Abstract
Replication of Streptomyces linear chromosomes and plasmids proceeds bidirectionally from a central origin, leaving recessed 5′ termini that are extended by a telomere binding complex. This complex contains both a telomere-protecting terminal protein (Tpg) and a telomere-associated protein that interacts with Tpg and the DNA ends of linear Streptomyces replicons. By using histidine-tagged telomere-associated protein (Tap) as a scaffold, we identified DNA polymerase (PolA) and topoisomerase I (TopA) proteins as other components of the Streptomyces telomere complex. Biochemical characterization of these proteins indicated that both PolA and TopA exhibit highly efficient reverse transcriptase (RT) activity in addition to their predicted functions. Although RT activity innate to other DNA-dependent DNA polymerases has been observed previously, its occurrence in a topoisomerase is unprecedented. Deletion mapping and sequence analysis showed that the RT activity of Streptomcyces TopA resides in a peptide region containing motifs that are absent from most bacterial topoisomerases but are highly conserved in a novel subfamily of eubacterial topoisomerases found largely in Actinobacteria. Within one of these motifs, and essential to the RT function of Streptomyces TopA, is an Asp-Asp doublet sequence required also for the RT activities of human immunodeficiency virus and eukaryotic cell telomerases.
Keywords: telomere replication, TopA, PolA
Because DNA replication requires both a primer and template and can proceed only in the 5′ to 3′ direction, replication at the telomeric ends of linear plasmids and chromosomes requires a specialized mechanism to ensure that full-length replicas of both DNA strands are produced (1). In eukaryotes, the preservation of telomere length involves the synthesis of tandem DNA repeats on an RNA template; telomerase, a ribonucleoprotein that functions as an RNA-dependent DNA polymerase (i.e., reverse transcriptase, RT), plays a crucial role in this process (2-5).
Streptomyces are eubacteria that contain linear plasmids and chromosomes, as well as circular replicons (6, 7). In contrast to eukaryotic cell adenoviruses and bacteriophage Ø29 of Bacillus subtilis, which replicate full-length linear DNA molecules by a strand displacement mechanism (8, 9), replication of linear DNAs in Streptomyces is initiated bidirectionally from an internal origin, generating 3′ leading strand overhangs on replication intermediates (10). Extension of the recessed 5′ ends is accomplished by a still-unknown mechanism that requires the functions of (i) telomeric inverted repeats (11, 12), (ii) a 21-kDa terminal protein (Tpg) that is attached covalently to 5′ DNA termini of postreplicative DNA molecules and is believed to act as a primer for lagging strand DNA replication (13, 14), and (iii) Tap, an 80-kDa telomere-associated protein that interacts with both Tpg and the overhanging 3′ strand sequence of replication intermediates (15). We report here that the telomere complex of Streptomyces linear replicons also includes DNA polymerase and DNA topoisomerase proteins essential to Streptomyces propagation and, remarkably, that both of these eubacterial enzymes can function efficiently as RTs in addition to having the biochemical properties predicted from their sequences. We further show that the Streptomyces coelicolor and Streptomyces lividans TopA proteins are prototypes for a subfamily of bacterial topoisomerases whose catalytic domains contain a unique Asp-Asp doublet motif that is required for their RT activity and which is essential also to the RNA-dependent DNA polymerase functions of HIV RT and eukaryotic cell telomerases (16, 17).
Materials and Methods
Plasmid and Bacterial Strains. S. coelicolor strains M145 and S. lividans 1326 (18) were kindly provided by D. A. Hopwood (John Innes Center, Norwich, U.K.). The Streptomyces-Escherichia coli shuttle plasmid pHZ1272, which replicates in both hosts and was used for gene expression in Streptomyces, was kindly provided by Z. Deng (Shanghai Jiaotong University, Shanghai, China) in a personal communication. S. lividans strain BKKO19, in which the originally linear chromosome had circularized (15), was used to generate deletions of the topA or polA gene as described in ref. 15.
Expression and Purification of Tap. The tap gene of S. coelicolor (15) was amplified by using a pair of primers: 5′-AAACATATGCATCATCATCATCATCATGTGTCCGGTAGAGGAGCGCAG-3′ and 5′-GCCGTTGCCGAACAGGCTCAT-3′, and the amplicon was inserted into the NdeI/EcoRI-digested pHZ1272 vector (Z. Deng, personal communication), generating plasmid pBC174. The correctness of the construct was confirmed by DNA sequence analysis. pBC174 was introduced by transformation into S. lividans 1326 for expressing Tap containing an NH2-terminal his6 tag encoded by the primer. Bacterial cultures were grown in yeast extract/malt extract (YEME) medium (19) for 12 h, and after treatment with 10 μg/ml thiostrepton at 30°C for 12 h, cells were harvested and resuspended in L buffer (50 mM NaH2PO4, pH 7.5/300 mM NaCl/10 mM imidazole/10% glycerol/0.2% Triton X-100/5 mM 2-mercaptoethanol/1 mM PMSF) before lysis by a French press (pressure at 1,000 kg/cm2). DNA in the lysis mixture was sheared by sonication, and cell debris was removed by centrifugation (39,000 × g for 20 min). The supernatant was incubated at 4°C for 90 min with Ni-NTA resin (Qiagen, Valencia, CA), which was then poured into a column and washed sequentially with L buffer containing increasing concentrations of imidazole. The Tap-His-6 fusion protein, which was detected in Ni-NTA column eluates by Western blot analysis using anti-His-tag antibody (Qiagen) was eluted from the column at ≈100 mM imidazole. Tap-containing fractions were dialyzed against S buffer (50 mM Tris·HCl, pH 8.0/100 mM NaCl/0.5 mM EDTA/0.5 mM DTT/10% glycerol) and stored at -80°C.
RT Activity Assays. RT assays were carried out in 20-μl reaction mixtures containing 20 mM Tris·HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, 50 ng/μl poly(rA)-(dT)12-18 (Amersham Pharmacia), 10 μCi (1 Ci = 37 GBq) of [32P]dTTP (Amersham Pharmacia), and 1-μl volumes of protein fractions. Reaction mixtures were incubated at 37°C for 45 min, stopped by addition of 10 μl of 200 mM EDTA, and purified by using MicroSpin S-200 HR columns (Amersham Pharmacia). Reaction products were analyzed by autoradiography after electrophoresis on 10% polyacrylamide, 8 M urea sequencing gels in buffer containing 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF. RT assays using mRNA isolated from a murine fibroblast 3T3 cell line as template were carried out as follows: poly(A)-containing RNA was isolated from cell cultures by using the FastTrack 2.0 kit (Invitrogen). RT reaction mixtures (20-μl volumes) reaction contained 20 mM Tris·HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, 50 μM dATP, dGTP, and dTTP, 10 μM dCTP, 10 μCi of [α-32P]dCTP (Amersham Pharmacia), 0.1-μg aliquots of poly(A)-mRNA, 50 ng of random (dNTP)6 primers (New England Biolabs), and 1-μl volumes of protein fractions. Assay for the specific activity of RT was performed according to Verma and Baltimore (20).
Protein Purification. Tap-containing fractions from Ni-NTA columns were further purified on a heparin-Sepharose column (Amersham Pharmacia) by using a linear gradient of NaCl (0.2-1 M). Two distinct Tap fractions associated with RT activity were eluted at 0.40 M NaCl and 0.65 M NaCl, whereas most of the 80-kDa Tap protein (as determined by gel electrophoresis) eluted at 0.55 M NaCl. By tracking RT activity, the fraction eluted before the bulk of Tap protein (i.e., at 0.40 NaCl) was purified by further fractionation on Mono Q, poly(rA)Sepharose 4Bm, and phenyl-Sepharose columns to a 99-kDa single protein band on an SDS/PAGE gel, as detected by Silver stain (Bio-Rad). The protein eluting at 0.65 M NaCl was purified as a single 104-kDa band through fractionation on Mono S, poly(rA)-Sepharose 4B, and cellulose phosphate P11 columns.
RT Activity in Renatured Protein Recovered from SDS/PAGE Gels. SDS loading buffer was added to gel slices containing the purified RT protein bands, and the protein was allowed to denature for 10 min at room temperature. The sample was loaded onto an 8% 1.5-mm-thick SDS-polyacrylamide preparative gel. The gel was run at 150 V for 1 h, washed briefly in water, and stained for 15 min in l liter of ice-cold staining buffer containing 250 mM KCl and 2 mM DTT. The gel was detained three times for 10 min in ice-cold water containing 2 mM DTT. The single band visible against a black background was excised and added to 0.5 ml of elution buffer (150 mM NaCl/0.1% SDS/50 mM Tris, pH 8.0/5 mM EDTA/5 mM DTT/100 μg/ml BSA) in an Eppendorf tube. The gel slices were crushed, and an additional 0.5 ml of elution buffer was added. The protein was eluted for 15 h at 4°C with agitation and separated from the acrylamide gel by centrifugation. The supernatant was precipitated with acetone, and the pellet was washed with 1 ml of -20°C acetone wash buffer (80% acetone/20% elution buffer), spun, and allowed to dry at room temperature for 5-15 min. The pellet was then resuspended in 100 μl of denaturing buffer (6 M guanidine-HCl/150 mM NaCl/50 mM Tris, pH 8.0/5 mM EDTA/5 mM DTT/100 μg/ml BSA/20% glycerol) for 30 min on ice and incubated at room temperature for 1 h. The denatured protein was then diluted with 0.55 ml of ice-cold dilution buffer (denaturing buffer without guanidine-HCl), thoroughly mixed, and diluted with another 0.55-ml aliquot dilution buffer. Proteins were allowed to refold for 15 h at 4°C and then dialyzed twice in 500 vol of dilution buffer for 6 h at 4°C. After dialysis, the samples were concentrated by ultrafiltration on a Centricon Plus-20 tube (Amicon) and then either assayed for RT activity or aliquoted and frozen.
Protein MS and Amino Acid Sequence Analysis. Protein MS and amino acid sequence analysis were performed at the Stanford Protein and Nucleic Acid facility and the Harvard Microchemistry Facility (Boston). Proteins eluted from SDS/PAGE gels were processed by proteolytic digestion, microcapillary HPLC, nano-Electrospray Ionization (ESI), and ion trap tandem MS analysis. For NH2-terminal sequence analysis, proteins were blotted onto poly(vinylidene difluoride) membrane according to the guidelines of the Stanford Protein and Nucleic Acid facility.
Cloning and Expression of PolA and TopA of Streptomyces and E. coli. The polA and topA genes of S. coelicolor M145 and E. coli N3433 were amplified by PCR and cloned into the pET28a (Novagen) expression vector. The His-tagged fusion proteins were purified by chromatography on Ni-NTA resin (Qiagen) and confirmed by Western blot analysis using anti-His antibody. The polyhistidine tag was removed from TopA by incubation with thrombin protease (Amersham Pharmacia) at 22°C for 2 h, after which the reaction mixture was passed over a second Ni-NTA column. Proteins were further purified by using heparin-Sepharose columns (Amersham Pharmacia).
Mutagenesis Analysis of Streptomyces DNA Topoisomerase I. PCR amplification of chromosomal DNA with relevant primer pairs was used to create His-tagged truncated forms of Streptomyces DNA topoisomerase I containing deletions of the NH2-terminal 140 amino acids, the COOH-terminal 353 amino acids, both of the above, or the COOH-terminal 400 amino acides alone. These were inserted into the pET28a vector and cloned and expressed in E. coli. The His-tag fusion truncated TopA proteins were purified by chromatography on Ni-NTA resin (Qiagen). The primer pair for full-length Streptomyces topA gene is 5′-A A ACATATGTCCCCGACCAGCGAGACC-3′ and 5′-GTGGAGACGTGCGGAGGAGCGGGC-3′. Other primer pairs were 5′-GCGCATATGGCCGCCGTCGCCA ACCCGCGC-3′ and 5′-GTGGAGACGTGCGGAGGAGCGGGC-3′ for NH2-terminal 140-aa truncation; 5′-AAACATATGTCCCCGACCAGCGAGACC-3′ and 5′-CGTTCAGTCGCCCTCGCCGAAGTAGAAGC-3′ for COOH-terminal 353-aa truncation; and 5′-GTGGTCAACCTCCTGGAGAAGCACTTC-3′ and 5′-GTGGAGACGTGCGGAGGAGCGGGC-3′ for COOH-terminal 400-aa peptide of TopA. The amino acid substitution of Asp-Asp doublet for Ala-Ala doublet of Streptomyces TopA was carried out by using a QuikChange II XL site-directed mutagenesis kit (Stratagene) and a pair of primers: topAS-mut (5′-GAGGGTGCCGCCGCCCCGA ACGCCGAGCTGGCCGCCCGCGAGCGG-3′) and topAS-mut-c (5′-CCGCTCGCGGGCGGCCAGCTCGGCGT TCGGGGCGGCGGCACCCTC-3′).
Gene Disruption. Direct gene disruptions of topA or polA in S. coelicolor M145 were carried out as described in ref. 15. A Streptomyces topA gene disruption plasmid pBC338 was constructed by replacing an apramycin resistance gene (amr) for topA ORF, which flanks ≈1.5-kb chromosomal DNA sequences at both sides in E. coli. To mutate the chromosomal topA gene of Streptomyces M145, protoplasts were transformed with pBC338 isolated from a Dam- Dcm-Hsd- derivative E. coli strain ET12567 (21), which lacks the ability to replicate in Streptomyces and contains a gene [aac (3)IV] encoding apramycin resistance, and transformants (amrspcr) were selected on R5 containing apramycin. These colonies were than transferred by replica plating to media containing either apramycin or spectinomycin to identify spectinomycin-sensitive derivatives that would putatively have undergone a double crossover event leading to replacement of chromosomal topA. An analogous gene-disruption construct (pBC353) was used to attempt deletion of the chromosomal polA gene in strain M145. However, no clone (amrspcs) undergoing double crossover was isolated from 2,000 single crossover transformants (amrspcr) examined in each case. Similar results were obtained during attempts to disrupt the topA or polA gene in S. lividans strain BKKO19, which contains a circular chromosome (15).
topA and polA gene conditional knockout experiments were carried out in S. lividans 1326 by introducing a functional polA or topA gene on a Streptomyces-E. coli shuttle vector pHZ132 (22) containing a temperature-sensitive Streptomyces replicon pSG5 (23). A functional Streptomyces promoter tipAp (thiostrepton inducible promoter, ref. 24) and topA ORF of S. coelicolor M145 were cloned into the BamHI site of pHZ132 as plasmid pBC307, which expresses TopAS in S. lividans 1326. Plasmid pBC316 was similarly constructed with polA ORF of S. coelicolor M145 for providing PolAS in S. lividans 1326. Protoplasts of S. lividans 1326(pBC307) were transformed with pBC338 for screening mutants with the replacement of chromosomal topAS for apramycin resistance gene. Five clones (amrspcs) with double crossover were isolated from 200 single crossover transformants (amrspcr). All five showed the desired chromosomal topAS allelic replacement confirmed by PCR analysis by using primer pair corresponding to flanking DNA sequences of topAS. Similar conditional polAS knockout experiment was carried out with S. lividans 1326(pBC316) and pBC353. Seven clones (amrspcs) with double crossover were isolated from 200 single crossover transformants (amrspcr) and showed the desired replacement of polAS for apramycin resistance gene. The conditional knockouts of TopA or PolA can grow normally at 30°C, but not at 39°C.
Results and Discussion
Earlier work has shown that the terminal proteins of Streptomyces plasmids and chromosomes contain a domain that resembles a DNA binding region of the HIV RT (13). Whereas the absence of a DNA polymerase domain in the 21-kDa Tpg proteins makes it unlikely that Tpg can act independently as a RT (13), the discovery of a region potentially capable of serving as an RT subunit within Tpg prompted us to search for RT activity in the Streptomyces telomere complex.
To isolate the telomere complex, the His-6-tagged Tap (15) was expressed in S. lividans and purified, together with S. lividans proteins that interact with it, by Ni-NTA resin column chromatography. The resulting preparation showed prominent RT activity, as indicated by the ability to incorporate dTTP into oligomers when assayed in reaction mixtures containing poly(A) as template; analysis by SDS/PAGE (Fig. 1A, lane b) indicated that the Tap-based telomere complex contained multiple Coomassie blue-staining bands (Fig. 1B, lane b), and enzymatic assay of protein eluted from different gel regions and then renatured as described in Materials and Methods indicated that RT activity was present in the gel segment containing and bracketing the His-6-tagged Tap protein. In contrast, no RT activity was detected in preparations of His-6-Tap, His-6-Tpg, or His-6-tagged Tap/Tpg mixtures expressed in and recovered from E. coli (data not shown), suggesting that the RT activity identified in the Tap-based S. lividans telomere complex does not reside in either Tpg or Tap proteins acting individually or in combination.
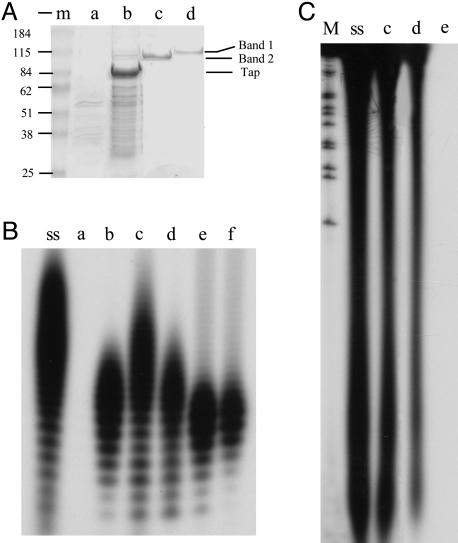
Fig. 1.
RT activity of components of the telomere replication complex of Streptomyces linear replicon. (A) SDS/PAGE analysis of components with RT activity in extracts of S. lividans transfected with pBC174 expressing His-6-tagged Tap protein. Samples were precipitated with 0.015% deoxycholate/5% trichloroacetic acid. Proteins were visualized by Coomassie blue staining. m, protein molecular mass marker (Bio-Rad) shown at left; the approximate mass of marker bands is indicated. Samples isolated at disparate steps in purification are shown. Lanes a and b, corresponding chromatography column fractions eluted from Ni-NTA resin that had been loaded with lysate containing only the expression vector (a) or the same vector expressing Tap. Fractions containing separate protein bands purified to apparent homogeneity by using RT activity as an assay are shown in lanes c and d. The positions of the two purified protein bands and of the 80-kDa Tap protein are indicated at right. (B) Lanes a-d show RT activity analyses of the fractions in A by using polyA as the template. ss, DNA synthesized on polyA template by SuperScript II reverse transcriptase (Invitrogen) was used as a positive control. Lanes e and f show the RT activity of proteins renatured from SDS/PAGE gel corresponding to the single protein band in lanes c and d of A. (C) RT activity analysis by using murine fibroblast cell 3T3 mRNA as template. Lane M, 1-kb DNA ladder labeled with 32P by using KinaseMax 5′ End-Labeling Kit (Ambion, Austin, TX). Lanes ss, c, and d show the DNA synthesized on 3T3 mRNA template by SuperScript II reverse transcriptase (Invitrogen) by using protein fractions c and d of A. Lane e, negative control in which no protein was added to the reaction mixture.
Fractionation of a His-6-tagged Tap preparation by chromatography on a heparin-Sepharose column revealed RT activity in two separate column fractions that flanked the Tap-containing eluate. The RT activity identified in the column fraction eluted before Tap was further purified by fractionation on Mono Q, poly(rA)-Sepharose 4B, and phenyl-Sepharose columns and found to migrate in agarose gels as a single protein band detected by silver staining (Fig. 1 A, Band 2, and B, lane c). A separate protein that contained RT activity was purified as a slower-migrating band (Fig. 1 A, Band 1, and B, lane d) by fractionation on Mono S, poly(rA)-Sepharose 4B, and cellulose phosphate P11 columns. Each purified protein retained its RT activity when eluted from SDS/PAGE gels and renatured. The oligodeoxynucleotide chains generated by both proteins were in the same size range as, but on average were slightly shorter than, cDNA generated by a commercial preparation of retroviral RT (SuperScript II, Invitrogen) (Fig. 1 B, lanes ss and c-f). Pretreatment of the poly(rA)-dAT12-18 substrate with pancreatic ribonuclease (1 mg/ml) for 30 min at 37°C abolished the incorporation of radioactively labeled dTTP by both proteins; additionally, dATP, dGTP, and dCTP were not used in the presence of the poly(rA)-oligo(dT) template, and no RNA-dependent RNA polymerase activity was detected for either protein (data not shown). The RT activity of these two protein preparations was confirmed by using mRNA isolated from a murine fibroblast cell line as template (Fig. 1C, lanes c and d).
MS analysis and amino acid sequencing performed as described in Materials and Methods indicated that the purified Band 2 and Band 1 proteins, respectively, were identical to 99- and 104-kDA proteins annotated as DNA polymerase I (PolA) and DNA topoisomerase I (TopA) (GenBank accession nos. CAB52059.1 and CAB38480.1) (25) of S. coelicolor strain M145, a microorganism that is closely related to S. lividans and whose genome sequence has been fully determined (25). The probabilities for identifying PolA and TopA by these analyses were 1.0e + 000; the probability of the next candidate was 2.1e - 036 for PolA and 2.9e - 036 for TopA.
The polA and topA genes of S. coelicolor M145 were cloned, expressed carrying NH2-terminal His-6 tag in E. coli, and purified; in parallel experiments, we cloned and purified the products of E. coli polA and topA genes. The S. coelicolor protein identified from sequence analysis as PolA, when expressed in E. coli from the cloned S. coelicolor gene had the predicted pI and size and showed not only the expected ability to synthesize DNA by using a DNA template (specific activities shown in Table 1) (26), but also RNA-dependent ability to incorporate deoxynucleotides into acid-insoluble oligomers (Table 1). In contrast, the only DNA polymerase activity detected for the cloned E. coli polA gene product similarly expressed and purified was DNA dependent. In analogous experiments we found that the His-tagged TopA proteins of both Streptomyces and E. coli showed DNA topoisomerase activity (Fig. 2A) but that only the Streptomyces TopA protein displayed RT activity (specific activity, 550 nmol·min-1·mg-1) (Fig. 2B). Neither the S. coelicolor TopA protein nor E. coli TopA fusion protein had detectable DNA-dependent DNA polymerase or RNA polymerase activity (data not shown).
Table 1. Catalytic activities of DNA polymerase I of E. coli and Streptomyces.
| Specific activity, units/mg
|
||
|---|---|---|
| Catalytic activities | E. coli | S. coelicolor |
| DNA polymerase | 5,900 | 4,200 |
| 3′ → 5′ exonuclease | 320 | 210 |
| 5′ → 3′ exonuclease | 850 | 730 |
| Reverse transcriptase | 0 | 2,900 |
One unit of enzyme is defined as conversion of 10 nmol of substrate to product in 30 min as described in Materials and Methods.
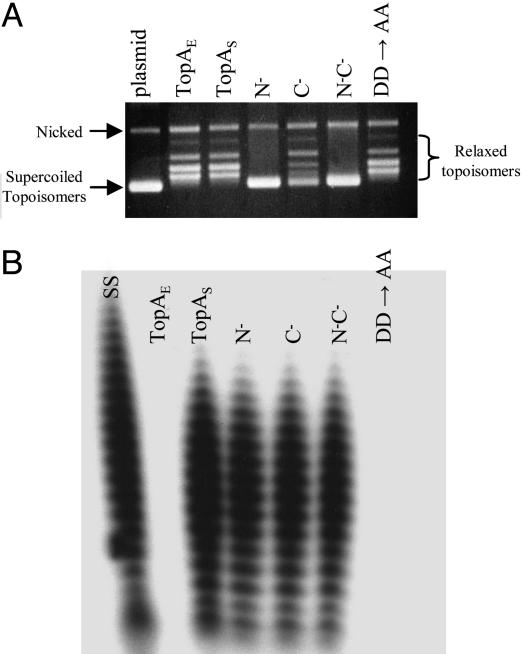
Fig. 2.
Analysis of catalytic activity of DNA topoisomerase I. Proteins are used for analyzed as follows: TopAE, E. coli TopA; TopAS, S. coelicolor TopA; N-, truncated S. coelicolor TopA proteins lacking the NH2-terminal 140 aa; C-, truncated S. coelicolor TopA proteins lacking the COOH-terminal 353 aa; N-C-, truncated S. coelicolor TopA proteins lacking both NH2-terminal and COOH-terminal segments; DD→AA, mutated S. coelicolor TopA protein containing amino acids substitutions of Asp-Asp with Ala-Ala doublet. (A) Assay of DNA topoisomerase activity. Negatively supercoiled plasmid pSP72 DNA (lane plasmid, isolated from E. coli by Qiagen miniprep kit) and the same DNAs relaxed by treatment with above proteins were resolved in a 0.7% agarose gel and stained with ethidium bromide. The relative positions of the negatively supercoiled DNA, and the relaxed plasmid DNA topoisomers ladder are as indicated. (B) RT activity analysis of proteins as indicated above by using a poly(A) template. Lane ss, SuperScript II RT (Invitrogen) used as a positive control.
The predicted amino acid sequence for the S. coelicolor DNA polymerase I protein (25), which carried out RNA-dependent DNA synthesis with approximately half the efficiency as DNA-dependent DNA synthesis (Table 1), revealed extensive homology to DNA polymerase I enzymes of the Archaea species Thermus aquaticus and Thermus thermophilus, which also have been reported to have RT activities (27, 28). Neither the S. coelicolor nor Archea DNA polymerase proteins (25, 27, 28) had detectable similarity to motifs of eukaryotic viral RTs or telomerases (17).
The occurrence of RT activity innate to a topoisomerase protein is unprecedented. Truncation experiments carried out to identify the protein regions containing the individual catalytic activities of the Streptomyces TopA protein localized the topoisomerase activity to NH2-terminal 600 aa and the RT activity to a segment between amino acid residues 141 and 600 (Figs. 2B and 3A). blast sequence analysis and a PSI-blast search of the National Center for Biotechnology Information protein database indicated that this region contains substantial homology to E. coli topoisomerases I and topoisomerase III proteins (56% and 40% similarity, respectively), but also showed that the S. coelicolor topoisomerase I belongs to a distinct subfamily of topoisomerases, which share two conserved segments: amino acid residues 221-264 and 441-498 of S. coelicolor TopA protein (Fig. 3B), with greatest sequence homology to DNA topoisomerase I of Streptomyces avermitilis (95% similarity), Thermobifida fusca (82% similarity), Mycobacterium tuberculosis (72% similarity), Mycobacterium leprae (72% similarity), Deinococcus radiodurans (71% similarity), and Nostoc punctiforme (67% similarity). Within the segment required for RT function of the S. coelicolor TopA protein is a 58-aa region shared by all of these topoisomerases, but not by E. coli TopA. The RT locus of S. coelicolor TopA, as well as the homologous regions of certain other bacterial topoisomerases in its subclass, includes a motif consisting of Asp-Asp (aspartic acid-aspartic acid) residue doublets spaced five amino acids apart (in the S. coelicolor enzyme at amino acid positions 464-465 and 471-472, Fig. 3 A and B). In some topoisomerases in its subclass, a motif consisting of an Asp-Glu (aspartic acid-glutamic acid) doublet spaced six amino acids from an Asp-Asp doublet exists in the conserved region (Fig. 3B).
Fig. 3.
Schematic diagram showing Streptomyces TopA regions identified by blast protein sequence search. An ≈600-aa segment corresponding to the TopA domain of multiple bacterial proteins annotated as topoisomerases contains two short segments (amino acid residues 221-264 and 441-498 of the S. coelicolor TopA protein) that are absent in most of these proteins. These segments are conserved in proteins annotated as topoisomerases in the subclass of enzymes shown in B. As shown, the 441-498 segment contains two Asp-Asp doublets. (B) Alignment of the conserved motif regions of the subfamily bacterial topoisomerases. The bold DD in S. coelicolor are the DD doublets that were mutated to Ala doublets. (C) Phylogenetic analysis of proteins annotated from DNA sequences of certain bacterial genomes as topoisomerase I enzymes. The subfamily identified by the analysis is discussed in the text. The Calculation Phylogeny Unweighted Pair Group Method with Arithmetic Mean Analysis (UPGMA) Tree program was used for analysis. The numbers indicate tree distance proportional to the amount of inferred evolutionary change.
Although no domain conserved between any of these topoisomerase proteins and known RT enzymes was found, we noted that an Asp-Asp doublet-encoding sequence is included in the consensus RT motif (as motif C) present in the catalytic subunit of eukaryotic telomerases and in the HIV-RT (16, 17); furthermore, mutation of this doublet abolishes the RNA-dependent DNA polymerase activity of both telomerase and HIV-RT (16, 17). Accordingly, we replaced the Asp-Asp doublets at positions 464-465 and 471-472 in the Streptomyces TopA protein with Ala-Ala doublets and, remarkably, found that the resulting protein failed to show detectable RT activity (Fig. 2B) while retaining the topoisomerase activity of the WT protein (Fig. 2 A). In the case of telomerase and HIV-RT, the Asp-Asp doublets in motif C coordinate active site metal ions and are essential for metal binding and catalysis (29); another conserved Asp residue in motif A of telomerase and HIV-RT is also required for metal binding (29), and mutation of this aspartic acid also prevents normal telomerase activity (16, 17). The aspartic acid residue at position 249 or 251 of S. coelicolor TopA protein, which are also conserved among this subfamily (Fig. 3B), may be the equivalent of the Asp residue in motif A of telomerase and HIV-RT. Together, conservation of Asp residues in other bacterial proteins in the subfamily of bacterial topoisomerases we have identified (Fig. 3 B and C) suggests that other molecules of this subfamily are likely to also have RT function.
Whereas the tpg or tap telomere-related genes are dispensable for replication of Streptomyces chromosomes and plasmids in a circular form (13, 15), we were unable to generate gene replacement knockouts (see Materials and Methods) of PolA or TopA in Streptomyces cells having either linear or circular chromosomes, suggesting that both enzymes are essential for replication in both the linear and circular modes. Consistent with this interpretation is the identification of only one PolA gene and one DNA topoisomerase I gene in the annotated S. coelicolor M145 chromosome (25).
The discovery in a telomere-bound bacterial topoisomerase of an RT activity that requires a motif that previously has been shown to be essential to the RT activity of a eukaryotic telomerase raises the prospect that telomere replication in Streptomyces may bear functional similarities to the replication of eukaryote telomeres. However, notwithstanding considerable efforts to do so, our analysis of the telomere complexes of Streptomyces replication intermediates has not identified a template RNA complementary in sequence to the 5′ DNA termini of Streptomyces linear replicons. Nevertheless, we have observed that the S. coelicolor chromosome includes a 13-nt intergenic sequence (between ORFSC6D7.26 and ORFSC6D7.27c) that corresponds exactly to the terminal 13-bp sequence of the 3′ telomeric DNA overhang shown previously (11) to be required for linear DNA replication in Streptomyces. Potentially, an RNA encoded by this sequence may serve as template for terminal protein-primed synthesis of the 5′ terminus of lagging DNA strands at telomeres. In this context, we note that the RTs of hepatitis B and other hepadnaviruses can initiate protein-primed synthesis of minus strands of viral DNA on an RNA template by covalent attachment of RT to the first deoxynucleotide of the nascent DNA chain (30). Interestingly, the viral DNA strand remains bound covalently to the polymerase, paralleling the covalent linkage of 5′ ends of Streptomyces linear plasmid lagging strand DNA to Tpg proteins.
Acknowledgments
We thank Z. Deng for providing Streptomyces gene expression vector pHZ1272 and D. A. Hopwood, K. Chater, and H. Kieser for providing Streptomyces strains and cosmids. This work was supported by National Institutes of Health Grant AI08619 (to S.N.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RT, reverse transcriptase; Tap, telomere-associated protein.
See Commentary on page 14307.
References
- 1.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York), 2nd Ed.
- 2.Greider, C. W. & Blackburn, E. H. (1987) Cell 51, 887-898. [DOI] [PubMed] [Google Scholar]
- 3.Lingner, J. & Cech, T. R. (1998) Curr. Opin. Genet. Dev. 8, 226-232. [DOI] [PubMed] [Google Scholar]
- 4.McEachern, M. J., Krauskopf, A. & Blackburn, E. H. (2000) Annu. Rev. Genet. 34, 331-358. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, E. H. (2001) Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- 6.Kinashi, H., Shimaji, M. & Sakai, A. (1987) Nature 328, 454-456. [DOI] [PubMed] [Google Scholar]
- 7.Lin, Y. S., Kieser, H. M., Hopwood, D. A. & Chen, C. W. (1993) Mol. Microbiol. 10, 923-933. [DOI] [PubMed] [Google Scholar]
- 8.Salas, M. (1991) Annu. Rev. Biochem. 60, 39-71. [DOI] [PubMed] [Google Scholar]
- 9.Hay, R. T. (1996) in DNA Replication in Eukaryotic Cells, ed. DePamphilis, M. L. (Cold Spring Harbor Lab. Press, Plainview, NY), p. 699.
- 10.Chang, P. C. & Cohen, S. N. (1994) Science 265, 952-954. [DOI] [PubMed] [Google Scholar]
- 11.Qin, Z. J. & Cohen, S. N. (1998) Mol. Microbiol. 28, 893-903. [DOI] [PubMed] [Google Scholar]
- 12.Huang, C., Lin, Y., Huang, S. & Chen, C. W. (1998) Mol. Microbiol. 28, 905-916. [DOI] [PubMed] [Google Scholar]
- 13.Bao, K. & Cohen, S. N. (2001) Genes Dev. 15, 1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, C., Huang, C., Li, C., Tsay, Y., Lee, S. & Chen, C. W. (2002) Mol. Microbiol. 43, 297-305. [PubMed] [Google Scholar]
- 15.Bao, K. & Cohen, S. N. (2003) Genes Dev. 17, 774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe, D. M., Parmar, V., Kemp, S. D. & Larder, B. A. (1991) FEBS Lett. 282, 231-234. [DOI] [PubMed] [Google Scholar]
- 17.Lingner, J., Hughes, T. R., Shevchenko, A., Mann, M., Lundblad, V. & Cech T. R. (1997) Science 276, 561-567. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood, D. A., Kieser, T., Wright, H. M. & Bibb, M. J. (1983) J. Gen. Microbiol. 129, 2257-2269. [DOI] [PubMed] [Google Scholar]
- 19.Kieser, T., Bibb, M. J., Chater, K. F., Butter, M. J. & Hopwood, D. A. (2000) Practical Streptomyces Genetics (John Innes Centre, Norwich, U.K.).
- 20.Verma, I. M. & Baltimore, D. (1974) Methods Enzymol. 29, 125-130. [DOI] [PubMed] [Google Scholar]
- 21.MacNeil, D. J., Gewain, K. M., Ruby, C. L., Dezeny, G., Gibbons, P. H. & MacNeil T. (1992) Gene 111, 61-68. [DOI] [PubMed] [Google Scholar]
- 22.Hu, Z., Bao, K., Zhou, X., Zhou, Q., Hopwood, D. A., Kieser, T. & Deng, Z. (1994) Mol. Microbiol. 14, 163-172. [DOI] [PubMed] [Google Scholar]
- 23.Muth, G., Nussbaumer, B., Wohlleben, W. & Puhler, A. (1989) Mol. Gen. Genet. 219, 341-348. [Google Scholar]
- 24.Takano, E., White, J., Thompson, C. J. & Bibb, M. J. (1995) Gene 166, 133-137. [DOI] [PubMed] [Google Scholar]
- 25.Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., et al. (2002) Nature 417, 141-147. [DOI] [PubMed] [Google Scholar]
- 26.Setlow, P. (1974) Methods Enzymol. 29, 3-12. [DOI] [PubMed] [Google Scholar]
- 27.Grabko, V. I., Chistyakova, L. G., Lyapustin, V. N., Korobko, V. G. & Miroshnikov, A. I. (1996) FEBS Lett. 387, 189-192. [DOI] [PubMed] [Google Scholar]
- 28.Myers, T. W. & Gelfand, D. H. (1991) Biochemistry 30, 7661-7666. [DOI] [PubMed] [Google Scholar]
- 29.Kohlstaedt, L. A., Wang, J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1992) Science 256, 1783-1790. [DOI] [PubMed] [Google Scholar]
- 30.Wang, G. H. & Seeger, C. (1992) Cell 71, 663-670. [DOI] [PubMed] [Google Scholar]