Abstract
The gene blaCARB-9 was located in the Vibrio cholerae super-integron, but in a different location relative to blaCARB-7. CARB-9 (pI 5.2) conferred β-lactam MICs four to eight times lower than those conferred by CARB-7, differing at Ambler's positions V97I, L124F, and T228K. Comparison of the genetic environments of all reported blaCARB genes indicated that the CARB enzymes constitute a family of cassette-encoded β-lactamases.
The clinical significance of Vibrio cholerae non-O1, non-O139 strains is being increasingly recognized since several outbreaks of diarrhea occurred in the early 1990s (3, 8, 29). A high prevalence of V. cholerae non=O1, non-O139 ampicillin (AMP)-resistant (Ampr) isolates from both clinical and environmental origins was reported (3, 7, 10, 19, 29). To date, only three β-lactamases have been characterized in V. cholerae non-O1, non-O139 strains: CARB-2 (PSE-1), CARB-6, and CARB-7, which belong to the CARB family (Table 1). This major group of carbenicillinases is characterized by an RSG amino acid triad in positions 234 to 236 instead of the K-T/S-G motif of other class A β-lactamases (15). The proposed ancestors of carbenicillinases are the recently designated RTG enzymes (Table 1), which have an RTG triad instead of the CARB family RSG motif and share low identity with the CARB enzymes (5).
TABLE 1.
Sequences used in the alignments of the bla genes and the flanking regions of the blaCARB genes
| bla gene | Source, location (reference) | GenBank (accession no.) | FR-CARB (nucleotide position)a |
|---|---|---|---|
| blaCARB | |||
| blaCARB-1b | Pseudomonas aeruginosa, CHR,k Tn2521, ln33 (22) | AF313471 | CARB-1 (1239-2450) |
| blaCARB-2c | P. aeruginosa, pRPL11, Tn1403, ln28 (23) | AF313472 | A.CARB-2 (1324-2535) |
| (blaCARB-2)d | Salmonella typhimurium, pMG217 (unpublished) | Z18955 | B.CARB-2 (1-1199) |
| (blaCARB-2)d | S. typhimurium, CHR, class 1 integron (2) | AF071555 | C.CARB-2 (11109-12320) |
| (blaCARB-2)d | V. choleraee plasmid, class 1 integron (8) | AF221899 | D.CARB-2 (1-1195) |
| blaCARB-3 | P. aeruginosa, CHR, Tn1408 (13) | S46063 | CARB-3 (1-1060) |
| blaCARB-4 | P. aeruginosa, pUD12, Tn1408, class 1 integron (27) | U14749 | CARB-4 (501-1734) |
| blaN29 | Proteus mirabilis, unknown location (11) | D86225 | N29 (50-1260) |
| blaCARB-6 | V. cholerae non-O1, non-O139, CHR (4) | AF030945 | CARB-6 (1-967) |
| blaCARB-7 | V. cholerae non-O1, non-O139, SI (19) | AF409092 | CARB-7 (697-1933) |
| blaP2f | S. typhimurium, pST2301, class 1 integron (21) | AY123251 | blaP2 (3432-4651) |
| blaCARB-9 | V. cholerae non-O1, non-O139, SI (this work) | AY248038 | CARB-9 (1881-3119) |
| blaRTG | |||
| blaRTG-1g | P. mirabilis, CHR (26) | D13209 | NCh |
| blaRTG-2i | Acinetobacter calcoaceticus, unknown location (5) | AF135373 | NC |
| blaRTG-3j | Oligella urethralis, CHR (16) | AY178993 | NC |
FR-CARB, flanking region of blaCARB gene. Nucleotide positions of aligned sequences in the corresponding GenBank reports are given in parentheses.
Formerly known as blaPSE-4. A shorter sequence containing blaCARB-1 in a Tn1405 chromosomal background was also reported from P. aeruginosa (accession no. J05162) and was identical to a subsequence of AF313471.
Formerly named as blaPSE-1.
Not included in the alignment of bla genes (sequences of blaCARB-2 in AF313472, Z18955, AF071555, and AF221899 shared 100% identity).
blaCARB-2 was detected in both V. cholerae O1 and non-O1, non-O139 isolates; the sequence under accession no. AF221899 corresponds to a V. cholerae non-O1, non-O139 strain.
Named as blaCARB-8 in the GenBank report.
Formerly known as the β-lactamase of P. mirabilis GN79.
NC, not considered; the blaRTG genes were not included in the analysis of flanking regions.
Formerly, blaCARB-5.
Also named as blaCARB-8.
CHR, chromosome.
The blaCARB genes have been broadly dispersed among distantly related bacteria, probably through mobile genetic elements (14, 18). Several blaCARB genes were found as cassettes of class 1 integrons (Table 1), which have already been identified in V. cholerae (8, 29). In addition, a 126-kb-long integron (named the super-integron [SI]), comprising 216 open reading frames (ORFs) mainly of unknown functions, was found in the V. cholerae chromosome 2 (9, 17). Approximately 179 of these ORFs were found as cassettes, flanked by highly conserved sequences of 123 to 126 bp showing imperfect symmetry, named V. cholerae repeats (VCRs) (6, 9, 24).
In Argentina, we had previously observed a similar prevalence of AMP-nonsusceptible phenotype (AmpNS; resistant plus intermediate categories) in clinical (29%) and environmental (32%) isolates of a sample of 669 V. cholerae non-O1, non-O139 strains. The analysis of a subset of 131 AmpNS isolates detected two carbenicillinases, with pIs at 5.4 (CARB-7; 77 isolates) and 5.2 (54 isolates). The blaCARB-7 gene was located in the VCR island (19). Here, in order to characterize the β-lactamase-encoding gene of the second pool (pI 5.2), we selected the representative strain BA5, recovered from water samples in Buenos Aires province (Argentina, December 1993). The β-lactam MICs for BA5, determined as reported previously (19), were four to eight times lower than the corresponding figures for the CARB-7-producing isolate ME11762 (19). The MICs for BA5 and ME11762, respectively, were as follows: AMP, 64 and 256 μg/ml; AMP-sulbactam, 4 and 16 μg/ml; ticarcillin, 128 and 512 μg/ml; ticarcillin-clavulanic acid, 1 and 8 μg/ml; piperacillin, 8 and 32 μg/ml; cephalothin, 0.25 and 2 μg/ml; and cefoxitin, 1 and 8 μg/ml. By biparental conjugation (19), this resistant phenotype could not be transferred to Escherichia coli.
Characterization of blaCARB-9 and phylogenetic analysis.
A Sau3AI-based genomic library of V. cholerae BA5 was prepared as described previously (19). An ∼4-kb HindIII fragment, conferring Ampr, was subcloned from a 9-kb Sau3AI insert and sequenced, resulting in a 3,985-bp-long sequence. Putative Sau3AI fragment rearrangements produced in the construction of the library were discarded by PCR-restriction fragment length polymorphism of two amplimers from genomic DNA of BA5 (primers P1/P2 [forward, 5′-CAGGTTGTCAGTTCTCTG; reverse, 5′-GCTAGCTAAAGGTTACTCG] and CARB-F/CARB-R [forward, 5′-CCATCTGTAGTTTTTGCAAGCAG; reverse, 5′-CAACGCGACTGTGATGTATAAAC]), which comprised all of the Sau3AI sites found in the sequenced region. The amplification conditions used were described previously (19), with annealing temperatures of 49°C (P1/P2) or 60°C (CARB-F/CARB-R).
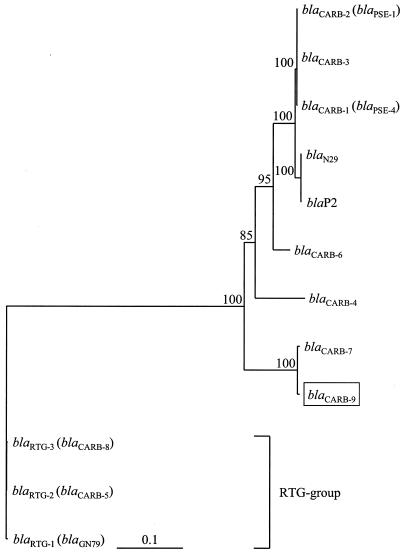
Sequence analysis revealed a major ORF of 867 bp, named blaCARB-9, as it shares 99 and ∼83% identities on both nucleotide and protein levels with blaCARB-7 and with other blaCARB genes, respectively. A neighbor-joining-based phylogenetic tree was constructed from multiple alignment of blaCARB and blaRTG genes, using the Clustal X program (ftp://ftp-igbmc.u-strasbg.fr/pub/). Among the blaCARB group, five highly related genes (i.e., blaCARB-1-2-3, blaN29, and blaP2 [named as the blaCARB-1/2 subgroup]) were tightly clustered (Fig. 1). The evolutionary distance between this subgroup and the remainder of the genes suggests a more recent divergence in the evolution of the CARB family. Conversely, blaCARB-7 and blaCARB-9 appear to have differentiated earlier.
FIG. 1.
Phylogenetic neighbor-joining tree of the blaCARB genes. Relevant features of compared sequences are in Table 1. The RTG cluster of presumptive ancestors of the carbenicillinases (5, 16) is indicated. The tree was rooted with blaRTG-1. Bootstrap percentages (based on 1,000 replicates) of 70% (or higher) of key nodes are shown.
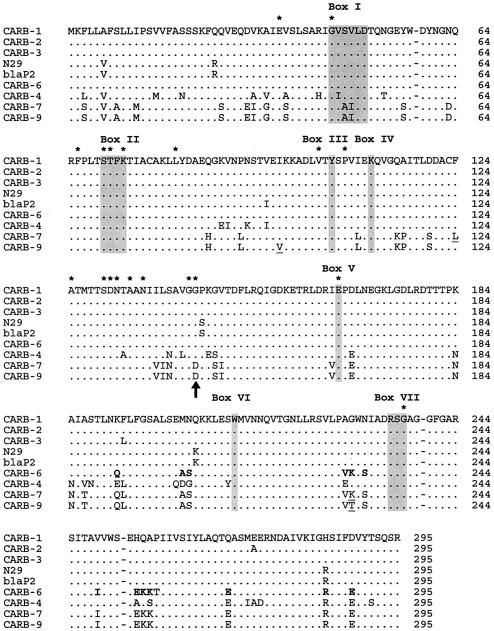
Besides three synonymous changes found between blaCARB-7and blaCARB-9, three nonsynonymous changes differentiated CARB-7 from CARB-9: I97V, F124L, and K228T, in the ABL numbering system (1) (Fig. 2). Although these changes are not located at the conserved positions found in class A β-lactamases, they may have a subtle effect on the activity of CARB-9 relative to CARB-7, which would explain the lower β-lactam MICs conferred by the former. In addition, the change K228T (a basic substituted for a noncharged amino acid) may explain the lower pI of CARB-9 (5.2) in comparison to CARB-7 (5.4) (19).
FIG. 2.
Alignment of CARB-9 (288 amino acids) with the CARB enzymes. CARB-1 and CARB-2 were formerly named PSE-4 and PSE-1, respectively. Identical residues are indicated by dots. Amino acid motifs conserved in all penicillin-recognizing enzymes (12) are signaled by shaded boxes (I to VII). Residues highly conserved among class A β-lactamases are indicated by asterisks. The unique change, G144D, between CARB-7 and CARB-9 and the remainder of class A β-lactamases reported to date is signaled by the arrow; this change would be located far from the active site, as inferred from the crystallographic structure reported for CARB-1 (15). The three residues that discriminate between CARB-7 and CARB-9 are underlined. Amino acids that differentiate CARB-6 from CARB-1, -2, and -3 and which are identical to residues in the CARB-7 sequence are shown in boldface. Sequences are numbered as described by Ambler et al. Gaps (dashes) at positions 58, 239, and 253 are indicated (1).
When comparing the blaCARB genes, we noticed that the sequence of about the first two-thirds of blaCARB-6 was identical to that of blaCARB-1-2-3, while the last third displayed a high identity with blaCARB-7. This observation was corroborated by constructing two neighbor-joining phylogenetic trees based on separate alignments of partial sequences: (i) nucleotides 1 to 546 (NH2 two-thirds) and (ii) nucleotides 547 to 867 (COOH third). The first tree showed that the NH2 two-thirds of blaCARB-6 clustered with that of the blaCARB-1/2 subgroup, while the COOH third of blaCARB-6 grouped with those of blaCARB-7 and blaCARB-9 in the second tree (data not shown). This uncommon nucleotide structure was reflected not only in the phylogenetic location of blaCARB-6, but also in its deduced amino acid sequence (Fig. 2), suggesting the occurrence of a recombination event (see below).
The flanking regions of blaCARB-9.
The analysis of 2,072 nucleotides upstream and 1,046 nucleotides downstream to blaCARB-9 revealed six sequences (five of 123 bp and one of 122 bp) which shared an overall 95% identity with the reported VCR consensus sequence (data not shown) (6, 24). These VCRs were in the same orientation relative to one another bordering four predicted ORFs. The upstream region showed high identity mainly with other VCRs when compared with sequences in the GenBank database. Interestingly, almost all of the downstream region (nucleotides 2940 to 3967) shared 96% identity with a fragment of section 35 of V. cholerae chromosome 2 (nucleotides 5105 to 6132; GenBank accession no. AE004378). In BA5, this region comprised two VCRs bordering an ORF (nucleotides 3068 to 3664) which showed 99.7% identity with the locus VCA0455 (9) (nucleotides 5232 to 5828; GenBank accession no. AE004378). This analysis, in addition to the fact that blaCARB-9 was not transferred to E. coli by conjugation, supports the assumption that blaCARB-9, like blaCARB-7, is placed within the V. cholerae SI. These results may help to explain the high prevalence of Ampr observed in clinical and environmental V. cholerae non-O1, non-O139 isolates. However, the comparison of the flanking regions of blaCARB-9 and blaCARB-7 (19) indicated that these genes share different VCRs (7% of genetic divergence) and that their locations inside the VCR island are clearly different. Moreover, the environments of the ORFs homologous to VCA0455 and VCA0424, in the isolates BA5 and ME11762 (CARB-7 producer) (19), respectively, are different from those found in V. cholerae El Tor N16961 (9). These facts constitute new evidence of plasticity in the VCR region (6).
The CARB enzymes constitute a family of cassette-encoded β-lactamases.
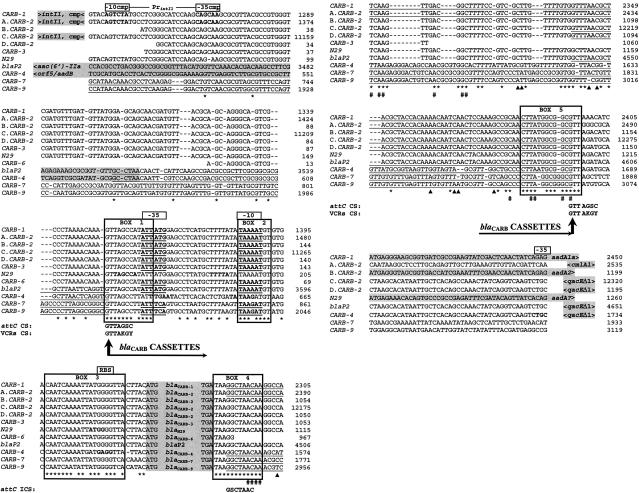
In order to analyze the genetic background of blaCARB genes, we compared 12 flanking regions reported to date, including four different environments of blaCARB-2 (Table 1). The upstream blaCARB flanking sequences were identical for CARB-1, all of the CARB-2 variants, CARB-3, N-29, and CARB-6 (only 34 bp are available) (Fig. 3). The 3′ terminus of the 5′-conserved segment of class 1 integrons was previously reported in three of these regions (CARB-1, A.CARB-2, and C.CARB-2) (2, 22, 23). By sequence identity, we inferred the presence of the 5′-conserved segment upstream to blaCARB-2 in plasmid pMG217 (B.CARB-2), blaCARB-3, blaN29, and blaCARB-6, which had not been previously associated with integrons. Despite the differences in genetic backgrounds, all of the blaCARB genes are contained in cassettes sharing a common array of genetic elements. These elements are located in highly conserved boxes and comprise sequences required for both cassette integration and transcription of blaCARB genes (Fig. 3): a unique core site (CS), putative promoters and ribosome-binding sites (RBS) (boxes 1 to 3); a double translation-stop signal (TGATAA), followed by a unique inverse CS (ICS) (box 4), and the 3′ terminus of attC and VCRs (box 5). In addition, sequences identical to the 105-bp-long attC previously reported downstream of blaCARB-1 and blaCARB-2 (A.CARB-2) (22, 23) were found downstream of the remainder of the blaCARB-1/2 genes (except blaCARB-3, where the sequence was unavailable). As this attC is predicted to form the typical stem-loop structure (DNASIS software v2.5; data not shown) previously reported for both attC and VCRs (24), we redefined the 5′ boundary of the attC reported for blaCARB-2 (C.CARB-2 and D.CARB-2) and blaP2 cassettes, proposing a longer attC identical to that of blaCARB-1. Thus, the CS and the ICS of blaCARB cassettes, with the recombination points defined as described previously (24, 28), were perfectly complementary in any circularized cassette (Fig. 3). Interestingly, the occurrence of promoters is very uncommon in the cassettes from class 1 integrons (20, 28), while it would be very unlikely that a single promoter could drive expression of the plethora of cassettes within an SI (25).
FIG. 3.
Flanking regions of blaCARB genes. Relevant features of compared sequences are in Table 1. Numbers on the left correspond to nucleotide positions in the GenBank reports. Gaps (dashes) were introduced in order to maximize the alignment (Clustal X). Codifying regions are signaled by shaded boxes, and the names of corresponding genes are indicated. The 5′ and/or 3′ ends not included in the alignment are represented by < and >, respectively. cmp, complementary sequence. The truncated codifying sequence (aadA?) at the end of N29 shared 97% identity with the integron-encoded gene aadA10 from the P. aeruginosa plasmid R388-R151 (GenBank accession no. U37105). Promoter (Pr) regions (−35 and −10) and the RBS reported previously are shown in boldface, and those identified in this work (the BPROM program, http://www.softberry.com/berry.phtml?topic=promoter) are additionally underlined. For clarity, VCRs and reported attC elements (underlined sequences) were defined from the third nucleotide upstream to the motif TAAC in the ICS, up to the G in the GTT of the CS. Consensus sequences for both ICS and CS are shown below the alignment. The identities among all available sequences (*) and positions highly conserved among Vibrio repeated sequences (VXRs; #) (24) are indicated. The regions of highest identity are boxed (boxes 1 to 5). Changes between the VCRs associated with blaCARB-7 and blaCARB-9 are signaled (▴). Horizontal arrows indicate the boundaries of the blaCARB cassettes (5′ to 3′), and vertical arrows indicate the putative recombination points (between G and T in the GTT of the CS).
The simplest explanation for to the aforementioned facts is that the blaCARB genes have evolved as cassette-encoded rather than “naked” genes, from a unique ancestral cassette. Additional evidence supports this assumption. First, a neighbor-joining tree based on cassette noncoding sequences showed essentially the same topology as that of the encoding sequences depicted in Fig. 1 (data not shown), suggesting that both regions have evolved in parallel. Second, there are several identities scattered along the fragment from box 4 to box 5, which include all the highly conserved positions previously found among Vibrio repeated sequences (VXRs) (Fig. 3) (24). Thus, under this rationale, it is hypothesized that the ancestral cassette could have originated in V. cholerae, probably as a unit of its SI, since the earliest differentiated blaCARB genes seem to be blaCARB-7 and blaCARB-9 (Fig. 1). The blaCARB-4 gene, with an attC only 3 bp shorter than the blaCARB-7-9-associated VCRs (68% identity with them), may have emerged after horizontal exchanges mediated by plasmids and transposons/integrons, as an integron-associated cassette in Pseudomonas aeruginosa (27). Finally, the blaCARB-1/2 group, showing a unique attC, can be considered as more recent cassette variants generated by nonsynonymous point mutations, while blaCARB-6 could be generated by recombination between blaCARB-7 and blaCARB-1, blaCARB-2, or blaCARB-3. Interestingly, blaCARB-1-2-3 are the most broadly dispersed genes among clinical isolates (8, 14, 18), while blaCARB-6 was only detected in a unique V. cholerae non-O1, non-O139 clinical strain (4). Further sequencing of the 3′-flanking region of blaCARB-6 will be useful to corroborate the recombination event. In summary, the CARB enzymes constitute the first family of cassette-encoded β-lactamases reported to date.
The data presented here also provide additional evidence to support the assumption of the in vivo capture of VCR cassettes by class 1 integrons, reinforcing the notion that these elements have evolved from SIs (25). Thus, the ORFs of SIs may constitute a big reservoir for horizontal gene transfer, including antibiotic resistance genes, such as blaCARB-7 and blaCARB-9.
Nucleotide sequence accession number.
The nucleotide sequence determined here will appear in the GenBank database under accession no. AY248038.
Acknowledgments
We thank Norma Binsztein for providing the V. cholerae non-O1, non-O139 collection and María I. Caffer for serotyping the strains. We thank Karen Brassinga for a critical reading of the manuscript.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tirabi, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty, S., P. Garg, T. Ramamurthy, M. Thungapathra, J. K. Gautam, C. Kumar, S. Maiti, S. Yamasaki, T. Shimada, Y. Takeda, A. Ghosh, and G. B. Nair. 2001. Comparison of antibiogram, virulence genes, ribotypes and DNA fingerprints of Vibrio cholerae of matching serogroups isolated from hospitalised diarrhoea cases and from the environment during 1997-1998 in Calcutta, India. J. Med. Microbiol. 50:879-888. [DOI] [PubMed] [Google Scholar]
- 4.Choury, D., G. Aubert, M.-F. Szajnert, K. Azibi, M. Delpech, and G. Paul. 1999. Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing β-lactamase from Vibrio cholerae. Antimicrob. Agents Chemother. 43:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choury, D., M.-F. Szajnert, M.-L. Joly-Gillou, K. Azibi, M. Delpech, and G. Paul. 2000. Nucleotide sequence of the blaRTG-2 (CARB-5) gene and phylogeny of a new group of carbenicillinases. Antimicrob. Agents Chemother. 44:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 7.Dalsgaard, A., A. Forslund, D. Bodhidatta, C. Serichantalergs, L. Pitarangsi, T. Pang, T. Shimada, and P. Echeverria. 1999. A. high proportion of V. cholerae isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogeneous non-O1, non-O139 O-serotypes. Epidemiol. Infect. 122:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg, J., J. Eisen, W. Nelson, R. Clayton, M. Gwinn, R. Dodson, D. Haft, E. Hickey, J. Peterson, L. Umayam, S. Gill, K. Nelson, T. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. Fleishmann, W. Nierman, O. White, S. Salzberg, H. Smith, R. Colwell, J. Mekalanos, J. Venter, and C. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaac-Márquez, A. P., C. M. Lezama-Dávila, C. Eslava-Campos, A. Navarro-Ocaña, and A. Cravioto-Quintana. 1998. Serotypes of Vibrio cholerae non-O1 isolated from water supplies for human consumption in Campeche, México and their antibiotic susceptibility patterns. Mem. Inst. Oswaldo Cruz 93:17-22. [DOI] [PubMed] [Google Scholar]
- 11.Ito, Y., and T. Hirano. 1997. Carbenicillin-hydrolysing penicillinase mediated by a plasmid of Proteus mirabilis and its relationship to the PSE-type enzymes of Pseudomonas aeruginosa. J. Appl. Microbiol. 83:175-180. [DOI] [PubMed] [Google Scholar]
- 12.Joris, B., J. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J. Frère, J. Kelly, J. Boyington, P. Moews, and J. Knox. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem. J. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachapelle, J., J. Dufresne, and R. C. Levesque. 1991. Characterization of the blaCARB-3 gene encoding the carbenicillinase-3 beta-lactamase of Pseudomonas aeruginosa. Gene 102:7-12. [DOI] [PubMed] [Google Scholar]
- 14.Levesque, R. C., and G. A. Jacoby. 1988. Molecular structure and interrelationships of multiresistance β-lactamase transposons. Plasmid 19:21-29. [DOI] [PubMed] [Google Scholar]
- 15.Lim, D., F. Sanschagrin, L. Passmore, L. De Castro, R. C. Levesque, and N. C. Strynadka. 2001. Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies. Biochemistry 40:395-402. [DOI] [PubMed] [Google Scholar]
- 16.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazel, D., B. Dychinco, V. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 18.Medeiros, A. A., R. W. Hedges, and G. A. Jacoby. 1982. Spread of a “Pseudomonas-specific” β-lactamase to plasmids of enterobacteria. J. Bacteriol. 149:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melano, R., A. Petroni, A. Garutti, H. A. Saka, L. Mange, F. Pasterán, M. Rapoport, A. Rossi, and M. Galas. 2002. New carbenicillin-hydrolyzing beta-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 46:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pai, H., J.-H. Byeon, S. Yu, B. K. Lee, and S. Kim. 2003. Salmonella enterica serovar Typhi strains isolated in Korea containing a multidrug resistance class 1 integron. Antimicrob. Agents Chemother. 47:2006-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe-Magnus, D. A., A.-M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe-Magnus, D. A., A.-M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai, Y., K. Tsukamoto, and T. Sawai. 1991. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J. Bacteriol. 173:7038-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanschagrin, F., N. Bejaoui, and R. C. Levesque. 1998. Structure of CARB-4 and AER-1 carbenicillin-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 42:1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 29.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]