Abstract
The evolution of antimicrobial resistance in Salmonella isolates causing traveler's diarrhea (TD) and their mechanisms of resistance to several antimicrobial agents were analyzed. From 1995 to 2002, a total of 62 Salmonella strains were isolated from stools of patients with TD. The antimicrobial susceptibility to 12 antibiotics was determined, and the molecular mechanisms of resistance to several of them were detected as well. The highest levels of resistance were found against tetracycline and ampicillin (21 and 19%, respectively), followed by resistance to nalidixic acid (16%), which was mainly detected from 2000 onward. Molecular mechanisms of resistance were analyzed in 16 isolates. In these isolates, which were resistant to ampicillin, two genes encoding β-lactamases were detected: oxa-1 (one isolate) and tem-like (seven isolates [in one strain concomitantly with a carb-2]). Resistance to tetracycline was mainly related to tetA (five cases) and to tetB and tetG (one case each). Resistance to chloramphenicol was related to the presence of the floR and cmlA genes and to chloramphenicol acetyltransferase activity in one case each. Different genes encoding dihydrofolate-reductases (dfrA1, dfrA12, dfrA14, and dfrA17) were detected in trimethoprim-resistant isolates. Resistance to nalidixic acid was related to the presence of mutations in the amino acid codons 83 or 87 of the gyrA gene. Further surveillance of the Salmonella spp. causing TD is needed to detect trends in their resistance to antimicrobial agents, as we have shown in our study with nalidixic acid. Moreover, such studies will lead to better treatment and strategies to prevent and limit their spread.
Traveler's diarrhea (TD) is the most frequent infection acquired by travelers to developing countries, affecting 20 to 50% of the 35 million travelers from industrialized countries each year. The most important risk factors are the destination of the traveler, host-associated factors, and exposure to contaminated food and water (19). Farm animals often carry salmonellas, affecting meat, dairy products and eggs. Thus, salmonella is one of the main etiological agents causing TD (7, 19, 36).
Salmonella usually produces a self-limited illness, although the duration or the severity of the symptoms may require antibiotic treatment. Salmonella spp. display high natural susceptibility levels to the most commonly used antibacterial agents (34). However, the occurrences of isolated Salmonella strains showing resistance to one or more antibacterial agent have steadily increased, probably due to continuous antibiotic pressure (3, 6, 18, 27, 35). This is an important public health problem that may be related to therapeutic failure (43).
This problem is especially relevant in developing areas, where the lack of economic resources does not allow a wide antibacterial armamentarium. Moreover, in some of these areas, both the social situation and the presence of other basal illnesses, such as malaria, favor the acquisition of systemic Salmonella infections (9). To date, only a few studies have extensively analyzed the levels of resistance to antimicrobial agents in Salmonella spp. isolated in developing areas (9, 15, 16, 21, 31). Moreover, none of them have analyzed in depth the mechanisms of resistance underlying the resistant phenotypes.
Analysis of bacterial infections in international travelers is an indirect source of information about these developing countries, providing information on both the characteristics of these particular infections and the current situation in some less-developed areas.
Thus, resistance to some antibiotics, such as β-lactam, tetracycline, chloramphenicol, or trimethoprim-sulfamethoxazole is being reported with increasing frequency (3, 12). Moreover, the development of quinolone resistance has been described not only in some clones of the widespread Salmonella enterica serovar Typhimurium definitive phage type 104 (8) but also in some Salmonella strains isolated from travelers returning from India and other areas due to the introduction of nalidixic acid for the treatment of some infections (4, 12).
The aim of our study was to analyze the evolution of antimicrobial resistance in Salmonella isolates causing TD and to characterize the mechanisms of resistance to several antimicrobial agents.
MATERIALS AND METHODS
Bacterial isolates.
From 1995 to 2002, a total of 2,216 travelers with TD presented at the Tropical Medicine Unit; feces samples were processed at the laboratory of clinical microbiology of the Hospital Clinic, Barcelona, Spain. In all cases, diarrhea commenced during the trip or no more than 3 days after return. Diarrheagenic pathogens were isolated and identified by conventional methods (20). All patients filled out a clinical and epidemiological protocol form.
Serotyping and phage typing.
Serotyping was performed with somatic and flagella antiserum. The flagellum phase was determined by the inversion of phase method as described by Kauffman and White and according to the recommendations of Edward and Ewing (20). We also determined the phage types of Salmonella enterica serovars Enteritidis, Typhimurium, Hadar, and Virchow (20).
Antimicrobial susceptibility testing.
The antimicrobial susceptibility test to 12 antimicrobial agents: ampicillin, amoxicillin-clavulanic acid, nalidixic acid, tetracycline, trimethoprim-sulfamethoxazole, chloramphenicol, gentamicin, amikacin, imipenem, norfloxacin, ciprofloxacin, and ceftazidime were performed by using the Kirby-Bauer method (2). Interpretation of results was performed according to NCCLS guidelines.
The MICs of ampicillin, chloramphenicol, nalidixic acid, tetracycline, trimethoprim-sulfamethoxazole, ciprofloxacin, and amoxicillin-clavulanic acid to the strains showing resistance was also determined by the agar dilution method according to NCCLS guidelines. Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 25922, Escherichia coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as controls (23).
Detection of the mechanism of resistance.
To determine the quinolone resistance mechanisms, mutations in the gyrA and parC genes were detected by PCR with the primers and conditions presented in Table 1. The presence of the cmlA and floR genes associated with chloramphenicol resistance was determined by PCR (Table 1) and electrophoresis in 2% agarose gels. A previously described colorimetric assay was performed to determine the presence of chloramphenicol acetyltransferase (CAT) activity (6, 11). The detection of genes encoding β-lactamases (tem-like, carb-like, shv-like, and oxa-1-like (24) was carried out by PCR (Table 1). The PCR products were detected by electrophoresis in 2% agarose gels. To detect the mechanism of tetracycline resistance, the presence of tetA, tetB, and tetG genes was determined by PCR and electrophoresis in 2% agarose gels (Table 1) (10, 11, 26). To determine the mechanism of trimethoprim resistance, the presence of dihydrofolate reductases was detected by PCR with generic primers (Table 1), and posterior restriction fragment length polymorphism with the appropriate restriction enzyme as described previously (25).
TABLE 1.
Sequences of primers for amplification
| Antibiotic | Gene | Primer sequence | Amplicon size (bp) | Annealing temp (°C) | Source or reference |
|---|---|---|---|---|---|
| Nalidixic acid | gyrA | 5′-AAATCTGCCCGTGTCGTTGGT-3′ | 343 | 55 | 30 |
| 5′-GCCATACCTACG GCGATACC-3′ | |||||
| Tetracycline | tetA | 5′-GATATTCTGAGCACTGTCGC-3′ | 950 | 55 | 10 |
| 5′-CTGCCTGGACAACATTGCTT-3′ | |||||
| tetB | 5′-TTGGTTAGGGGCAAGTTTTG-3′ | 600 | 55 | 26 | |
| 5′-GTAATGGGCCAATAACACCG-3′ | |||||
| tetG | 5′-GCTCGGTGGTATCTCTGC-3′ | 500 | 55 | 26 | |
| 5′-AGCAACAGAATCGGGAAC-3′ | |||||
| Trimethoprim | dfrA1 | 5′-GTGAAACTATCACTAATGG-3′ | 474 | 55 | 25 |
| 5′-TTAACCCTTTTGCCAGATTT-3′ | |||||
| dfrA14 | 5′-GAGCAGCTICTITTIAAAGC-3′ | 393 | 60 | 25 | |
| 5′-TTAGCCCTTTIICCAATTTT-3′ | |||||
| dfrA12 | 5′-GGTGSGCAGAAGATTTTTCGC-3′ | 319 | 60 | 25 | |
| 5′-TGGGAAGAAGGCGTCACCCTC-3′ | |||||
| dfrA7 | 5′-TTGAAAATTTCATTGATTG-3′ | 474 | 55 | 25 | |
| 5′-TTAGCCTTTTTTCCAAATCT-3′ | |||||
| dfrB | 5′-GATCACGTGCGCAAGAAATC-3′ | 141 | 60 | 25 | |
| 5′-AAGCGCAGCCACAGGATAAAT-3′ | |||||
| Chloramphenicol | floR | 5′-CACGTTGAGCCTCTATAT-3′ | 868 | 55 | 26 |
| 5′-ATGCAGAAGTAGAACGCG-3′ | |||||
| cmlA | 5′-TGTCATTTACGGCATACTCG-3′ | 435 | 55 | This study | |
| 5′-ATCAGGCATCCCATTCCCAT-3′ | |||||
| Ampicillin | tem | 5′-TTGGGTGCACGAGTGGGTTA-3′ | 503 | 55 | 6 |
| 5′-GACAGTTACCAATGCTTAATCA-3′ | |||||
| carb | 5′-AATGGCAATCAGCGCTTCCC-3′ | 586 | 55 | 6 | |
| 5′-GGGGCTTGATGCTCACTCCA-3′ | |||||
| oxa-1-like | 5′-ACCAGATTCAACTTTCAA-3′ | 598 | 55 | 6 | |
| 5′-TCTTGGCTTTTATGCTTG-3′ | |||||
| shv | 5′-ATGCGTTATATTCGCCTGTG-3′ | 841 | 55 | This study | |
| 5′-TTAGCGTTGCCAGTGCTCG-3′ |
DNA sequencing of the PCR products.
The purified PCR products visualized in gels were processed for DNA sequencing and analyzed in an automatic DNA sequencer (ABI 377; Perkin-Elmer, Emeryville, Calif.) by using the BigDye terminator cycle sequencing kit (v3.1; Perkin-Elmer).
RESULTS AND DISCUSSION
From 1995 to 2002, 2,216 patients with TD were treated in the Tropical Medicine Unit of our hospital. Salmonella spp. were identified in 62 patients (3.8%) in as described in previous studies developed for Spanish travelers (37), showing that the relevance of this pathogen remains unaltered as a cause of TD. The distribution of the Salmonella spp. clinical isolates according to serovar and geographical origin is shown in Table 2. Overall, 20 different serovars were identified, with serovar Enteritidis being the most prevalent, followed by serovar Typhimurium. Similar results were reported by Hakanen et al. (12) for Salmonella spp. isolated from travelers to Southeast Asia. The remaining isolates belonged to a wide variety of serovars (Table 2).
TABLE 2.
Distribution of Salmonella clinical isolates according to serovar and geographical origin
| No. of strains | Serovar | Phage type | Country(ies) visited (n)b |
|---|---|---|---|
| 4 | Enteritidis | 4 | Senegal, Ecuador, Thailand, ND |
| 10 | Enteritidis | 1 | Kenya, Ecuador, Australia, India, Mexico/Guatemala, Peru, Morocco, ND (3) |
| 1 | Enteritidis | 20 | Tanzania |
| 4 | Enteritidis | Turkey, Nepal, Dominican Republic, SEA | |
| 5 | Typhimurium | Maldivas, Mali, Cuba, India/Nepal, Gambia | |
| 1 | Typhimurium | 104B | Ivory Coast |
| 1 | Braenderup | Nicaragua | |
| 1 | Tenesse | Egypt | |
| 1 | Newport | Mexico/El Salvador | |
| 1 | London | ND | |
| 1 | Vinohrady | Mali | |
| 1 | Paratyphi A | India | |
| 1 | Carno | Ivory Coast | |
| 1 | Typhi | Philippines | |
| 1 | Kiambu | Mali/Burkina-Faso | |
| 1 | Muenchen | Cameroon | |
| 1 | Hadar | NRPa | Bolivia |
| 1 | Agona | India | |
| 1 | Infantis | Equatorial Guinea | |
| 2 | Kentucky | Mali/Burkina-Faso, Senegal | |
| 1 | Cholerasuis | Mexico | |
| 1 | Haifa | Egypt | |
| 1 | Wangata | Equatorial Guinea | |
| 1 | Lexington | Uganda | |
| 1 | Montevideo | Senegal | |
| 1 | Goldcoast | Senegal | |
| 1 | Risseu | Mali/Burkina-Faso | |
| 1 | Virchow | 31 | India |
| 14 | Salmonella spp. | India (3), Mali (2), SEA, Morocco, South Africa, Peru, Cuba, Vietnam, Nepal, Indonesia (2) |
NPR, nonrecognized pattern.
ND, not determined; SEA, Southeast Asia. n = number of isolates.
According to the geographical origins, 16 isolates (25.8%) were from Western Africa and 8 (12.9%) were from the Indian subcontinent. Meanwhile, other geographical areas showed <10% of the total isolates (Table 2), and six isolates were of unknown origin. No relationship was observed between the serotypes found and the geographical origin of the samples.
In some of these patients other microorganisms, such as Shigella sp. (one case), Campylobacter spp. (two cases), Aeromonas sp. (one case), diarrheagenic Escherichia coli (four cases), and Giardia lamblia (one case) were isolated.
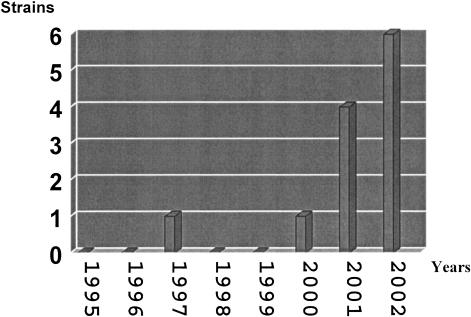
Twenty-eight strains showed resistance to at least one of the antibacterial agents analyzed. Seven of these strains showed resistance to three or more unrelated antibacterial agents being considered as multiresistant strains. The antimicrobial resistance observed mainly affected five antibacterial agents: ampicillin, trimethoprim-sulfamethoxazole, chloramphenicol, tetracycline, and nalidixic acid. Similar results have been observed in analyses of clinical isolates in developing areas (9, 16, 21, 31, 39). The highest levels of resistance, 21 and 19%, were found against tetracycline and ampicillin, respectively, followed by nalidixic acid (16%). Resistance to the other antimicrobial agents tested was <10% (Table 3). The incidence of nalidixic acid-resistant Salmonella isolates causing TD has been steadily rising since 2000 (Fig. 1) and has mainly been observed in isolates belonging to serotype Enteritidis. The wide use of quinolones such as nalidixic acid and ciprofloxacin for the treatment of infections in regions such as India and Central America has been correlated with the increase in resistance to these antimicrobial agents (4, 39, 40). In our study, two nalidixic acid-resistant strains were isolated from feces of travelers to India, and two strains were from Central America. The remaining nalidixic acid-resistant isolates were from Egypt, Morocco, Peru, and Kenya, with two strains of unknown origin. These results suggest that quinolone resistance is not limited to aforementioned areas but is widespread.
TABLE 3.
Antimicrobial susceptibility of Salmonella clinical isolatesa
| Antimicrobial agent | Susceptible
|
Intermediate
|
Resistant
|
|||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Ampicillin | 49 | 79 | 1 | 2 | 12 | 19 |
| Amoxicillin-clavulanic acid | 57 | 92 | 5 | 8 | 0 | 0 |
| Imipenem | 62 | 100 | 0 | 0 | 0 | 0 |
| Ceftazidime | 62 | 100 | 0 | 0 | 0 | 0 |
| Norfloxacin | 62 | 100 | 0 | 0 | 0 | 0 |
| Nalidixic acid | 51 | 82 | 1 | 2 | 10 | 16 |
| Ciprofloxacin | 62 | 100 | 0 | 0 | 0 | 0 |
| Gentamicin | 59 | 95 | 0 | 0 | 3 | 5 |
| Amikacin | 62 | 100 | 0 | 0 | 0 | 0 |
| Trimethoprim-sulfamethoxazole | 56 | 91 | 0 | 0 | 6 | 9 |
| Tetracycline | 47 | 76 | 2 | 3 | 13 | 21 |
| Chloramphenicol | 57 | 92 | 0 | 0 | 5 | 8 |
n, number of strains.
FIG. 1.
Evolution of nalidixic acid resistance in Salmonella spp. causing TD from 1995 to 2002. The resistance to nalidixic acid represented in this figure was established according to the number of strains isolated in each year.
Antibiotic treatment (mainly ciprofloxacin) was required in 73% of the patients. In all cases the treatment was done empirically prior to obtaining laboratory results. The analysis of the outcome of these patients showed that treatment with ciprofloxacin was effective in all patients with nalidixic acid-resistant isolates except one, in whom a change in the treatment to amoxicillin-clavulanic acid was required.
Reports of different microorganisms, such as Shigella flexneri or Salmonella enterica serovar Typhi, showed high levels of therapeutic failure in treatments with fluoroquinolones in patients infected with isolates showing nalidixic acid resistance but fluoroquinolone susceptibility (17, 41). Thus, although fluoroquinolones might be used as treatment in diarrhea by nalidixic acid-resistant Salmonella strains, this option should be carefully chosen, as shown by the therapeutic failure reported in the present study.
Only 44 of the 62 Salmonella strains were able to grow from the frozen stock, and these strains were therefore serotyped again and phage typed in a reference laboratory (Table 2). Of these 44 isolates, 16 isolates showed resistance to any antibiotic and were further investigated to determine the mechanisms of antibacterial resistance.
Single amino acid substitutions in both positions 83 and 87 of GyrA have been described as a cause of nalidixic acid resistance in Salmonella spp., whereas substitutions at both positions plus substitutions in ParC have been detected in fluoroquinolone-resistant isolates (1, 11, 13, 22, 30, 32). In the present study 9 of these 16 isolates showed resistance to nalidixic acid and decreased susceptibility to ciprofloxacin and norfloxacin. Seven were identified as serovar Enteritidis; the rest were identified as serovar Virchow and serovar Haifa. Eight of these nine nalidixic-resistant Salmonella strains, including all of the serovar Enteritidis strains, presented a mutation in codon 87 of the gyrA gene, resulting in the amino acid change Asp to Tyr, whereas the other strain (serovar Haifa) presented a mutation in amino acid codon 83 (Ser to Tyr). No mutation was found in the parC gene.
Eight isolates were resistant to ampicillin. This resistance was associated with the presence of β-lactamases. The oxa-1 gene was detected in one strain, whereas a tem-like gene was found in the other seven strains, in one case concomitantly with a carb-2 (pse-1) gene. This result is in agreement with previously developed studies (5, 6, 29, 42). Although in some of these studies, such as that performed by Roy et al. (29), the percentage of TEM-like β-lactamase-producing strains was higher than in ours, this may be explained by the diversity of the geographical origin of the strains collected in our study. In some of the strains producing a TEM-like β-lactamase an intermediate level of susceptibility to amoxicillin plus clavulanic acid was found. It has been shown that an overproduction of these enzymes may result in decreased susceptibility or resistance to amoxicillin plus clavulanic acid, which may explain the above-mentioned situation (33).
The main mechanism of resistance to trimethoprim is the presence of integron-borne dihydrofolate reductases. Only four isolates resistant to trimethoprim were studied, showing the presence of four different dihydrofolate reductase genes (dfrA1, dfrA12, dfrA14, and dfrA17) (Table 4). Despite the greater use of cotrimoxazole in developing countries, in our study only four strains showed resistance to this antimicrobial agent. This finding is in contrast to the high percentages of trimethoprim resistance in Shigella or diarrheagenic E. coli isolates causing TD (38, 39, 40). To our knowledge, this is the first time that dfrA17 has been found in a Salmonella strain of African origin.
TABLE 4.
Mechanisms of resistance of Salmonella spp. causing TD
| Strain | Serovar | Origin | Resistanceb (MIC [μg/ml]) | Mechanism of resistance
|
||||
|---|---|---|---|---|---|---|---|---|
| Q (GyrA) | CHL | AMP | TET | SXT | ||||
| 30922 | Virchow | India | NAL (≥256), SXT (≥32) | Tyr-87 | dfrA14 | |||
| 62155 | Enteritidis | Mexico | AMP (≥256), NAL (≥256) | Tyr-87 | tem | |||
| 57360 | Haifa | Egypt | TET (≥256), NAL (≥256) | Tyr-83 | tetA | |||
| 37758 | Enteritidis | Morocco | NAL (≥256) | Tyr-87 | ||||
| 13472 | Enteritidis | Peru | NAL (≥256) | Tyr-87 | ||||
| 14089 | Enteritidis | NDa | NAL (≥256) | Tyr-87 | ||||
| 49297 | Enteritidis | ND | NAL (≥256) | Tyr-87 | ||||
| 33568 | Enteritidis | India | NAL (≥256) | Tyr-87 | ||||
| 86 DV | Enteritidis | Kenya | NAL (≥256) | Tyr-87 | ||||
| 36452 | Typhimurium | Ivory Coast | TET (128), AMP (≥256) | tem | tetA | |||
| 21340 | Typhimurium | Gambia | TET (≥256), AMP(≥256), SXT (≥32), CHL (128) | cmlA | tem | tetA | dfrA12 | |
| 27976 | Goldcoast | Senegal | TET (128), AMP (≥256), SXT (≥32), CHL (32) | tem | tetA | dfrA17 | ||
| 29839 | Risseu | Mali | TET (128), AMP (≥256) | tem | tetA | |||
| 36498 | Paratyphi | India | TET (≥258), AMP (≥256), SXT (≥32), CHL (≥128) | cat | oxa-1 | tetB | dfrA1 | |
| 13805 | Kiambu | Mali | TET (64), AMP (≥256), CHL (≥256) | floR | tem, carb | tetG | ||
| 15095 | Hadar | Bolivia | TET (128), AMP (≥256) | tem | tetA | |||
ND, not determined.
NAL, nalidixic acid; SXT, trimethoprim-sulfamethoxazole; AMP, ampicillin; TET, tetracycline; CHL, chloramphenicol; Q, quinolones.
Resistance to chloramphenicol was determined in four cases and was associated with the presence of floR and cmlA genes and CAT activity in three different cases. These cases showed chloramphenicol MICs higher than 128 μg/ml, whereas no specific mechanism was identified in the remaining isolate (chloramphenicol MIC of 32 μg/ml) (Table 4).
Eight isolates were resistant to tetracycline; the tetA gene was detected in six cases and the tetB and tetG genes were detected in one case each (Table 4). The intestinal tract is a suitable habitat for the acquisition of the tetA and tetB genes by horizontal gene transfer, since these tetracycline determinants are common in Enterobacteriaceae (10, 14, 28). In a recent study developed in Shigella spp. and enteroinvasive Escherichia coli of different geographical origins, the tetB gene was more prevalent than the tetA gene (14). Our results show that the most prevalent mechanism of resistance to tetracycline among the strains we tested was tetA. This difference may be explained by the fact that Shigella and Salmonella spp. may have different ecological niches.
In summary, our results show the level of antimicrobial resistance of Salmonella spp. from different geographical origins and although, overall, this resistance is lower than that presented by Shigella spp. or diarrheagenic Escherichia coli, the increasing resistance to nalidixic acid in compared to the above-mentioned microorganisms is of special concern and may result in a loss of therapeutic usefulness of fluoroquinolones. Moreover, high heterogeneity of the mechanisms of resistance to ampicillin, trimethoprim, and tetracycline was observed. Surveillance of the Salmonella spp. causing TD is necessary in order to have current information on the resistance levels and the mechanism of resistance present in these strains, which will in turn help to provide better treatment and to develop strategies to prevent and limit their spread.
Acknowledgments
This study was funded by grant FIS02/0353 from the Spanish Ministry of Health and grant 2002 SGR00121 from the Department d'Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya of Spain. R.C. has a fellowship from Fundación Carolina and BBVA (Spain). J.R. and I.O. have RICET fellowships.
REFERENCES
- 1.Baucheron, S., H. Imberechts, E. Chlaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrugs transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT 204. Microb. Drug Resist. 8:285-289. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, A., W. M. M. Kirby, J. C. Sherris, and M. Truck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 3.Brisabois, A., I. Cazin, J. Breuil, and E. Collatz. 1997. Vigilancia de la resistencia a antibióticos de Salmonella. Eurosurv. Month 2:19-20. [Google Scholar]
- 4.Brown, J. C., C. J. Thomson, and S. G. Amyes. 1996. Mutation of the gyrA gene of clinical isolates of Salmonella typhimurium and three other Salmonella species leading to decreased susceptibilities to 4-quinolone drugs. Antimicrob. Agents Chemother. 37:351-356. [DOI] [PubMed] [Google Scholar]
- 5.Bush, K. 2001. New β-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 6.Gallardo, F., J. Ruiz, F. Marco, K. J. Towner, and J. Vila. 1999. Increase in incidence to resistance to ampicillin, chloramphenicol, and trimethoprim in clinical isolates of Salmonella serotype Typhimurium with investigation of molecular epidemiology and mechanisms of resistance. J. Med. Microbiol. 48:367-374. [DOI] [PubMed] [Google Scholar]
- 7.Gascón, J., J. Vila, M. E. Walls, J. Ruiz, J. Vidal, M. Corachan, G. Prats, and M. T. Jiménez de Anta. 1993. Etiology of traveller's diarrhea in Spanish travellers to developing countries. Eur. J. Epidemiol. 9:217-223. [DOI] [PubMed] [Google Scholar]
- 8.Giraud, E., A. Cloeckaert, S. Baucheron, C. Mouline, and E. Chaslus-Dancla. 2003. Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 52:697-703. [DOI] [PubMed] [Google Scholar]
- 9.Graham, S. M., A. L. Walsh, E. M. Molyneux, A. J. Phiri, and M. E. Molineux. 2000. Clinical presentation of non-typhoidal Salmonella bacteriaemia in Malawian children. Trans R. Soc. Trop. Med. Hyg. 94:310-314. [DOI] [PubMed] [Google Scholar]
- 10.Guardabassi, L., L. Dijkshoorn, J. M. Collard, and J. E. Olsen Dalsgaard. 2000. Distribution and in vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 11.Guerra, B., B. Malorny, A. Schroeter, and R. Helmuth. 2003. Multiple resistance mechanisms in fluoroquinolone-resistant Salmonella isolates from Germany. Antimicrob. Agents Chemother. 47:2059.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakanen, A., P. Kotilainen, P. Huovinen, H. Helenius, and A. Siitonens. 2001. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerg. Infect. Dis. 7:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, H., and P. Heisig. 2003. Topoisomerase IV mutations in quinolone-resistant salmonellae selected in vitro. Microb. Drug Resist. 9:25-32. [DOI] [PubMed] [Google Scholar]
- 14.Hartman, A. B., I. I. Essiet, D. W. Isenbarger, and L. E. Linder. 2003. Epidemiology of tetracycline resistance determinants in Shigella spp. an enteroinvasive Escherichia coli: characterization and dissemination of Tet (A)-1. J. Clin. Microbiol. 41:1023-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoge, C. W., J. M. Gambel, A. Srijan, C. Pitarangsi, and P. Echeverría. 1998. Trends in antimicrobial resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin. Infect. Dis. 26:341-345. [DOI] [PubMed] [Google Scholar]
- 16.Isenbarger, D. W., C. W. Hoge, A. Srijan, C. Pitarangsi, N. Vithayasai, L. Bodhidatta, K. W. Hickey, and P. Dac Cam. 2002. Comparative antibiotic resistance of diarrheal pathogens from Vietnam and Thailand, 1996-1999. Emerg. Infect. Dis. 8:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan, W. A., C. Seas, U. Dhar, M. A. Salam, and M. L. Bennish. 1997. Treatment of shigellosis. V. Comparison of azithromycin and ciprofloxacin. Ann. Intern. Med. 126:697-703. [DOI] [PubMed] [Google Scholar]
- 18.Lawson, A. J., M. U. Dassama, L. R. Ward, and E. J. Threlfall. 2000. Multiply resistant (MR) Salmonella enterica serotype Typhimurium DT 12 and DT 120: a case of MR DT 104 in disguise? Emerg. Infect. Dis. 8:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreira Lima, A. A. 2001. Tropical diarrhoea: new developments in traveller's diarrhoea. Curr. Opin. Infect. Dis. 14:547-552. [DOI] [PubMed] [Google Scholar]
- 20.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1995. Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 21.Mwansa, J., K. Mutela, I. Zulu, B. Amadi, and P. Kelly. 2002. Antimicrobial sensitivity in enterobacteria from AIDS patients, Zambia. Emerg. Infect. Dis. 8:92-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaya, H., A. Yasuhara, K. Yoshimura, Y. Oshihoi, H. Izumiya, and H. Watanabe. 2003. Life-threatening infantile diarrhea from fluoroquinolone-resistant Salmonella enterica Typhimurium with mutation in both gyrA and parC Emerg. Infect. Dis. 9:255-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing, 9th ed. Approved standard M100-S13. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Navia, M. M., L. Capitano, J. Ruiz, M. Vargas, H. Urassa, D. Schellemberg, J. Gascón, and J. Vila. 1999. Typing and characterization of mechanisms of resistance Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J. Clin. Microbiol. 37:3113-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navia, M. M., J. Ruiz, J. Sánchez Céspedes, and J. Vila. 2003. Detection of dihydrofolate reductase genes by PCR and RFLP. Diagn. Microbiol. Infect. Dis. 46:295-298. [DOI] [PubMed] [Google Scholar]
- 26.Ng, L.-K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orman, Be., S. A. Piñeiro, S. Arduino, M. Galas, R. Melano, M. I. Caffer, D. O. Sordelli, and D. Centrón. 2002. Evolution of multiresistance in no typhoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microb. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 29.Roy, C., A. Foz, C. Segura, M. Tirado, C. Fuster, and R. Reig. 1983. Plasmid-determined β-lactamases identified in a group of 204 ampicillin-resistant Enterobacteriaceae. J. Antimicrob. Chemother. 12:507-510. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, J., D. Castro, P. Goñi, J. A. Santamaría, J. J. Borrego, and J. Vila. 1997. Analysis of the mechanism of quinolone-resistance in nalidixic acid-resistance clinical isolates of Salmonella serotype Typhimurium. J. Med. Microbiol. 46:623-628. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro, R. L., L. Kumar, P. Phillips-Howard, J. G. Wells, P. Adcock, J. Brooks, M. L. Ackers, J. B. Oshieng, E. Mintz, S. Wahlquist, P. Waiyaki, and L. Slutsker. 2001. Antimicrobial resistance bacterial diarrhea in rural Western Kenya. J. Infect. Dis. 183:1701-1704. [DOI] [PubMed] [Google Scholar]
- 32.Soto, S. M., J. Ruiz, M. C. Mendoza, and J. Vila. 2003. In vitro fluoroquinolone-resistant mutants of Salmonella enterica serotype Enteritidis: analysis of mechanisms involved in resistance. Int. J. Antimicrob. Agents. 22:537-540. [DOI] [PubMed] [Google Scholar]
- 33.Stapleton, P., P. Wu, A. King, K. Shanon, G. French, and I. Phillips. 1995. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob. Agents Chemother. 39:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock, I., and B. Wiedemann. 2000. Natural antibiotic susceptibility of Salmonella enteric strains. Int. J. Antimicrob. Agents 16:211-217. [DOI] [PubMed] [Google Scholar]
- 35.Szych, J., A. Cieslik, J. Pacioreck, and S. Kaluzewski. 2001. Antibiotic resistance in Salmonella enterica subsp. enterica strains isolated in Poland from 1998 to 1999. Int. J. Antimicrob. Agents. 18:37-42. [DOI] [PubMed] [Google Scholar]
- 36.Venolakis, E. N., A. Markogiannakis, L. Kondili, E. Varjioti, Z. Mahera, E. Dedouli, A. Karaitianou, N. Vakalis, and K. Bethimouti. 2001. Evolución de la resistencia a antibióticos de Salmonellas no tifoideas en Grecia durante el período comprendido entre 1990 y 1997. Eurosurv. Month 6:117-120. [Google Scholar]
- 37.Vila, J., J. Gascón, S. Abdalla, J. Gómez, A. Moreno, M. Corachán, and M. T. Jimenez de Anta. 1995. Antimicrobial resistance of nontyphoidal Salmonella isolates in traveller's diarrhea. J. Travel Med. 2:45-47. [DOI] [PubMed] [Google Scholar]
- 38.Vila, J., J. Gascón, S. Abdalla, J. Gómez, A. Moreno, M. Corachán, and M. T. Jimenez de Anta. 1994. Antimicrobial resistance of Shigella isolates causing traveler's diarrhoea. Antimicrob. Agents Chemother. 38:2668-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vila, J., M. Vargas, J. Ruiz, M. Corachán, M. T. Jiménez de Anta, and J. Gascón. 2000. Quinolone resistance in enterotoxigenic Escherichia coli causing diarrhea in travelers to India in comparison with travelers to other geographical areas. Antimicrob. Agents Chemother. 44:1731-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vila, J., M. Vargas, J. Ruiz, M. Espasa, M. Pujol, M. Corachán, M. T. Jiménez de Anta, and J. Gascón. 2001. Susceptibility patterns of enteroaggregative Escherichia coli associated to traveller's diarrhoea: emergence of quinolone resistance. J. Med. Microbiol. 50:996-1000. [DOI] [PubMed] [Google Scholar]
- 41.Wain, J., N. T. T. Hoa, N. T. Chinh, H. Vinh, M. J. Everett, T. S. Diep, N. P. J. Day, T. Solomon, N. J. White, L. J. V. Piddock, and C. M. Parry. 1997. Quinolone-resistant Salmonella typhi in Vietnam: molecular basis of resistance and clinical response to treatment. Clin. Infect. Dis. 25:1404-1410. [DOI] [PubMed] [Google Scholar]
- 42.Walker, R. A., E. Lindsay, M. J. Wooward, L. R. Ward, and J. Threlfall. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype Typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 7:13-21. [DOI] [PubMed] [Google Scholar]
- 43.Zahurul Haque Asna, S. M., J. Ashraful Haq, and M. D. Mushfequr Rahman. 2003. Nalidixic acid-resistant Salmonella enterica serovar Typhi with decreased susceptibility to ciprofloxacin caused treatment failure: a report from Bangladesh. Jpn. J. Infect. Dis. 56:32-33. [PubMed] [Google Scholar]