Abstract
Glycine is the simplest amino acid and is used as a metabolic product in some bacteria. However, an excess of glycine inhibits the growth of many bacteria, and it is used as a nonspecific antiseptic agent due to its low level of toxicity in animals. The effect of glycine on Helicobacter pylori is not precisely known. The present study was conducted to investigate (i) the effect of glycine on clarithromycin (CLR)-resistant and -susceptible strains of H. pylori, (ii) the effect of glycine in combination with amoxicillin (AMX), and (iii) the postantibiotic effect (PAE). The MIC at which 90% of strains are inhibited for glycine was almost 2.5 mg/ml for 31 strains of H. pylori, including CLR-resistant strains. We constructed isogenic CLR-resistant mutant strains by natural transformation and investigated the difference between clinical wild-type strains and isogenic mutants. There were no differences in the MICs between CLR-resistant and -susceptible strains or between clinical wild-type and mutant strains. The combination of AMX and glycine showed synergistic activity, with the minimum bactericidal concentration of AMX with glycine decreasing to 1/10 that of AMX alone. Glycine showed no PAE against H. pylori. These results suggest that glycine may be a useful antimicrobial agent against H. pylori not only alone but also in combination with antibacterial drugs for the treatment of H. pylori-associated diseases. Glycine may represent a component of a new type of eradication therapy for CLR-resistant H. pylori.
Helicobacter pylori is a microaerophilic, gram-negative spiral bacterium that is recognized as a pathogenic bacterium (9, 26). In humans, H. pylori is associated with peptic ulcer, malignant lymphoma, and gastric cancer (39). It infects approximately 80% of the population in many developing countries (40) and about 30% of the population in developed countries (12). Although the mechanism of the pathogenicity of this bacterium has not been clearly explained, eradication of H. pylori greatly contributes to an eminent improvement in these diseases (7, 34, 39). In Japan, the present first-line treatment for H. pylori eradication is a triple-therapy combination with a proton pump inhibitor, clarithromycin (CLR), and amoxicillin (AMX). This combination achieves clinical cure rates of more than 80% (5). The prevalence of CLR resistance varies with geographical location and is generally estimated to be about 10% in Japan (21). Given that CLR has a stronger antibacterial effect on H. pylori than other agents (24), the presence of resistant microbes may result in eradication failure (1). Although various other agents such as metronidazole or tetracycline are candidates for eradication of CLR-resistant strains (17), their clinical use is limited because of side effects or the presence of strains resistant to those drugs as well (36). The identification of new antibacterial agents is therefore strongly desirable.
Glycine is the simplest amino acid and has been used as an antibacterial agent in foods due to its low toxicity in animals. Although glycine is toxic to humans when it is given in large amounts, it also has antibacterial potential (11, 13, 33). For example, Lactococcus lactis subspecies failed to grow in medium containing more than 2% glycine (15). Furthermore, glycine concentrations of 1.5 to 6% resulted in 70 to 90% reductions in growth of Enterococcus faecalis (10). Glycine is known to inhibit the synthesis of a peptidoglycan component of the bacterial cell wall (16). Although the bacterial cell wall is thinner in gram-negative bacteria than in gram-positive bacteria, it is thought that the amount of glycine required to suppress bacillus proliferation is lower than that required to suppress gram-positive bacteria (30). However, the effect of glycine against H. pylori has not been well studied in detail.
The postantibiotic effect (PAE) is a phenomenon by which the inhibition of bacterial growth continues after exposure to an antimicrobial agent (8, 19, 25). It is classically defined as the period of bacterial growth suppression that persists after limited exposure of organisms to antimicrobials. The PAE may remain even after drug levels are no longer detectable. The duration of the PAE is of important clinical interest in establishing dosing schedules, particularly given that longer intervals between intermittent dosage schedules may reduce the toxicities of antibiotics (25). To date, however, the PAE of glycine against H. pylori has not been well studied and little information is available.
AMX, a beta-lactam antibiotic which inhibits the synthesis of a peptidoglycan of the cell wall, is also effective against H. pylori (6), albeit with an eradication rate when it is used alone that is lower than those of the other drugs used in combination (23), and AMX-resistant H. pylori are rare (1). The present study was conducted to determine the effect of glycine on H. pylori, especially CLR-resistant strains, and to evaluate the PAE of glycine and the effect of glycine in combination with AMX. The results obtained lead us to propose a new therapy for H. pylori eradication.
MATERIALS AND METHODS
Agents.
Glycine (assay [after drying] minimum, 99.0%; soluble in water; maximum amount of chloride, 0.02%, maximum amount of sulfate, 0.005%; maximum amount of heavy metal [as Pb], 0.001%; maximum amount of iron, 0.0005%; maximum amount of ammonium, 0.02%; total nitrogen, 18.5 to 18.8%; Wako Pure Chemical Industries, Ltd., Osaka, Japan) and AMX (Sigma Chemical Co., St. Louis, Mo.) were purchased, and CLR (Taisho Pharmaceutical Co., Ltd., Osaka, Japan) was kindly provided as a gift. Glycine, CLR, and AMX were dissolved in distilled water to a final concentration of 20% (wt/vol), methanol, and 70% ethanol, respectively.
Bacterial strains and culture conditions.
The susceptibilities to glycine of 31 H. pylori strains, including three standard strains (strains 26695 [37] and J99 [2] and a quality control strain [ATCC 43504] [27]), and clinical isolates (4, 28, 35, 38) were measured. The clinical isolates were from biopsy specimens of lesions obtained by endoscopy from different patients with gastroenterological diseases at Nagoya University Hospital and a regional health care center. Stock cultures of H. pylori were grown for 4 days on brucella broth agar (Difco, Detroit, Mich.) plates supplemented with 7% heat-inactivated fetal calf serum (FCS; Gibco BRL, Rockville, Md.) (BB agar) at 37°C in a microaerophilic atmosphere. Broth cultures of H. pylori were prepared from subcultures of colonies from freshly cultured agar plates placed into brucella broth (Difco) supplemented with 7% FCS (BB) for 3 days at 37°C in a microaerophilic atmosphere. The identification of H. pylori was confirmed by characteristic colony morphology, Gram staining, and positive reactions by urease, catalase and oxidase tests.
DNA techniques.
Standard molecular biology-based techniques were used (29). H. pylori chromosomal DNA was prepared from cells of each strain after 48 h of growth on two agar plates and was extracted with a Wizard Genomic DNA Purification kit (Promega, Madison, Wis.), according to the instructions of the manufacturer.
Construction of isogenic mutant strains.
CLR-resistant isogenic mutants were constructed by natural transformation methods (3, 29). Domain V of the 23S rRNA gene in H. pylori, which is associated with CLR resistance (32), was amplified by PCR. Template DNA was extracted from CLR-resistant strains 628 and 535 and was found to possess A-to-G point mutations at either position 2143 (strain 628) or position 2144 (strain 535) in domain V of the 23S rRNA gene. The PCR products from donor DNA samples were purified. Briefly, 1 ng of DNA was amplified with 100 pmol of the sense and antisense primers (sense primer, 5′-CCACAGCGATGTGGTCTCAG-3′ [positions 1820 to 1839 of 23S rRNA sequence]; antisense primer, 5′-CTCCATAAGAGCCAAAGCCC-3′ [positions 2244 to 2225 of the 23S rRNA sequence]) in a 50-μl reaction mixture containing 0.25 μl of Ex Taq polymerase (Takara Biomedicals, Ohtsu, Japan) for 25 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C in a DNA thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, Foster City, Calif.). The presence of a single 425-bp band was verified on a 1% agarose gel, and the PCR products were purified with a QIAquick PCR Purification kit (Qiagen, Valencia, Calif.).
H. pylori strain 26695 was used as the transformation recipient. After 48 h of growth, recipient H. pylori cells were harvested from one BB agar plate and placed into 1 ml of phosphate-buffered saline (PBS) and then centrifuged at 8,500 × g for 5 min. The pellet was resuspended in 300 μl of PBS. Each transformation mixture, consisting of 25 μl of recipient cells and 1 μg of donor DNA, was spotted onto a BB agar plate (approximately 600 ng of DNA/25 μl of cells is a saturating amount of DNA). The plates were incubated overnight at 37°C in a microaerophilic atmosphere. After 18 h of incubation, the transformation mixture was spread onto BB agar containing 4 μg of CLR/ml. All plates were incubated for 5 days at 37°C in a microaerophilic atmosphere to select transformants (3). The presence of point mutations in the transformants was confirmed by PCR and direct sequencing (CEQ2000XL; Beckman Coulter Inc., Fullerton, Calif.).
Susceptibility testing.
The susceptibilities of the CLR-resistant strains were determined by the agar dilution method (27). An MIC of more than 1 μg/ml was considered CLR resistance. The MIC of glycine was defined as the lowest concentration that inhibited the visible growth of isolates completely by the agar dilution method on Mueller-Hinton agar (Becton Dickinson and Company, Paramus, N.J.) plates supplemented with 5% aged sheep blood (Nippon Bio-Test Laboratories Inc., Tokyo, Japan) (M-H agar) (27). Briefly, all isolates were incubated for 4 days on BB agar. After this incubation, inocula were prepared by suspending growth from the BB agar plates with antimicrobial agents in saline to achieve a suspension equivalent to a 2.0 McFarland standard. Final inocula of 106 CFU/spot were applied to M-H agar or BB. All plates were incubated for 3 days at 35°C in a microaerophilic atmosphere, and the number of CFU was counted. In the liquid culture study, aliquots from each culture were applied to BB agar after 3 days of incubation of H. pylori at 37°C under microaerophilic conditions. After 4 days of incubation, the number of CFU was counted. The mean value for the log number of CFU per milliliter was plotted. By a second method, the minimal bactericidal concentrations (MBCs), defined as the lowest concentration of each antibiotic that completely killed the isolates, were determined by using liquid cultures with antimicrobial agents by a modification of the method of Sjöström et al. (31). The MBC of AMX with glycine was determined by serial dilution of AMX and glycine in BB in the presence of approximately 106 CFU/ml in 25-ml tissue culture flasks (Becton Dickinson and Company). After 3 days of incubation of H. pylori at 37°C under microaerophilic conditions, aliquots from each culture were applied to BB agar for determination of the MBC. After 4 days of incubation, the number of CFU was counted. AMX and glycine were tested at concentrations ranging from 0 to 75 ng/ml and 0 to 2.5 mg/ml, respectively. The MBCs were measured to study the effect of the interaction between AMX and glycine against strain 26695.
Determination of PAE.
The PAE on bacteria was measured by a modification of the method of Ibrahim et al. (19). After 4 days of growth of strain 26695 on BB agar, the bacteria were harvested, placed into PBS, and diluted with BB to ∼108 CFU/ml; and a final inoculum of 106 CFU/ml was applied to BB agar. Glycine was added to BB in a 25-ml tissue culture flask at a concentration of 1.25 mg/ml (five times the MIC). After incubation for 8 h, the antibiotics were removed by dilution 1:103 into fresh BB and the cultures were incubated for 24 h. Samples were collected for viable counts (numbers of CFU per milliliter) every 4 h, plated onto BB agar plates, and incubated for 4 days. Viability curves were then determined. The control culture, which was not exposed to any antimicrobial agents, was treated similarly. For quantification of the PAE, viable counts were determined before and after antibiotic exposure. The T − C value was calculated as follows (8): T was the time required for the CFU count in the test culture to increase 10-fold above the count observed immediately after antibiotic removal, and C was the time required for the CFU count in an untreated control culture to increase 10-fold above the count observed immediately after completion of the same procedure for antibiotic removal used for the test culture.
Time-kill curve test.
The time course of the antibacterial effects was determined with strain 26695 (18). Briefly, bacteria cultured for 4 days on BB agar were diluted with fresh BB to ∼108 CFU/ml. A final inoculum of 106 CFU/ml was placed into a 25-ml tissue culture flask containing 10 ml of BB with glycine or both glycine and AMX. Broth cultures with glycine were incubated for 2 days at 37°C in a microaerophilic atmosphere. At 0, 12, 24, 36, and 48 h, samples were removed and 0.1 ml of 10-fold serial dilution was plated onto BB agar. Broth cultures with both glycine and AMX were incubated for 1 day at 37°C in a microaerophilic atmosphere. At 0, 6, 12, 18, and 24 h, samples were removed and 0.1 ml of 10-fold serial dilutions was plated onto BB agar. The number of colonies growing on the plates after 4 days of incubation was counted in both studies.
Statistical analysis.
The degree of significance between means was determined by the paired t test. A P value of <0.01 was regarded as significant. These studies were repeated at least five times to confirm the reliability of the data.
RESULTS
Construction of CLR-resistant isogenic mutants and MIC determination.
CLR-resistant isogenic mutants with a single point mutation in domain V of the 23S rRNA gene were created from strain 26695 by natural transformation (3, 29). Transformants were isolated, and the MICs of CLR were determined by the disk dilution method. Strain 26695:628 possessed an A-to-G point mutation at position 2143 in domain V of the 23S rRNA gene, and strain 26695:535 possessed an A-to-G point mutation at position 2144. The MIC of CLR was dependent on that for the donor strain (for strain 628, MIC = 32 μg/ml; for strain 535, MIC = 4 μg/ml; for strain 26695:628; MIC = 32 μg/ml; for strain 26695:535, MIC = 4 μg/ml).
Glycine MICs.
The effect of glycine on 31 bacterial strains, including 10 CLR-resistant H. pylori strains, was examined. The glycine MICs for the 31 strains ranged from 1 to 2.5 mg/ml, while the concentrations required to inhibit 50 and 90% of the strains (MIC50 and MIC90, respectively) were 1.5 and 2.5 mg/ml, respectively. Glycine inhibited the growth of all strains tested, and no colonies were detected in M-H agar with glycine at more than 2.5 mg/ml. The MIC of glycine for the quality control strain (ATCC 43504) was same as that for strain 26695 (2.5 mg/ml). There was no difference in the glycine MIC for CLR-susceptible and -resistant strains. Furthermore, there was no association between the MIC of CLR and the MIC of glycine. The MIC of glycine for the experimental strain was dependent on that for the recipient strain (strain 26695), and the MICs for the isogenic mutant strains were the same as the MIC for strain 26695.
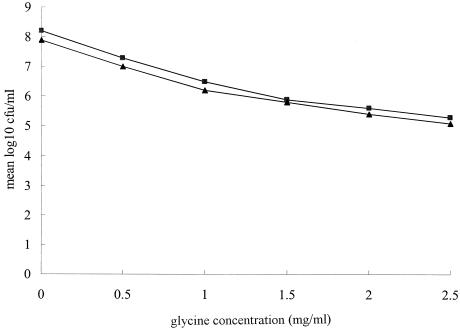
These data strongly suggest that the antibacterial mechanisms of glycine and CLR are independent. The addition of glycine resulted in a significant decrease in the number of bacteria. Furthermore, significant concentration-dependent suppression of bacterial growth was observed for both strains. However, there was no difference in the suppression of growth between the CLR-susceptible and -resistant strains, including the isogenic mutant strains (strains 26695:628 and 26695:535) (Fig. 1). In other words, glycine acted similarly against both CLR-resistant and -susceptible H. pylori.
FIG. 1.
Comparison of the inhibitory effects of glycine against CLR-susceptible and CLR-resistant H. pylori strains. H. pylori strains were treated with the indicated concentrations of test compound for 3 days. The mean value for the log number of CFU per milliliter of all CLR-susceptible and all CLR-resistant H. pylori strains at each concentration was plotted. Glycine was used at concentrations of 0, 0.5, 1, 1.5, 2.0, and 2.5 mg/ml. Representative results from five independent experiments are shown. Symbols: ▪, CLR-susceptible strains; ▴, CLR-resistant strains.
Time-kill curve experiments with glycine.
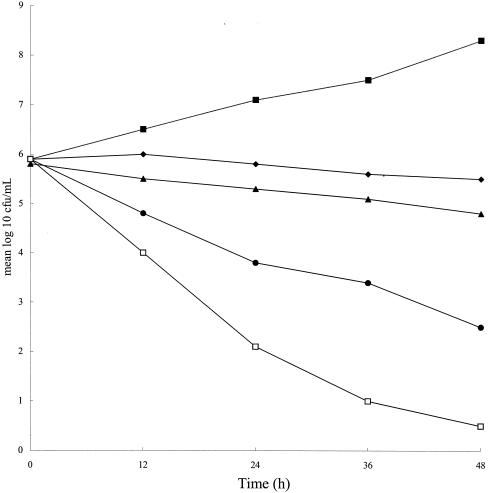
The results of the time-kill curve experiments with glycine alone at a concentration of 0 to 10 mg/ml are shown in Fig. 2. The number of CFU increased over time when no glycine was present in the culture medium. When glycine was present, however, a significant decrease in the number of CFU was seen at 48 h with all concentrations (P < 0.01). This decrease was significant (P < 0.01) at as early as 24 h with a concentration of 5 mg/ml and as early as 12 h with a concentration of 10 mg/ml. These results show that the effect of glycine is bactericidal, because the number of CFU was not stable but was decreased by glycine administration.
FIG. 2.
Time-kill curve experiment with glycine and H. pylori 26695. H. pylori was inoculated into BB with glycine and incubated at 37°C for 2 days. Glycine in BB was added to the cultures at the start of incubation. The mean values for the log number of CFU per milliliter versus time for H. pylori were tested with different concentrations of glycine. At 12-h intervals after drug addition, aliquots of each culture were spread onto BB agar for determination of the number of CFU. Representative results from five independent experiments are shown. Symbols: ▪, glycine at 0 mg/ml; ♦, glycine at 1 mg/ml; ▴, glycine at 2.5 mg/ml; •, glycine at 5 mg/ml; □, glycine at 10 mg/ml.
Effect of combination of glycine with AMX.
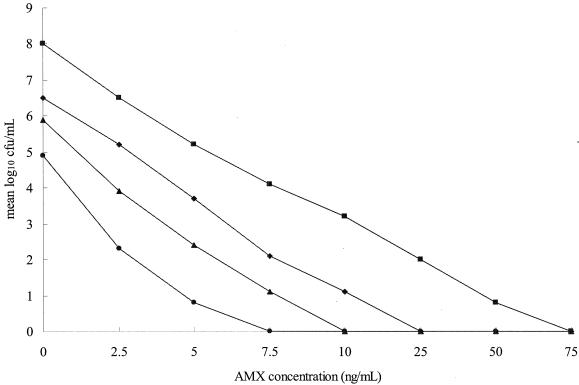
We next determined the effect of the combination of glycine with AMX against H. pylori. The MBC of AMX with glycine was determined by the liquid culture method with the following doses: AMX at 7.5 ng/ml with glycine at 2.5 mg/ml, AMX at 10 ng/ml with glycine at 1 mg/ml, AMX at 25 ng/ml with glycine at 0.5 mg/ml, and AMX at 75 ng/ml with glycine at 0 mg/ml (Fig. 3). The MBC of AMX and glycine in combination was less than that of AMX alone (P < 0.01). It was found that the combination of AMX and glycine reduced the viable count for strain 26695, even at concentrations that had subinhibitory effects when the agents were used alone. Their combined effect against other H. pylori strains was the same as that against strain 26695 (data not shown). Incubation with AMX and glycine yielded significant inhibition of growth for strain 26695. The combination of AMX and glycine enhanced the killing effect against strain 26695 and resulted in a dramatic decrease in the number of viable cells that was greater than that achieved with glycine or AMX alone.
FIG. 3.
Inhibitory effects of various concentrations of glycine and AMX on H. pylori 26695. H. pylori was treated with the indicated concentrations of the test compounds for 3 days. AMX was used at concentrations of 0 to 75 ng/ml. Glycine was used at concentrations of 0, 0.5, 1, and 2.5 mg/ml. The mean value for the log number of CFU per milliliter at each concentration was plotted. Representative results from five independent experiments are shown. Symbols: ▪, glycine at 0 mg/ml with AMX; ♦, glycine at 0.5 mg/ml with AMX; ▴, glycine at 1 mg/ml with AMX; •, glycine at 2.5 mg/ml with AMX.
Time-kill curve experiments with glycine and AMX.
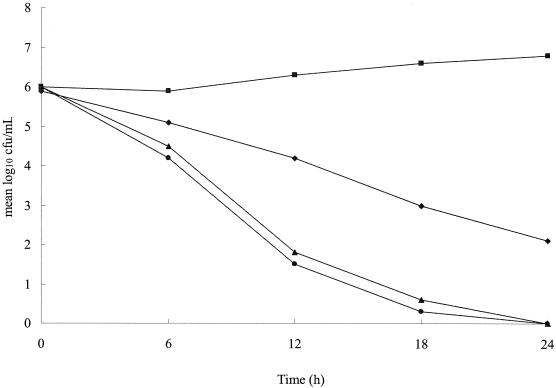
A time-kill curve experiment was performed to investigate the rate at which the viable cell count decreased after treatment with glycine and AMX (Fig. 4). The number of bacterial colonies increased in the absence of glycine. However, when a combination of glycine at 2.5 mg/ml and AMX at 7.5 ng/ml (the MBCs) was used, a significant decrease in the number of bacteria was seen after 6 h (P < 0.01). Moreover, the rate of decrease in cell numbers achieved with the combination of glycine and AMX was greater than that achieved with glycine at 10 mg/ml alone and was as fast as that achieved with AMX at 75 ng/ml alone, with no viable cells found at 24 h when either AMX alone at 75 ng/ml or the combination of glycine and AMX was used. Furthermore, strain 26695 was not detected at 24 h when the other combinations of glycine and AMX were used (AMX at 10 ng/ml plus glycine at 1 mg/ml and AMX at 25 ng/ml plus glycine at 0.5 mg/ml). These findings confirm the synergistic activity of glycine and AMX against H. pylori strain 26695.
FIG. 4.
Time-kill curve experiments with glycine-AMX and H. pylori 26695. H. pylori was inoculated into BB with glycine and incubated at 37°C for 24 h. Glycine and AMX in BB were added to cultures at the start of incubation. The mean values for the log of the number of CFU per milliliter versus time for H. pylori were tested with different concentrations of glycine and AMX. At 6-h intervals after drug addition, aliquots from each culture were spread onto BB agar plates for determination of the numbers of CFU. Representative results from five independent experiments are shown. Symbols: ▪, glycine at 0 mg/ml; ♦, glycine at 10 mg/ml; ▴, at glycine at 2.5 mg/ml with AMX at 7.5 ng/ml; •, AMX at 75 ng/ml.
PAE of glycine against H. pylori.
The PAE regrowth curves for glycine showed growth rates which, after correction for the dilution rate, were close to the growth rate of the control culture measured for this organism. The T − C value was almost zero (data not shown).
DISCUSSION
Glycine is the simplest amino acid, and its structure has no asymmetric carbon atom. As it is a metabolite of serine, it is not an essential amino acid for bacteria (11). The major mechanism of glycine's antimicrobial activity is its inhibition of cell wall synthesis. High concentrations of glycine induce bacteriolysis or morphological alterations in various bacteria (11, 13, 16, 30, 33). A possible mechanism of this effect may be inhibition of the reaction in which l-alanine is added to UDP-acetylmuramic acid by UDP-N-acetylmuramate-alanine ligase (20). The UDP-N-acetylmuramate-alanine ligase of Bacillus subtilis is inhibited in vitro by glycine (13). Glycine may also act on the replacement of l-alanine residues as an analogue of either l- or d-alanine, which may weaken the structure of the cell wall (16). H. pylori strain 26695 was reported to have d-alanine:d-alanine ligase A (HP0738) and UDP-N-acetylmuramate-alanine ligase (HP0623) (22), which supports our hypothesis.
The inhibition of proliferation of H. pylori was dependent on the glycine concentration. Furthermore, the MBC of AMX decreased to 1/10 when it was used together with glycine, suggesting that glycine may have a synergistic effect with AMX. Cell wall synthesis involves a number of available active enzymes, such as dd- and dl- carboxypeptidases, which are important for the formation of cell wall-bound peptidoglycan. Because glycine inhibits ld-carboxypeptidase, the modifying effect of glycine on cell wall synthesis was explained to be largely due to this mechanism. Beta-lactam antibiotics are capable of inhibiting not only dl-carboxypeptidases but also dd-carboxypeptidases, which are required for the synthesis of cross-linked peptidoglycan in Gaffkya homari (14). Given these observations, we assume that the synergism of glycine in increasing the antimicrobial efficacies of beta-lactam antibiotics is mainly due to its inhibition of this enzyme system (11). However, H. pylori murein lacks ld-cross-linked-carboxypeptidase muropeptides, and H. pylori may not have ld-carboxypeptidases (22). We therefore hypothesize that the synergism of AMX and glycine against H. pylori involves AMX inhibition of dl- or dd-carboxypeptidases and glycine mainly inhibits UDP-N-acetylmuramate-alanine ligase.
Appropriate antibiotic use remains central to achieving the optimum outcomes for infected patients and infection control. PAE and postexposure events are clearly relevant to achieving these goals. In this study, glycine showed no PAE against H. pylori, and in general, beta-lactam antibiotics are not thought to possess PAEs (8). Glycine's lack of a PAE may be related to the similarity of its antibacterial mode of action to those of beta-lactams, namely, its inhibition of cell wall synthesis. The full clinical relevance of PAE studies and how they may influence glycine dosing regimens for H. pylori eradication therapy remain matters for future research. No reports of notably glycine-tolerant strains have been presented, and no strains for which glycine MICs varied were found in the present study. Glycine significantly reduced the number of both CLR-sensitive and -resistant H. pylori isolates. This result shows that the mechanism of resistance to CLR, namely, point mutation in the V domains (36), is not associated with the antibacterial mechanism of glycine and suggests that glycine has effective activity against CLR-resistant strains.
A full understanding of the antibacterial mechanism of glycine and the establishment of a new eradication therapy based on that mechanism await further investigations. However, the results presented here open new avenues toward more effective eradication therapy for H. pylori and suggest that the administration of a combination of a proton pump inhibitor, AMX, and glycine deserves further evaluation.
Acknowledgments
We thank Teruko Nakazawa for many suggestions and encouragement, as well as the members of the Department of Molecular Bacteriology of Nagoya University Graduate School of Medicine for technical advice. We also thank Martin J. Blaser for reading the manuscript and giving us helpful suggestions.
REFERENCES
- 1.Adamek, R. J., S. Suerbaum, and B. Pffaffenbach. 1998. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole, and amoxicillin—influence on treatment outcome. Am. J. Gastroenterol. 93:386-389. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ando, T., Q. Xu, M. Torres, K. Kusugami, D. A. Israel, and M. J. Blaser. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37:1052-1065. [DOI] [PubMed] [Google Scholar]
- 5.Asaka, M., T. Sugiyama, M. Kato, K. Satoh, H. Kawayama, Y. Fukuda, T. Fujioka, T. Takemoto, K. Kimura, T. Shimoyama, K. Shimizu, and S. Kobayashi. 2001. A multi center, double-blind study on triple therapy with lansoprazole, amoxicillin and clarithromycin for eradication of Helicobacter pylori in Japanese peptic ulcer patients. Helicobacter 6:254-261. [DOI] [PubMed] [Google Scholar]
- 6.Bayerdörffer, E., G. Mannes, and A. Sommer. 1992. High dose omeprazole treatment combined with amoxicillin eradicates Helicobacter pylori. Eur. J. Gastroentoerol. Hepatol. 4:697-702. [Google Scholar]
- 7.Bayerdörffer, E., A. Neubauer, B. Rudolph, C. Thiede, N. Lehn, S. Eidt, and M. Stolte. 1995. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet 345:1591-1594. [DOI] [PubMed] [Google Scholar]
- 8.Craig, W. A., and S. Gudmundsson. 1996. Postantibiotic effect, p. 296-329. In V. Lorian (ed.), Antibiotics in laboratory medicine. The Williams & Wilkins Co., Baltimore, Md.
- 9.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gary, M. D., N. L. Linda, and J. L. Donald. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillissen, G., M. Schumacher, and M. Breuer-Werle. 1991. Modulation of antimicrobial effects of beta-lactams by amino acids in vitro. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 275:223-232. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. Y. 1991. Helicobacter pylori: its epidemiology and its role in duodenal ulcer disease. J. Gastroenterol. Hepatol. 6:105-113. [DOI] [PubMed] [Google Scholar]
- 13.Hammes, W., K. H. Schleifer, and O. Kandler. 1973. Mode of action of glycine on the biosynthesis of peptidoglycan. J. Bacteriol. 116:1029-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammes, W. P. 1978. The ld-carboxypeptidase activity in Gaffkya homari. The target of the action of d-amino acids or glycine on the formation of wall-bound peptidoglycan. Eur. J. Biochem. 91:501-515. [DOI] [PubMed] [Google Scholar]
- 15.Helge, H., and F. N. Ingolf. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hishinuma, F., K. Izaki, and H. Takahashi. 1969. Effects of glycine and d-amino acids on growth of various microorganism. Agric. Biol. Chem. 33:1577-1586. [Google Scholar]
- 17.Hojo, M., H. Miwa, A. Nagahara, and N. Sato. 2001. Pooled analysis on the efficiency of the second-line treatment regimens for Helicobacter pylori infection. Scand. J. Gastroenterol. 36:690-700. [DOI] [PubMed] [Google Scholar]
- 18.Horii, T., T. Kimura, K. Sato-Kawamura, T. Nada, K. Shibayama, and M. Ohta. 1999. Beta-lactamase inhibitors have antibacterial activities against Helicobacter pylori. J. Infect. Chemother. 5:206-207. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim, J., R. M. Hassan, J. Stark, M. Greenman, and R. Millar. 1998. Absence of a post-antibiotic effect (PAE) of beta lactams against Helicobacter pylori NCTC 11637. J. Antimicrob. Chemother. 42:661-663. [DOI] [PubMed] [Google Scholar]
- 20.Ito, E., and J. L. Strominger. 1962. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. J. Biol. Chem. 237:2689-2703. [PubMed] [Google Scholar]
- 21.Kato, M., Y. Yamaoka, R. R. Kim, and M. Asaka. 2000. Regional differences in metronidazole resistance and increasing clarithromycin resistance among Helicobacter pylori isolates from Japan. Antimicrob. Agents Chemother. 44:2214-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katyssulla, C., B. Gerold, A. Günter, G. D. Maria, E. Lars, F. Per, A. D. P. Miguel, and G. D. P. Francisco. 1999. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 181:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labenz, J., E. Gyenes, G. H. Rühl, and G. Börsch. 1993. Omeprazole plus amoxicillin: efficacy of various treatment regimens to eradicate Helicobacter pylori. Am. J. Gastroenterol. 88:491-495. [PubMed] [Google Scholar]
- 24.Logan, R. P. H., P. A. Gummett, B. T. Hegarty, M. M. Walker, J. H. Baron, and J. J. Misiewicz. 1992. Clarithromycin and omeprazole for Helicobacter pylori. Lancet 340:239. [DOI] [PubMed] [Google Scholar]
- 25.Majcherczyk, P. A. 1996. The issue of the true post antibiotic effect. J. Antimicrob. Chemother. 37:188-189. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 8390:1311-1315. [DOI] [PubMed] [Google Scholar]
- 27.NCCLS. 2003. Performance standards for antimicrobial susceptibility testing: M100-S13. NCCLS, Wayne, Pa.
- 28.Nobata, K., K. Ina, M. Ohta, K. Kawamura-Sato, T. Tsuzuki, T. Ando, and K. Kusugami. 2002. Lower concentrations of clarithromycin suppress urease activity, motility, and binding to gastric epithelial cells in Helicobacter pylori isolates. Dig. Liver Dis. 34:489-497. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schwartz, A. C., W. Quecke, and G. Brenschede. 1979. Inhibition by glycine of the catabolic reduction of proline in Clostridium sticklandii: evidence on the regulation of amino acid reduction. Z. Allg. Mikrobiol. 19:211-220. [DOI] [PubMed] [Google Scholar]
- 31.Sjöström, J. E., J. Fryklund, T. Kühler, and H. Larsson. 1996. In vitro antibacterial activity of omeprazole and its selectivity for Helicobacter spp. are dependent on incubation conditions. Antimicrob. Agents Chemother. 40:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone, G. G., D. Shortridge, and J. Versalovic. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stronminger, J. L., and C. H. Birge. 1965. Nucleotide accumulation induced in Staphylococcus aureus by glycine. J. Bacteriol. 89:1124-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, J. J. Y., S. Chung, T. K. W. Ling, M. Y. Yung, V. K. S. Leung, E. K. W. Ng, M. K. K. Li, A. F. B. Cheng, and A. K. C. Li. 1995. Antibacterial treatment of gastric ulcers associated with Helicobacter pylori. N. Engl. J. Med. 332:139-142. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, H., M. Shirai, J. K. Akada, M. Tsuda, and T. Nakazawa. 1998. Nucleotide sequence and characterization of cdrA, a cell division related gene of Helicobacter pylori. J. Bacteriol. 180:5263-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teresa, A., D. Diego, P. Nuria, and L. B. Manuel. 2000. Clarithromycin resistance stability in Helicobacter pylori: influence of the MIC and type of mutation in the 23S rRNA. J. Antimicrob. Chemother. 46:613-616. [DOI] [PubMed] [Google Scholar]
- 37.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 38.Tsuzuki, T., K. Ina, M. Ohta, T. Hasegawa, T. Nagasaka, N. Saburi, M. Ueda, T. Konagaya, H. Kaneko, A. Imada, T. Nishiwaki, K. Nobata, T. Ando, and K. Kusugami. 2002. Clarithromycin increases the release of heat shock protein B from Helicobacter pylori. Aliment. Pharmacol. Ther. 16(Suppl. 2):217-228. [DOI] [PubMed] [Google Scholar]
- 39.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Tanaiyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 40.Yakoob, J., F. Xuegong., H. Guoling, L. Li, and Z. Zheng. 2001. Antibiotic susceptibility of Helicobacter pylori in the Chinese population. J. Gastroenterol. Hepatol. 16:981-985. [DOI] [PubMed] [Google Scholar]