be attributed solely to hepatitis virus infection, we hypothesized that the genes frequently mutated in HCC may also have functions of maintaining genomic stability and examined the functions of the frequently mutated genes and pathways from the standpoint of their relations to genomic instability.
Contemporary omics approaches and tumor mass-sequencing have identified various gene mutations, genome alterations and epigenetic misregulations in several tumors, including those in liver cancer (12–22,50–52). Additionally, technological advances that enabled omics approaches with smaller number of samples (e.g. single-cell sequencing) led to mutational linage analyses in one tumor or among multiple tumors (5–8). Demonstrating cancer development (or mutational ‘evolution’ during the cancer development) and increasing degree of heterogeneity has become possible through phylogenic analysis (5–8).
A characteristic trait in genetic/genomic alterations in liver cancer is the broad spectrum of apparent mutations and limited number of encompassing mutations. For example, 88–100% of pancreatic cancers show k-ras mutation, which strongly suggests that k-ras mutation may serve as a critical ‘bottle neck’ initiator in pancreatic carcinogenesis. In contrast, except for telomerase reverse transcriptase (TERT) promoter (60% mutation rate), TP53/p53 (35–50% mutation rate) and CTNNB1/beta-catenin (19–40%), mutation rates for the most frequently mutated genes in HCC are relatively low [e.g. AXIN1 (13%), ARID1A (12%) and WWP1 (9%)]. Although there is some variation among studies, most of the ‘most frequently mutated genes’ were mutated in <10% of HCC samples examined (12–22,50–52). In addition, microRNA expressions and epigenetic factors, such as EZH2, were determined to play a role in HCC, adding more complexity to HCC tumor heterogeneity (50).
In the following sections, we will discuss genes that are frequently mutated in human HCC, identified by recent genomic analyses based on next-generation sequencing (NGS). The genes are selected based on higher mutation rates in genome sequencing data [compiled from references (12–22, 50–52); for most recent review, see ref. (14)] and presented in order from higher mutation rate to lower rate. The overview of the genes that are frequently mutated in human HCC will focus on most heavily mutated genes, with cutoff at approximately 2–3% mutation rate (12–22,50–52).
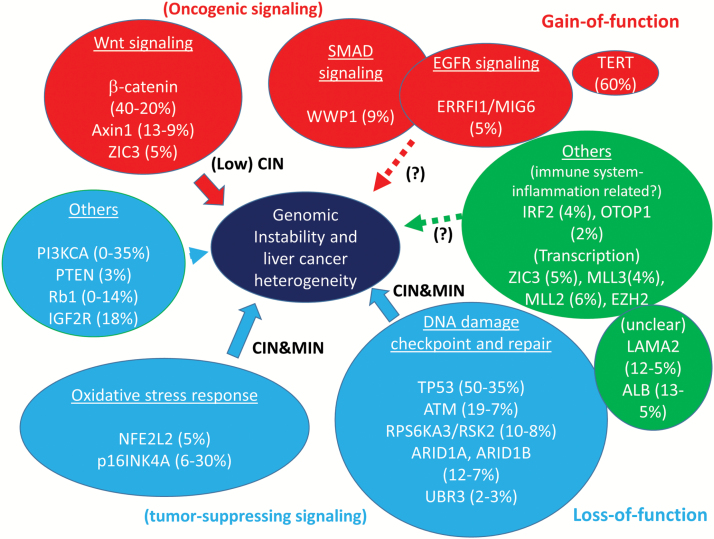
By collating the genes and identifying the pathways to which they belong, several signaling pathways that may contribute to HCC development were identified: (i) oncogenic pathways (Wnt, Smad and EGFR), (ii) DNA damage checkpoint and repair pathway, (iii) oxidative stress response pathway, (iv) cell cycle pathway and (v) immune function pathways (Figure 2).
Figure 2.
Genes that are most frequently mutated in HCC, involved pathways, and their effects on genomic instability. Mutation rate estimates are based on NGS studies on HCC (12–22,50–52). Mutation rates and functions of individual gene are well-summarized in a table in ref. (14). Wnt signaling, Smad signaling and EGFR oncogenic signaling are influenced by gain-of-function mutations. Activation of Wnt signaling can lead to a low degree of CIN. Tumor suppressor pathways show loss of function. Oxidative stress response and DNA damage checkpoint and repair pathways are also affected. Notably, mutations in these pathways can cause severe DNA damage and genomic instability through CIN and MIN, especially when combined.
Then, we investigated how the pathways may be involved in maintaining genomic integrity. Some mutations could cause genomic instability by itself (e.g. TP53, Wnt signaling genes including CTNNB1, AXIN1; chromatin modulators including ARID1, MLL2/KMT2D; and DNA damage checkpoint or repair genes including ATM). In addition, there are pathways whose simultaneous loss can cause additive or synergistic damage to genome integrity. Namely, when the DNA damage checkpoint and repair and oxidative stress response pathways are simultaneously impaired, extensive DNA damage or proneness to DNA damage can occur. Unrepaired or misrepaired DNA double-strand breaks lead to the formation of chromosome aberrations. Replication stress (53) and inappropriate activation of the DNA damage repair pathway during mitosis (54) have also been shown to cause CIN.
Genes that are frequently mutated in liver cancer: their role and involvement in genomic instability
In this section, we will discuss genes that are frequently mutated in HCC individually. The genes are selected based on higher mutation rates in genome sequencing data [compiled from references (12–22,50–52)] and presented in the order from higher mutation rate to lower rate. Genes with <2% mutation rate are not discussed in this review.
TERT promoter (gain of function, 60%)
This gene (including the promoter region) has a mutation rate of 60% in HCC. It is associated with increased telomerase expression/gain of function. Mutations in TERT promotor created an E-twenty-six family/T-cell factor transcription factor binding site and induced telomerase promoter activity and TERT transcription. They were among those most frequently found in liver cancer, leading to overactive telomerase (52). Reduction of telomerase and telomere shortening is associated with DNA double-strand breaks, genomic instability and senescence. In contrast, overexpression of telomerase elongates telomeres and aids immortalizing cells, benefiting cancer cell survival.
TP53 (loss of function, 35–50%)
TP53 encodes p53, one of most investigated tumor suppressors with multiple functions. Wild-type p53 suppresses the accumulation of aneuploid cells (55) and plays a major role in maintaining genomic stability. Loss of p53 function can permit survival or propagation of aneuploid cells, especially when combined with mitotic error-generating mutations, such as spindle checkpoint defects and/or Rb defect (56,57). p53 mutation can produce centrosome amplification and CIN (58). Laurent-Puig et al. reported that high CIN in HCC is associated with p53 mutation and hepatitis virus infection, whereas low CIN is associated with beta-catenin mutation (59). The association is consistent with the notion that p53 mutation can lead to CIN. However, p53 mutation alone may not be a strong driver of CIN, and additional mutation(s) may be needed to develop cancer.
CTNNB1 (gain of function, 20–40%)
Beta-catenin (gene: CTNNB1) is the effector of Wnt signaling. The C-terminus region is found to be heavily mutated in cancers including HCC. Since the amino terminus is involved in degradation of β-catenin, the deletion likely results in its stabilization, leading to activation of Wnt signaling (gain of function) (19). Its gain of function activates various pro-growth genes as a transcription factor. Overactive Wnt signaling is observed in a variety of cancers, and Wnt signaling is considered oncogenic. β-Catenin–induced T-cell lymphoma by promoting genomic instability. Activated beta-catenin altered double-strand break repair and increased survival of thymocytes with damaged DNA, which promoted genomic instability and formation of T-cell lymphomas (60). Inhibitors of a Wnt signaling component glycogen synthase kinase 3 (GSK-3) induced chromosome instability (61). Overall, these reports suggest that beta-catenin and Wnt signaling activation can cause genomic instability, especially when combined with an increase in DNA damage or mismatch repair defects, which often occur in the process of HCC development.
AXIN1 (Loss of function, mutation rate, 9–13%)
AXIN1 is also a component of Wnt signaling, and the protein interacts with other Wnt signaling proteins, adenomatosis polyposis coli (APC), β-catenin (CTNNB1), GSK3β, protein phosphatase 2 and itself. However, despite belonging to the same pathway, genetic alterations in CTNNB1 and AXIN1 are mutually exclusive, possibly because they carry opposite roles in terms of pathway activation (14). AXIN1 is a negative regulator of Wnt signaling, and AXIN1 loss of function activates Wnt signaling. Ectopic expression of Axin1 paralog Axin2 or its up-regulation through small interfering RNA-mediated knockdown of adenomatosis polyposis coli, led to CIN in chromosomally stable colon cancer cells, indicating the capability of Wnt signaling to induce genomic instability (62).
LAMA2 (loss of function, mutation rate, 5–12%)
LAMA2 gene encodes Laminin subunit alpha 2, a major component of the muscle basement membrane, and mutations in this gene have been identified as the cause of congenital merosin-deficient muscular dystrophy, an autosomal recessive disease typically presenting as a severe, early onset congenital muscular dystrophy (63). Laminin-α2 is expressed in skeletal muscle myoblasts and myotubes where it promotes the survival of satellite cells as well as myoblast fusion and myotube formation (64). The mutation in HCC was identified in a 2014 study with a high frequency (6 of 42, 12%), and retrospective analysis in other HCC studies also reported 6% and 5% mutation rates (19). LAMA2 is frequently mutated in other cancers, including lung adenocarcinoma (11%), lung squamous cell carcinoma (13%), uterine corpus endometrioid carcinoma (13%) and head and neck squamous cell carcinoma (10%) (19). LAMA2 is suggested to play a role as a tumor suppressor (19). LAMA2−/− mice were viable, but showed signs of muscle dystrophy and died by 5 weeks of age (65). LAMA2−/− cells showed a higher percentage of mononucleic population (65). However, its link to genomic instability has not been demonstrated.
ARID1A and ARID2 (ARID1A: loss of function, 12%. ARID2: loss of function, 7%)
ARID1A (AT-rich interactive domain-containing protein 1, aka BAF250a) and ARID2/ARID1B (BAF200) are members of the switch (SWI)/sucrose non-fermenting (SNF) family and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. As the loss of ARID1A increases cancer incidence in several organs (e.g. breast, gatrointestinal tract and ovary) and is a marker for poor prognosis, it is considered to be a tumor suppressor. ARID1A function is also implicated in regulation of mismatch repair, as its loss is associated with MIN. For example, in microsatellite unstable colorectal cancer, frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A were observed (66). Hepatocyte-specific Arid1a deficiency initiated steatohepatitis and HCC in mice (67). Reduced expression of ARID1A is associated with poor prognosis and promotes HCC metastases. Consistently, siRNA-targeted loss of function of SWI/SNF chromatin remodeling genes led to genomic instability with increased mutation rates in human lung cancer cells (68). In Japanese study, inactivating mutations were significantly enriched in non-HBV and non-HCV patients, suggesting a key tumor suppressor function of SWI/SNF complexes in metabolic/toxic rather than virus-related HCC (12).
WWP1 [9%: functional link unclear (possibly, gain of function)]
WWP1 (WW domain-containing Protein 1/NEDD4-like E3 ubiquitin-protein ligase) is a E3 ubiquitin ligase that can target multiple substrates, including KLF2, TGF beta, tumor suppressor smad4, p27, ErbB4, ErbB2 and EGFR. Involvement of WWP1 in regulations of oncogenic signaling with Smad4 and EGFR appears particularly relevant in hepatic carcinogenesis. Overexpression of WWP1 promoted tumorigenesis and predicted unfavorable prognosis in patients with HCC (69).
Some WWP1 targets are involved in genomic instability. For example, Smad4 loss in mice caused spontaneous head and neck cancer with increased genomic instability and inflammation (70). p27kip1 Deficiency impaired G2/M arrest in response to DNA damage, leading to an increase in genetic instability (71). Therefore, WWP1 misregulation, especially gain of function, can indirectly cause genomic instability.
RPS6KA3 (90 kDa, polypeptide 3/RSK2; likely loss of function, mutation rate, 8%)
This gene encodes a member of the ribosomal S6 kinase (RSK) family of serine/threonine kinases. Mutations in RPS6KA3 cause Coffin–Lowry syndrome, a rare X-linked dominant disorder characterized by intellectual disability, craniofacial abnormalities, short stature, tapering fingers, hypotonia and skeletal malformations (72). RSK2 affects p53-mediated downstream cellular events in response to DNA damage. RSK2 knockout relieves cell cycle arrest at the G2/M phase and an increased number of γH2AX foci, which are associated with defects in DNA repair. Therefore, RSK2 plays an important role in the DNA damage pathway that maintains genomic stability by mediating cell cycle progression and DNA repair (73).
ATM (Likely loss of function, mutation rate, 7%)
Ataxia telangiectasia mutated (ATM) is a kinase involved in DNA damage checkpoint and repair. Loss of ATM reduced hepatocellular apoptosis and fibrosis in a high fat-fed mouse model of nonalcoholic fatty liver disease, indicating that activation of ATM in response to oxidative stress plays a role in development of hepatic fibrosis (74).
Further work with mouse models indicated that ATM plays a role in suppressing intestinal carcinogenesis when combined with apc; therefore, ATM is a tumor suppressor. Defects in ATM inactivate p53BP1-MDC1-nbs-mediated DNA checkpoint and repair and sensitize cells to DNA damage and oxidative stress. ATM and p21 cooperate to suppress aneuploidy and subsequent tumor development (75). ATM defects alone may not be strong drivers for genomic instability but with other mutations (e.g. p21), genomic instability can manifest. Dramatically increased CIN was observed in brains of human ATM patients (76). As in these examples, ATM defects can make cells prone to increased MIN and CIN.
CDKN2A [loss of function (most cases), mutation rate, 6–30%]
CDKN2A encodes p16INK4A, which is a CDK inhibitor, a senescent protein, and a tumor suppressor. p16 suppresses cell cycle progression and inducing senescence. siRNA-mediated inhibition of p16 led to an increase in oxidative stress, that is, ROS and oxidative (8-oxoguanine) DNA damage (77), suggesting that loss of p16 can worsen genomic instability if combined with DNA damage checkpoint and repair defects. Consistently, p16INK4A-silencing augments DNA damage-induced apoptosis in cervical cancer cells (78). Oxidative stress can cause genomic instability by both CIN and MIN.
MLL2/KMT2D (loss of function, mutation rate, 6%)
KMT2D encodes lysine methyltransferase 2D and functions as a histone methyltransferase. Truncating mutations in the KMT2D gene have been identified in people with Kabuki syndrome, a disorder characterized by distinctive facial features, intellectual disability and abnormalities affecting other parts of the body (79). KMT2D mutation can be causal to B-cell leukemia, thus KMT2D is considered a tumor suppressor (80,81). Increasing evidence supports that KMT2D is involved in the regulation of gene expression. In B cells, KMT2D sustains gene expression program that represses B-cell lymphoma development (80). Disruption of KMT2D perturbs germinal center B-cell development and promotes lymphomagenesis (81,82). Kantidakis et al. identified KMT2D as an interacting protein with RECQL5, which associates with RNAPII subunits and several transcription-related factors and also with genomic instability. The authors suspected involvement of KMT2D in transcription. Although showing only small transcriptional changes, mouse cells in which KMT2D gene deletion can be induced, along with human cells with KMT2D knockout, display elevated levels of sister chromatid exchange, gross chromosomal aberrations, 53BP1 foci and micronuclei. KMT2D mutation gives rise to significant genomic instability in areas overlapping with early replicating fragile sites. This may be explained by the involvement of KMT2D in transcript elongation and specifically in mediating elongation-associated histone H3K4 methylation (82). Thus, KMT2D mutation can cause transcriptional stress, creation of fragile sites and genomic instability.
NFE2L2 (loss of function, mutation rate, 5%)
Nuclear factor [erythroid-derived 2]-like 2 (NFE2L2), or Nrf2, is a transcription factor with basic leucine zipper (bZIP). Nrf2 regulates the expression of antioxidant proteins that protect against oxidative damage triggered by injury and inflammation. Several drugs that stimulate the NFE2L2 pathway are being studied for the treatment of diseases that are caused by oxidative stress. In myelofibrosis and related blood cancers, Nrf2 expression is downregulated. Nrf2 knockout enhances intestinal tumorigenesis in Apc (min/+) mice due to attenuation of the antioxidative stress pathway, which potentiates inflammation (83). These findings illustrate the importance of Nrf2-mediated oxidative stress management in preventing carcinogenesis. Although evidence for Nrf2 defects directly causing genomic instability has not been found, Nrf2 defects may make cells prone to oxidative stress-mediated DNA damage. If combined with DNA damage checkpoint and repair defects, such as ATM or TP53 defect, genomic instability would follow under oxidative challenge.
ERBB receptor feedback inhibitor 1/MIG6 [loss of function (which activates EGFR pathway), mutation rate, 5%]
MIG6 is a multifunctional adaptor protein and binds, in particular, to the epidermal growth factor receptor (EGFR) kinase domain and inhibits signaling. The EGRF-mitogen-inducible gene 6 (MIG6) signaling axis plays a role in lung cancer (84). Mig6 is a sensor of EGFR inactivation that directly activates c-Abl to induce apoptosis during epithelial homeostasis. Hepatocyte growth factor (HGF)/Met signaling can also be inhibited by Mig6 in cells of hepatic origin and neurons, affecting cell migration, in a Cdc42/Rac-dependent manner. Thus, MIG6 is involved in the inhibition of the EGFR and HGF signaling pathways, and loss of function in MIG6 activates these oncogenic pathways. When MIG6 gene expressions were compared in HCC and hepatoblastomas, hepatoblastomas showed 3.88-fold higher expression. As hepatoblastomas tend to harbor fewer genomic alterations, low expression of MIG6 may be involved in genomic instability. Although circumstantial evidence exists, a direct link between misregulation of MIG6-EGFR or HGF and genomic instability in the liver remains to be found.
ZIC3 (mutation rate, 5%)
ZIC3 is a member of the zinc finger of the cerebellum (ZIC) protein family and is a part of the transcription factor complexes involved in development, especially of the heart, and morphogenesis. During gastrulation and neurulation, Zic3 acts by binding the distal regulatory regions associated with control of gene transcription in the Nodal and Wnt signaling pathways. Zic3 is also involved in regulation of stem cells and generation/conversion to pluripotent progenitor cells. In a zebrafish model, Zic3 was implicated in the regulation of BMP, Wnt and FGF (85). Consistent with the ability of Zic3 to control multiple development or oncogenic pathways, mutations and expression abnormalities of Zic3 have been found in other cancers, including lung, blood and brain cancer (86).
ALB (serum albumin; functional link unclear, mutation rate, 5%)
Serum albumin is synthesized in the liver and is involved in the transport of various biomolecules, including hormones, fatty acids, unconjugated bilirubin, drugs and ions. In an in vitro model with rat primary neuronal cultured cells, serum albumin-attenuated DNA damage, and the effect was suggested to be due to antioxidant properties through catalase activation. However, analbuminemia alone did not significantly influence hepatocarcinogenesis when F344 rats were compared with a congenic line carrying the analbuminemic mutation (87). Thus, alb mutation may indirectly contribute to oxidative stress management and affect the microenvironment.
MLL3/KMT2C (loss of function, mutation rate, 4%)
This gene is a member of the myeloid/lymphoid or mixed-lineage leukemia (MLL) family and encodes a nuclear protein with an AT hook DNA-binding domain, a DHHC-type zinc finger, six PHD-type zinc fingers, a SET domain, a post-SET domain, and a RING-type zinc finger. MLL3 is a member of the ASC-2/NCOA6 histone–methyltransferase complex (ASCOM) and is involved in transcriptional coactivation and epigenetic regulation. MLL3 can serve as a p53 coactivator and functions as tumor suppressor (88). Mutations in MLL3 were found in various tumors, including bladder, colon, lung and pancreas tumors. Low expression of MLL3 was linked with a low survival rate compared with positive MLL3 expression in patients with gastric cancer. MLL3 may function like aforementioned MLL2/KMT2D and may be involved in chromatin positioning and genomic instability (89). Additionally, MLL3 may affect chemoresistance. Loss of the MLL3/4 complex protein, PTIP, protects Brca1/2-deficient cells from DNA damage and rescues the lethality of Brca2-deficient embryonic stem cells (90).
IRF2 (likely loss of function, mutation rate, 4%)
Interferon regulatory factor 2 (IRF2) is thought to be an oncoprotein. IRF2 competitively inhibits the IRF1-mediated transcriptional activation of interferons alpha and beta and presumably other genes that employ IRF1 for transcription activation. Although siRNA-mediated knockdown of IRF2 in leukemic TF-1 cells resulted in growth inhibition associated with G2/M arrest as well as induction of polyploidy, differentiation and apoptosis (91), forced expression of the IRF2 caused polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells (92). Thus, IRF2 misregulation may result in cell cycle miscoordination and polyploidy, which may promote genomic instability and transformation.
BAZ2B (Functional link unclear, mutation rate, 3%)
BAZ2B (bromodomain adjacent to zinc finger domain 2B) is a bromodomain-containing protein whose function remains to be determined. Bromodomain proteins are thought to interact with acetylated lysine and are found in many chromatin remodeling complexes (93).
UBR3 (functional link unclear, mutation rate, 2%)
Ubr3 (ubiquitin protein ligase E3 component N-recognin 3) E3 ubiquitin ligase regulates apoptosis by controlling the activity of DIAP1 in Drosophila. UBR3 regulates cellular levels of APE1/Ref-1, a protein essential for DNA damage repair and transcription, thus indirectly affect DNA damage repair pathway. Knockout of Ubr3 in MEF led to genomic instability and cell death (94). Recent report identified Ubr3 as a novel, positive regulator of Hedgehog (Hh) signaling in Drosophila and vertebrates through Ubiquitylation of Costal2/Kif7, a central component of Hh signaling (95). As haploinsufficiency of Patched-1, a gene that encodes a repressor of the Hh pathway, dysregulates the Hh pathway and increases genomic instability, it was suggested that inappropriate Hh pathway activation may promote tumorigenesis by disabling a key signaling pathway that helps maintain genomic stability and inhibits tumorigenesis (96).
Mutated genes for which little information is available
Limited information is available for the functions of IGSF10 and ZNF226. IGSF10 (immunoglobulin superfamily member 10) and ZNF226 (zinc finger protein 226) are involved in loss of function and both have a mutation rate of 5% in HCC.
Summary
After examining the functions of the frequently mutated genes from a genomic instability standpoint, a picture emerged. First, HCC development is facilitated by the activation of oncogenic pathways, such as the Wnt (beta-catenin, Axin1 and ZIC3), Smad (WWP1) and EGFR (MIG6) pathways. Further, maintaining both the oxidative stress response (i.e. NFE2L2) and efficient DNA damage checkpoint and repair (i.e. TP53, ATM, RPS6KA3, ARID1A and ARID1B) pathways is important to prevent the genomic instability that leads to HCC. Combined defects in the oxidative stress response and DNA damage checkpoint and repair pathways in particular result in high DNA damage and genomic instability. Thus, we suggest a few approaches that may be administered individually or simultaneously: (i) to attenuate Wnt and/or EGFR signaling, (ii) to reduce ROS through antioxidants supplementation and (iii) to reduce DNA damage by controlling exposure to DNA-damaging agents or other DNA damaging events, such as inflammation or alcohol consumption. Alternatively, biomarker identification and immunotargeting or small molecule-mediated approaches may be used to eliminate cells with defects in these pathways. The pathway analysis should be refined and risk factors should be considered to optimize prevention approaches.
Funding
This work was supported by grants from the U.S. NIH NCI (no. R01CA094962 to C.V.R), the U.S. V.A. merit (grant no. 1I01BX003198-01 to C.V.R), and the Stephenson Cancer Center (H.Y.Y.).
Acknowledgements
We thank Kathy Kyler for editorial help.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATM
Ataxia telangiectasia mutated
- CIN
chromosomal instability
- EGFR
epidermal growth factor receptor
- HBV
hepatitis virus B
- HBC
hepatitis virus C
- HCC
hepatocellular carcinoma
- IL
interleukin
- IRF2
interferon regulatory factor 2
- MIG6
mitogen-inducible gene 6
- MLL
mixed-lineage leukemia
- NFE2L2
nuclear factor [erythroid-derived 2]-like 2
- MIN
microsatellite instability
- RSK
ribosomal S6 kinase
- SASP
senescence-associated secretory phenotype
- ZIC
zinc finger of the cerebellum.
References
- 1. American Cancer Society. (2016) http://www.cancer.org/cancer/livercancer/detailedguide/liver-cancer-what-is-key-statistics (9 December 2016, date last accessed)
- 2. Aerts M., et al. (2016) Current status and perspectives of immune-based therapies for hepatocellular carcinoma. World J. Gastroenterol., 22, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kanda M., et al. (2015) Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J. Gastroenterol., 21, 10584–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeng K.S., et al. (2015) Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit. Rev. Oncol. Hematol., 94, 337–347. [DOI] [PubMed] [Google Scholar]
- 5. Xue R., et al. (2016) Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology, 150, 998–1008. [DOI] [PubMed] [Google Scholar]
- 6. Gerlinger M., et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med., 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerlinger M., et al. (2014) Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet., 46, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexandrov L.B., et al. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Martel C., et al. (2015) World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology, 62, 1190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mittal S., et al. (2015) Temporal trends of nonalcoholic fatty liver disease-related hepatocellular carcinoma in the veteran affairs population. Clin. Gastroenterol. Hepatol., 13, 594–601.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singal A.G., et al. (2015) Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin. Gastroenterol. Hepatol., 13, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujimoto A., et al. (2012) Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet., 44, 760–764. [DOI] [PubMed] [Google Scholar]
- 13. Fujimoto A., et al. (2016) Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet., 48, 500–509. [DOI] [PubMed] [Google Scholar]
- 14. Schulze K., et al. (2016) Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol., 65, 1031–1042. [DOI] [PubMed] [Google Scholar]
- 15. Schulze K., et al. (2015) Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet., 47, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tao Y., et al. (2011) Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc. Natl Acad. Sci. USA, 108, 12042–12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang J., et al. (2012) Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet., 44, 1117–1121. [DOI] [PubMed] [Google Scholar]
- 18. Miao R., et al. (2014) Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J. Hepatol., 61, 840–849. [DOI] [PubMed] [Google Scholar]
- 19. Jhunjhunwala S., et al. (2014) Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol., 15, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li S., et al. (2013) Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett., 340, 247–253. [DOI] [PubMed] [Google Scholar]
- 21. Marquardt J.U., et al. (2012) Next-generation sequencing: application in liver cancer-past, present and future? Biology (Basel), 1, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z. (2012) Genomic landscape of liver cancer. Nat. Genet., 44, 1075–1077. [DOI] [PubMed] [Google Scholar]
- 23. Kondo Y., et al. (2000) Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis—a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology, 32, 970–979. [DOI] [PubMed] [Google Scholar]
- 24. Nishida N., et al. (2003) Chromosomal instability and human hepatocarcinogenesis. Histol. Histopathol., 18, 897–909. [DOI] [PubMed] [Google Scholar]
- 25. Burrell R.A., et al. (2013) The causes and consequences of genetic heterogeneity in cancer evolution. Nature, 501, 338–345. [DOI] [PubMed] [Google Scholar]
- 26. Zhang C.Z., et al. (2015) Chromothripsis from DNA damage in micronuclei. Nature, 522, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foijer F., et al. (2008). Studying chromosome instability in the mouse. Biochim. Biophys. Acta, 1786, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ricke R.M., et al. (2008). Whole chromosome instability and cancer: a complex relationship. Trends Genet., 24, 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rao C.V., et al. (2009) Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis, 30, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schvartzman J.M., et al. (2010) Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer, 10, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rao C.V., et al. (2013) Genomic instability and colon carcinogenesis: from the perspective of genes. Front. Oncol., 3, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sotillo R., et al. (2011) Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature, 464, 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamada H.Y., et al. (2015) Tumor-promoting/progressing role of additional chromosome instability in hepatic carcinogenesis in Sgo1 (Shugoshin 1) haploinsufficient mice. Carcinogenesis, 36, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weaver B.A., et al. (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell, 11, 25–36. [DOI] [PubMed] [Google Scholar]
- 35. Silk A.D., et al. (2013) Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl Acad. Sci. USA, 110, E4134–E4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zasadil L.M., et al. (2016) High rates of chromosome missegregation suppress tumor progression but do not inhibit tumor initiation. Mol. Biol. Cell, 27, 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao C.V., et al. (2016) Systemic Chromosome instability resulted in colonic transcriptomic changes in metabolic, proliferation, and stem cell regulators in Sgo1-/+ mice. Cancer Res., 76, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamada H.Y., et al. (2016) Systemic chromosome instability in Shugoshin-1 mice resulted in compromised glutathione pathway, activation of Wnt signaling and defects in immune system in the lung. Oncogenesis, 5, e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L.H., et al. (2015) Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget, 6, 2023–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheltzer J.M., et al. (2012) Transcriptional consequences of aneuploidy. Proc. Natl Acad. Sci. USA, 109, 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ovadya Y., et al. (2014) Senescent cells: SASPected drivers of age-related pathologies. Biogerontology, 15, 627–642. [DOI] [PubMed] [Google Scholar]
- 42. Irvine K.M., et al. (2014) Senescent human hepatocytes express a unique secretory phenotype and promote macrophage migration. World J. Gastroenterol., 20, 17851–17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoare M., et al. (2016) NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol., 18, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Serra M.P., et al. (2014) Hepatocyte senescence induced by radiation and partial hepatectomy in rat liver. Int. J. Radiat. Biol., 90, 876–883. [DOI] [PubMed] [Google Scholar]
- 45. Borghesan M., et al. (2016) DNA hypomethylation and histone variant macroh2a1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res., 76, 594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rao C.V., et al. (2016) Emerging links among Chromosome Instability (CIN), cancer, and aging. Mol. Carcinog., doi:10.1002/mc.22539. [DOI] [PubMed] [Google Scholar]
- 47. Kim S., et al. (2008) HBV X protein targets hBubR1, which induces dysregulation of the mitotic checkpoint. Oncogene, 27, 3457–3464. [DOI] [PubMed] [Google Scholar]
- 48. Baek K.H., et al. (2006) Overexpression of hepatitis C virus NS5A protein induces chromosome instability via mitotic cell cycle dysregulation. J. Mol. Biol., 359, 22–34. [DOI] [PubMed] [Google Scholar]
- 49. Decorsière A., et al. (2016) Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature, 531, 386–389. [DOI] [PubMed] [Google Scholar]
- 50. Ozen C., et al. (2013) Genetics and epigenetics of liver cancer. N. Biotechnol., 30, 381–384. [DOI] [PubMed] [Google Scholar]
- 51. van Malenstein H., et al. (2011) Molecular classification of hepatocellular carcinoma Anno 2011. Eur J Cancer, 47, 1789–1797. [DOI] [PubMed] [Google Scholar]
- 52. Zucman-Rossi J., et al. (2015) Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology, 149, 1226–1239. [DOI] [PubMed] [Google Scholar]
- 53. Burrell R.A., et al. (2013) Replication stress links structural and numerical cancer chromosomal instability. Nature, 494, 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bakhoum S.F., et al. (2014) DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov., 4, 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thompson S.L., et al. (2010) Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol., 188, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schvartzman J.M., et al. (2011) Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell, 19, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manning A.L., et al. (2014) Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation. Oncogene, 33, 2487–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kawamura K., et al. (2004) Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res., 64, 4800–4809. [DOI] [PubMed] [Google Scholar]
- 59. Laurent-Puig P., et al. (2001) Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology, 120, 1763–1773. [DOI] [PubMed] [Google Scholar]
- 60. Dose M., et al. (2014) β-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl Acad. Sci. USA, 111, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tighe A., et al. (2007) GSK-3 inhibitors induce chromosome instability. BMC Cell Biol., 14, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hadjihannas M.V., et al. (2006) Aberrant Wnt/beta-catenin signaling can induce chromosomal instability in colon cancer. Proc. Natl Acad. Sci USA, 103, 10747–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Helbling-Leclerc A., et al. (1995) Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat. Genet., 11, 216–218. [DOI] [PubMed] [Google Scholar]
- 64. Vachon P.H., et al. (1996) Merosin and laminin in myogenesis; specific requirement for merosin in myotube stability and survival. J. Cell Biol., 134, 1483–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miyagoe Y., et al. (1997) Laminin alpha2 chain-null mutant mice by targeted disruption of the Lama2 gene: a new model of merosin (laminin 2)-deficient congenital muscular dystrophy. FEBS Lett., 415, 33–39. [DOI] [PubMed] [Google Scholar]
- 66. Cajuso T., et al. (2014) Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer, 135, 611–623. [DOI] [PubMed] [Google Scholar]
- 67. Fang J.Z., et al. (2015) Hepatocyte-specific Arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS One, 10, e0143042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang H.T., et al. (2015) Loss of function of SWI/SNF chromatin remodeling genes leads to genome instability of human lung cancer. Oncol. Rep., 33, 283–291. [DOI] [PubMed] [Google Scholar]
- 69. Zhang X.F., et al. (2015) Overexpression of WWP1 promotes tumorigenesis and predicts unfavorable prognosis in patients with hepatocellular carcinoma. Oncotarget, 6, 40920–40933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bornstein S., et al. (2009) Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J. Clin. Invest., 119, 3408–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Payne S.R., et al. (2008) p27kip1 deficiency impairs G2/M arrest in response to DNA damage, leading to an increase in genetic instability. Mol. Cell. Biol., 28, 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nishimoto H.K., et al. (2014) The historical Coffin-Lowry syndrome family revisited: identification of two novel mutations of RPS6KA3 in three male patients. Am. J. Med. Genet. A, 164A, 2172–2179. [DOI] [PubMed] [Google Scholar]
- 73. Lim H.C., et al. (2013) Ribosomal S6 Kinase 2 (RSK2) maintains genomic stability by activating the Atm/p53-dependent DNA damage pathway. PLoS One, 8, e74334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Daugherity E.K., et al. (2012) The DNA damage checkpoint protein ATM promotes hepatocellular apoptosis and fibrosis in a mouse model of non-alcoholic fatty liver disease. Cell Cycle, 11, 1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shen K.C., et al. (2005) ATM and p21 cooperate to suppress aneuploidy and subsequent tumor development. Cancer Res., 65, 8747–8753. [DOI] [PubMed] [Google Scholar]
- 76. Iourov I.Y., et al. (2009) Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum. Mol. Genet., 18, 2656–2669. [DOI] [PubMed] [Google Scholar]
- 77. Jenkins N.C., et al. (2011) The p16(INK4A) tumor suppressor regulates cellular oxidative stress. Oncogene, 30,265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lau W.M., et al. (2007) p16INK4A-silencing augments DNA damage-induced apoptosis in cervical cancer cells. Oncogene, 26, 6050–6060. [DOI] [PubMed] [Google Scholar]
- 79. Guo C., et al. (2013) KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget, 4, 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ortega-Molina A., et al. (2015) The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med., 21, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang J., et al. (2015) Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med., 21, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kantidakis T., et al. (2016) Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev., 30, 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cheung K.L., et al. (2014) Nrf2 knockout enhances intestinal tumorigenesis in Apc(min/+) mice due to attenuation of anti-oxidative stress pathway while potentiates inflammation. Mol. Carcinog., 53, 77–84. [DOI] [PubMed] [Google Scholar]
- 84. Izumchenko E., et al. (2015) Understanding the MIG6-EGFR signaling axis in lung tumorigenesis. Cancer Discov., 5, 472–474. [DOI] [PubMed] [Google Scholar]
- 85. Garnett A.T., et al. (2012) BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development, 139, 4220–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Aruga J., et al. (2010) Expression of ZIC family genes in meningiomas and other brain tumors. BMC Cancer, 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ohta T., et al. (1994) Analbuminemia does not significantly influence hepatocarcinogenesis on comparing F344 rats and a congenic line carrying the analbuminemic mutation. Carcinogenesis, 15, 227–231. [DOI] [PubMed] [Google Scholar]
- 88. Lee J., et al. (2009) A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl Acad. Sci. USA, 106, 8513–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cheng J., et al. (2014) A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell, 53,979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chaudhuri A.R., et al. (2016) Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature, 535, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Choo A., et al. (2008) siRNA targeting the IRF2 transcription factor inhibits leukaemic cell growth. Int. J. Oncol., 33, 175–183. [PubMed] [Google Scholar]
- 92. Xie R.L., et al. (2002) Forced expression of the interferon regulatory factor 2 oncoprotein causes polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells. Cancer Res., 62, 2510–2515. [PubMed] [Google Scholar]
- 93. Steiner S., et al. (2013) Does bromodomain flexibility influence histone recognition? FEBS Lett., 587, 2158–2163. [DOI] [PubMed] [Google Scholar]
- 94. Meisenberg C., et al. (2012) Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res., 40, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li T., et al. (2016) Ubr3, a novel modulator of Hh signaling affects the degradation of Costal-2 and Kif7 through poly-ubiquitination. PLoS Genet., 12, e1006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Leonard J.M., et al. (2008) Sonic Hedgehog signaling impairs ionizing radiation-induced checkpoint activation and induces genomic instability. J. Cell Biol., 183, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]