Abstract
Sampling methods and results of a gene flow study are described that will be of interest to plant scientists, evolutionary biologists, ecologists, and stakeholders assessing the environmental safety of transgenic crops. This study documents gene flow on a landscape level from creeping bentgrass (Agrostis stolonifera L.), one of the first wind-pollinated, perennial, and highly outcrossing transgenic crops being developed for commercial use. Most of the gene flow occurred within 2 km in the direction of prevailing winds. The maximal gene flow distances observed were 21 km and 14 km in sentinel and resident plants, respectively, that were located in primarily nonagronomic habitats. The selectable marker used in these studies was the CP4 EPSPS gene derived from Agrobacterium spp. strain CP4 that encodes 5-enol-pyruvylshikimate-3-phosphate synthase and confers resistance to glyphosate herbicide. Evidence for gene flow to 75 of 138 sentinel plants of A. stolonifera and to 29 of 69 resident Agrostis plants was based on seedling progeny survival after spraying with glyphosate in greenhouse assays and positive TraitChek, PCR, and sequencing results. Additional studies are needed to determine whether introgression will occur and whether it will affect the ecological fitness of progeny or the structure of plant communities in which transgenic progeny may become established.
We developed sampling methods and describe results of a gene flow study that will be of interest to plant scientists, evolutionary biologists, ecologists, and stakeholders assessing the environmental safety of transgenic crops. Creeping bentgrass (Agrostis stolonifera L.) is one of the first wind-pollinated, perennial, and highly outcrossing transgenic crops being developed for commercial use. Unlike currently commercialized transgenic crops in the U.S., which have no synchronously flowering relatives in areas of commercial production, the cosmopolitan genus A. stolonifera has compatible relatives in a broad variety of habitats. The methods and results of using herbicide resistance as a selectable marker from a genetically modified (GM) crop to measure gene flow will be useful for assessing the potential for GM crops to transfer their novel genes to compatible relatives.
More data are available on gene flow from cultivated crops to other crops than from crops to resident (native, naturalized, or weedy) species (1). Typically, gene flow distances are reported on the scale of meters, much less often on the scale of kilometers. Maximum reported distance for gene flow between radish and wild radish (2) and between cultivated and wild sunflowers is 1,000 m (3); distances of 1,300 m have been reported between cultivated and wild squash (4). In an Australian study, crop-to-crop transfer distance of 3,000 m has been reported from source fields of nonGM herbicide-resistant canola to fields of herbicide-sensitive canola cultivars (5).
In this study, we present evidence that documents multiple instances at numerous locations of long-distance viable pollen movement from multiple source fields of GM creeping bentgrass. We used the CP4 EPSPS gene that encodes 5-enol-pyruvylshikimate-3-phosphate synthase from Agrobacterium spp. strain CP4 as a selectable marker to track gene movement. This gene confers resistance to glyphosate (N-phosphono methyl-glycine), the active ingredient in RoundUp herbicide (Monsanto, St. Louis, MO). Herbicide resistance as a result of expression of the engineered CP4 EPSPS gene was observed in seedling progeny of sentinel A. stolonifera and resident Agrostis spp. located at distances up to 21 km and 14 km, respectively, from the crop fields. Eight source fields totaling ≈162 hectares (ha) were located on an irrigated plateau above the Deschutes River in central Oregon. The fields were contained within a 4,453-ha GM bentgrass control district (http://arcweb.sos.state.or.us/rules/OARS_600/OAR_603/603_052.html; ref. 6) located ≈144 km east of commercial nonGM bentgrass seed production areas in Oregon's Willamette Valley. When the source fields of GM creeping bentgrass flowered for the first time during the summer of 2003, they presented a unique opportunity to use the CP4 EPSPS gene as a marker to quantify viable GM pollen movement and potential gene flow to compatible resident and sentinel plants located in areas beyond the crop source fields. Results presented here use multiple lines of evidence based on assays of seedlings germinated from seed harvested from sentinel and resident plants. These assays include tests in greenhouse settings for survival after spraying with RoundUp and tests for presence and expression of the CP4 EPSPS marker.
A. stolonifera is a cool season, wind-pollinated perennial grass used on golf courses around the world (7). It also is of interest as a forage crop (8), for phytoremediation of heavy metals in soils (9), and for water quality improvement by biofiltration (10). The taxonomically uncertain genus Agrostis is estimated to include >200 species worldwide (11, 12). In North America, 26 species of Agrostis are considered native, including 14 species found in Oregon (http://plants.usda.gov). Agrostis is found in riparian habitats, agronomic and urban settings, mountain meadows and woodlands, coastal sand dunes, fresh and salt water marshes, ditches, pastures, grasslands, and roadsides (13, 14). The small seeds of A. stolonifera (up to 6 × 106 per pound) are readily dispersed by wind, water, and animals (13, 15). Introduced and widespread in the U.S., A. stolonifera is sometimes considered an economic weed, e.g., as a volunteer in grass seed or other agronomic production fields and as a colonizer of nonagricultural habitats; it has been reported as weedy in Japan, Australia, New Zealand, Chile, Germany, Denmark, the United Kingdom, and Canada (16).
A. stolonifera is generally considered to be an obligate out-crosser (17); however, self-fertility also has been reported (18). The species is most typically an allotetraploid (19, 20) and has cytotypes of higher ploidy (21). Naturally forming interspecific F1 hybrids generally are low in fertility or sterile; in favorable habitats, some hybrids (e.g., F1 hybrids of A. stolonifera and Agrostis capillaris L.) have been reported to out-compete both parents (22). There are few clear examples of F2 hybrids (23) or of backcrosses of F1 hybrids to a parental species (18). Although native or naturalized hybrids may be sterile, they can constitute a significant component of plant communities because of vegetative spread by means of stolons (24).
Field studies of hybridization between A. stolonifera and other species of Agrostis or between A. stolonifera and closely related Polypogon spp. (18, 25, 26) have produced similar findings on outcrossing ability. In a field study that included several hundred plants as sources of pollen from bentgrass engineered to be resistant to glufosinate herbicide, a gene flow distance of 298 m was reported (25). Natural hybrids of A. stolonifera have been reported with six other native species.: Agrostis canina L., A. capillaris L., Agrostis castellana Boissier and Reuter, Agrostis gigantea Roth, Agrostis mertensii Trinius, and Agrostis vinealis Schreber (www.essentialbiosafety.info/docroot/articles/02-281-009.pdf). A computer model (27) found that pollen dispersal and gene introgression would be limited at some sites and extensive at others, depending on local wind conditions.
Materials and Methods
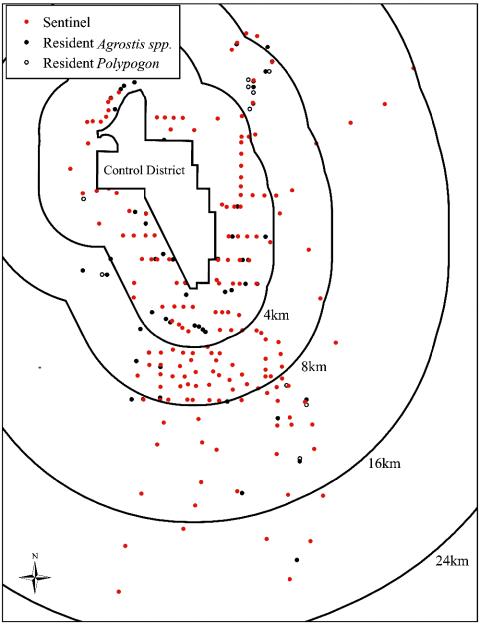
Sampling Design. A sampling grid (Fig. 1) was designed to determine the extent of viable GM pollen flow based on the temporary deployment of 178 compatible A. stolonifera sentinel plants and the monitoring of naturally occurring compatible resident plants. Critical assumptions in the sampling design included a maximal pollen viability of up to 3 h (28) and prevailing winds of 10 km/h from the north and northwest (data are from the Pacific Northwest Cooperative Agricultural Weather Network weather data archive, see www.usbr.gov/pn/agrimet/) during the expected period and hours of anthesis (e.g., mid-June to early July, from 11 a.m. to 2 p.m.) of the source GM creeping bentgrass crop fields. Thirty locations with resident plants of A. stolonifera, 39 locations with resident plants of A. gigantea, and 10 locations with resident plants of Polypogon monspeliensis (L.) Desfontaines also were included in the study. Plants of A. stolonifera (experimental population no. 1 CRBP, Seed Research of Oregon, Corvallis, OR) cultivated in a field in the Willamette Valley of western Oregon were transplanted to 23-cm diameter pots and used as sentinel plants. Before their transport to central Oregon, each of the sentinel plants tested negative for CP4 EPSPS by the TraitChek immunological lateral flow test strip method (Strategic Diagnostics, Newark, DE). Each of the 69 resident Agrostis plants and the 10 P. monspeliensis resident plants were tested by using the TraitChek method to ensure that they were negative for the CP4 EPSPS protein that confers resistance to glyphosate. In mid-June, sentinel plants were deployed to field positions at times of day when anthesis from the source fields was considered unlikely (i.e., before 8 a.m. and after 6 p.m.). Additional steps taken to minimize incidental pollination of sentinel plants included bagging each plant during transit and distribution of the plants by geographic sector. Within sectors, the first plants that were put in place were those at the greatest distance from the perimeter of the control district; the last plants that were placed within a sector were those closest to the control district perimeter. In mid-July, after anthesis in the source fields had ended, panicles were bagged in the field. Bagged sentinel plants with bagged panicles and bagged panicles from resident populations were collected several weeks later. These measures allowed for in situ seed fill and for temporal separation with seed harvesting activities on the GM bentgrass fields. An additional precaution taken to prevent dissemination of any potentially transgenic F1 seedling progeny from the field collections was the use of sealed boxes to transport the doubly bagged sentinel plants and the bagged resident plant panicles during their transport to greenhouses.
Fig. 1.
Sampling design to determine gene flow from source fields within the control district to potentially compatible plants outside the control district. A total of 178 sentinel A. stolonifera plants (red circles) were placed outside the control district (6) near accessible public roads spaced 1.6 km apart in the north-south direction and 0.8 km apart in the east-west direction. Given a prevailing wind of 10 km/h from the north or northwest, 76 sentinel plants were located downwind from the control district in a 9.6-km-wide by 3.2-km-deep grid with ≈0.8-km spacing. Remaining sentinel plants were placed at 1.6-km intervals for the next 4.8 km and 3.2-km intervals for the next 6-10 km out to a distance of 16-21 km along six transects corresponding to major highways. In addition to the sentinel plants, 69 compatible resident Agrostis plants (black circles) of A. stolonifera and A. gigantea, plus 10 P. monspeliensis (open circles) located primarily along waterways and in moist soils, were included in the study.
Greenhouse Assays. Seeds harvested from sentinel and resident plants were chilled at 5°C for 7-10 days in moist sand and grown in trays of a peat-based potting medium (Seedling Mix no. 1, OBC Northwest, Canby, OR) in the greenhouse until the two-leaf stage and then sprayed with the field rate (2.3 liters/ha) of RoundUp herbicide by using a track sprayer (model RC-500-100-EP, Mandel, Guelph, ON, Canada). Seedlings that survived the initial spraying with the field rate of RoundUp or emerged after the spray event were subjected to spraying with herbicide at twice the field rate (4.6 liters/ha) ≈2 weeks later. Survivors of the second cycle of herbicide spraying identified as presumptive positives were confirmed by means of the TraitChek test.
Molecular Characterization. DNeasy Plant Mini kits (Qiagen, Valencia, CA) were used to extract genomic DNA from leaves of seedling progeny derived from 130 sentinel and 45 resident plants that were both herbicide resistant and TraitChek-positive for CP4 EPSPS. Primers for amplification and sequencing of a 1,050-bp segment of the A. stolonifera CP4 EPSPS coding region were designed with PrimerSelect (DNASTAR, Madison, WI) based on Glycine max (L.) Merr. CP4 EPSPS (GenBank accession no. AF464188.1). Amplifications with P217F (5′-ACTATGGGCCTCGTCGGGGTCTA-3′) and P218R (5′-GGCAGCCTTCGTATCGGAGAG-3′) were conducted for 40 cycles of 94°C for 30 sec, 64°C for 30 sec, and 72°C for 90 sec. PCR products were purified with QIAquick Gel Extraction kits (Qiagen). Cycle-sequencing reactions used BigDye v3.1 chemistry and the standard thermal profile suggested by the manufacturer (Applied Biosystems). Labeled fragments were purified with CleanSeq kits (AgenCourt Bioscience, Beverly, MA). Sequence data were collected on a Prism 3100 Genetic Analyzer (Applied Biosystems). Sequences were then compared with matching GenBank accessions by using blastn searches (29).
Statistical Analyses. The percentage of positive seedling progeny was calculated as the number of seedlings that survived two sprays with RoundUp and had positive TraitChek tests for the CP4 EPSPS gene divided by the total estimated number of seedlings germinated in the greenhouse. Maximum likelihood estimation was used to fit the two-parameter gamma distribution (30), f(x) = x(α- 1) exp(-x/β)/(Γ(α)βα), where α and β are the model parameters and Γ(·) is a complete gamma function, to the observed distances from the control district perimeter at which positive seedling progeny were found. The adequacy of the gamma distribution was tested by using the one-sample Kolmogorov-Smirnov test (31, 32). The two-sample Kolmogorov-Smirnov test (33) was used to compare the probability distributions of the positive seedling progeny of sentinel and resident plants. Nonparametric kernel smoothing (34) was applied to percent positive seedling progeny to generate spatial maps of gene flow transfer for sentinel and resident plants separately. The estimation and hypothesis testing of the gamma distributions were performed by using s-plus v6.01 (Mathsoft, Cambridge, MA). Kernel smoothing and spatial maps were undertaken by using arcmap v8.3 and the arcgis spatial analyst 8.3 (Environmental Systems Research Institute, Redlands, CA).
Results
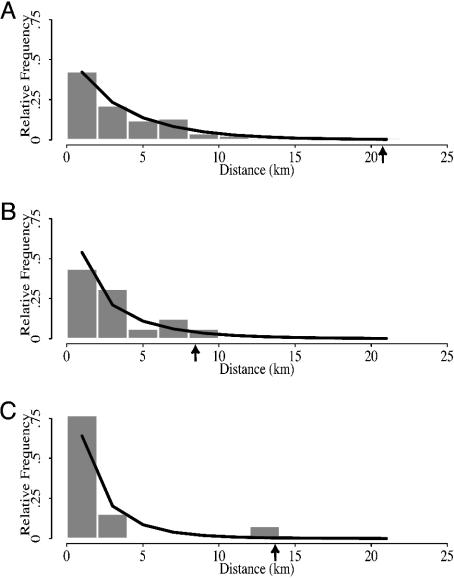
Gene Flow to Sentinel and Resident Agrostis. Molecular analyses by PCR (see Fig. 2) and by sequencing (sequence data not shown) confirmed the presence of the CP4 EPSPS marker in seedling progeny that had survived two cycles of spraying with RoundUp herbicide. The sequence matched that of GenBank accession AF464188.1 for a CP4 EPSPS construct in glyphosate-resistant soybean (G. max). The highest relative frequencies of pollen-mediated gene flow to A. stolonifera sentinel and A. stolonifera and A. gigantea resident plants were observed within 2 km of the control district perimeter. Maximal distances at which gene flow was observed in sentinel and resident A. stolonifera and resident A. gigantea plants were ≈21 km, 8 km, and 14 km, respectively. Viable pollen dissemination distances for sentinel plants may be biased low because this distance of 21 km represented the limit of the sampling design (Fig. 1). An additional source of bias is that distances from source fields to the control district perimeter were unknown. Based on the one-sample Kolmogorov-Smirnov goodness-of-fit test, the empirical distribution of minimum distances of the 75 positive sentinel A. stolonifera plant locations (Fig. 3A) was adequately described by a gamma distribution with α = 0.93 and β = 4.1 (P = 0.67); that of 16 positive resident A. stolonifera plant locations (Fig. 3B) was adequately described by a gamma distribution with α = 0.74 and β = 4.0 (P = 0.77); and that of 13 positive resident A. gigantea plant locations (Fig. 3C) was adequately described by a gamma distribution with α = 0.74 and β = 2.8 (P = 0.42). The mean (αβ) and variance (αβ2) of a gamma distribution decrease monotonically with respect to α and β. Consequently, higher α and β values indicate density distributions of viable pollen that hybridized with sentinel or resident plants that were spread farther from source fields. The gamma distributions for sentinel and resident A. stolonifera locations were not significantly different at the 0.05 level based on the two-sample Kolmogorov-Smirnov test (P = 0.63) but were significantly different from that for resident A. gigantea locations (P = 0.031 and 0.047, respectively). The mean distance from the perimeter of the control district ranged from 2.1 km for resident A. gigantea plant locations to 3.8 km for sentinel plant locations.
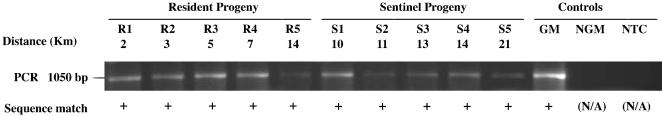
Fig. 2.
Molecular confirmation of the presence of the engineered CP4 EPSPS herbicide-resistance gene. The presence of the CP4 EPSPS gene as verified in a subsample of TraitChek-positive progeny from resident (R1-R5) and sentinel (S1-S5) plants located at various distances from the control district perimeter. All PCR products had the same size and DNA sequence as that amplified from the GM-positive control (A. stolonifera, designated event ASR368). blastn searches (29) revealed that the DNA sequences also matched GenBank accessions AF464188.1, Glycine max CP4 EPSPS (score = 1,271, E = 0.0), and AY125353.1, a synthetic CP4 EPSPS construct. Negative controls included DNA from nonGM (NGM) A. stolonifera, variant Penncross, and a nontemplate control (NTC). +, positive sequence matches; N/A, not applicable.
Fig. 3.
Skewed distribution of GM bentgrass pollen-mediated gene flow to sentinel and resident plants in 2003. Based on the presence and expression of the CP4 EPSPS gene for herbicide resistance, relative frequencies of gene flow among sentinel and resident plant seedling progeny were highest within the first 2 km from the perimeter of the control district and decreased with distance. Arrows depict maximal gene flow distances that were observed. A, B, and C represent locations of sentinel A. stolonifera, resident A. stolonifera, and resident A. gigantea plants, respectively.
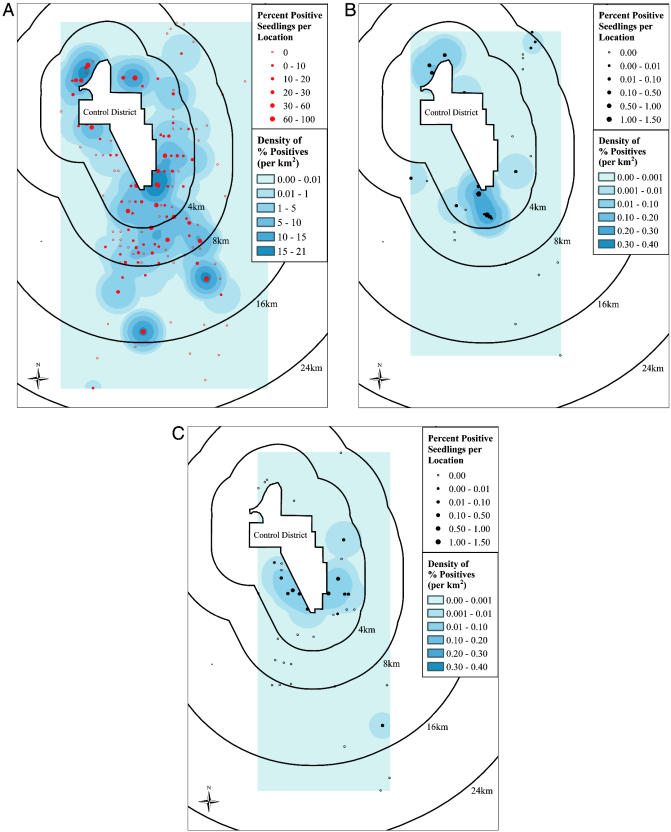
Spatial Patterns of Gene Flow. For both sentinel and resident A. stolonifera plants, the greatest spatial density of percent positive seedlings was found southeast and south of the control district in the direction of prevailing winds (Fig. 4 A and B). Positive seedlings derived from resident A. gigantea were found primarily east and west of the southern portion of the control district (Fig. 4C). In addition, some CP4 EPSPS-positive A. stolonifera seedling progeny were obtained from seeds harvested from plants near and below the northwest section of the control district perimeter (Fig. 4 A and B). This finding may be due to localized temperature gradients and wind conditions near the rim of the Deschutes River canyon, which brought viable pollen down to the canyon floor. In comparison, the percentage of positive sentinel plants was about an order of magnitude higher than that for resident plants.
Fig. 4.
Prevalence of gene flow based on percent positive seedling progeny of sentinel and resident plants at various distances from the control district perimeter. Kernel smoothing (34) was applied to percent positive seedling progeny (filled circles) of sentinel A. stolonifera plants (A), resident A. stolonifera plants (B), and resident A. gigantea plants (C) to generate spatial maps of the density of percentage positives. Open circles indicate locations where no positive seedling progeny were found. The highest densities of percent positive seedling progeny of sentinel and resident A. stolonifera plants occurred southeast and/or due south of the perimeter of the control district, in the direction of the prevailing winds.
Resident Agrostis typically were found in moist soils, e.g., riparian areas and along irrigation or drainage ditches. Most of the A. stolonifera resident plants with positive seedling progeny were located in sagebrush steppe or other nonagronomic land use areas (50% and 25%, respectively), whereas the majority (78%) of positive A. gigantea plants were located in agronomic production areas. Forty of 178 sentinel plants were lost to various causes, e.g., transplant shock and grazing. As shown in Table 1, hundreds of CP4 EPSPS-positive seedling progeny were found among A. stolonifera sentinel and resident plants and A. gigantea residents.
Table 1. Prevalence and incidence of CP4 EPSPS-positive plants and seedling progeny.
| Species | Plants with positive seedling progeny,* % | No. tested† | No. positive seedling progeny (%) |
|---|---|---|---|
| Sentinel | 54 | 32,000 | 625 (2.00) |
| A. stolonifera | (75/138) | ||
| Resident | 53 | 565,000 | 157 (0.03) |
| A. stolonifera | (16/30) | ||
| Resident | 33 | 397,000 | 159 (0.04) |
| A. gigantea | (13/39) | ||
| Resident | 0 | 190,000 | 0 (0.00) |
| P. monspeliensis | (0/10) |
Values in parentheses represent the ratio of plants with positive seedling progeny to the total number of plants.
Number of seedling progeny tested in greenhouse.
Discussion
Our multiple lines of evidence from greenhouse and laboratory tests document movement of viable GM creeping bentgrass pollen on a landscape level that encompassed ≈310 km2. The gene flow evidence presented here contrasts quantitatively with previous studies with A. stolonifera (18, 25, 26) with significantly higher numbers of occurrences and maximally observed linear distances. The higher number of observed occurrences may reflect greater total acreage of source fields in this study (162 ha) as compared with much smaller experimental field plots of previously reported studies with Agrostis in which only several hundreds of plants served as pollen donors. The long period of flowering (estimated at 4-5 weeks rather than a more typical flowering period of 2-3 weeks for creeping bentgrass in the Willamette Valley), may have been due to asynchronous flowering of GM crop source fields. Potential causes of floral asynchrony include differences in cultivars, soil characteristics, and microclimates among source fields. The long gene flow distances we observed may, in part, reflect our sampling design, which purposefully looked at a range of distances in directions guided by historic information on prevailing winds (www.usbr.gov/pn/agrimet) as well as a 3-h window of assumed pollen viability (28). Our landscape level sampling design was distinct from “wagon-wheel” designs typically used for gene flow determinations in agronomic settings; i.e., with regard to its geographic scale of several hundreds of kilometers-squared rather than linear meters, in the broad variety of nonagronomic as well as agronomic habitats that it encompassed, and in the use of both sentinel and resident plants.
Lower frequencies of gene flow observed in resident Agrostis as compared with sentinel plants are likely primarily due to initiation of flowering of resident plants 2-3 weeks later than crop source fields. Pollen competition, i.e., pollen loads in the vicinity of patches of resident plants were higher than around individual sentinel plants, may also have reduced the relative availability of stigma sites and GM pollen in resident plants. Diverse factors (35) may have resulted in our lack of observations of gene flow to P. monspeliensis resident plants; two reasons we consider most likely are flowering of P. monspeliensis residents 2-3 weeks later than the bentgrass fields and their generally upwind locations.
Our results clearly document pollen movement and gene flow from large source populations of GM creeping bentgrass into much smaller numbers of resident Agrostis plants and individual sentinel plants of A. stolonifera. Conceivably, gene flow to resident plants from small-scale field trials of GM creeping bentgrass initiated within the control district before 2003 (www.agcomm.ads.orst.edu/agcomwebfile/edmat/html/sr/sr1046.9htm; ref. 36), e.g., by wind-dispersed pollen or seeds, may have contributed to the observations we report here. However, all tests done to date on leaf and panicle tissue samples of resident plants that produced CP4 EPSPS-positive seedling progeny in our greenhouse assays have proven negative for the marker. Efforts will continue over the next few years to identify potential establishment and recruitment of resident Agrostis that express the CP4 EPSPS marker. More detailed molecular analyses of positive seedlings and of maternal or paternal crop or resident plant parents are planned to distinguish hybridization events between GM crop and resident plants from GM crop seed dispersal. Multiyear sampling to monitor potential introgression of the CP4 EPSPS marker into resident populations and for potential effects on plant community structure and the ecological fitness of progeny also is planned.
In competitor-stress tolerator-ruderal characterization of plant functional types (13, 37), A. stolonifera is considered to have both competitive and ruderal features; thus, its invasive root and stolon growth can contribute to weediness, and new plants can be established either by seeds or by dispersal of stolon pieces (13, 15, 38, 39). The particular engineered trait for herbicide resistance (CP4 EPSPS) that we used as a selectable marker would not be anticipated per se to confer a selective advantage in the absence of herbicide selective pressure. However, in areas where weed control or restoration efforts are being practiced, hybrid Agrostis progeny resistant to glyphosate herbicide might be expected to have a selective advantage. Further studies should continue over the next few years within resident plant populations to monitor for introgression, spread, or extinction of the engineered CP4 EPSPS gene, and for potential effects on ecological fitness of progeny and plant community structure in various, largely nonagronomic habitats.
Biological confinement strategies (e.g., male sterility, gene insertion into organelles or into targeted chromosomes or chromosome sites) are of interest to try to restrict gene flow; however, recent reports (40, 41) suggest that gene leakiness may make fully effective, long-term containment of transgenes unlikely. Studies, such as the one reported here, that use both sentinel- and resident-compatible plants in an appropriately large sampling design that includes nonagronomic and agronomic habitats may be useful to quantify potential rates of gene exchange between GM or conventional crops and nonagricultural resident plants when conducting assessments of ecological risks (35) and evaluating potential mitigation technologies (41). Similar approaches could be used to develop sampling designs to test for potential long-distance wind dispersal of GM seeds. Our methods and findings contribute significantly to the ongoing discussion about potential risks of gene flow from GM crops and thus are anticipated to be of interest to plant scientists, evolutionary biologists, ecologists, policy makers, and regulators.
Acknowledgments
We thank Scotts (Marysville, OH) and Monsanto for providing leaf tissue samples and molecular information; Oregon State University, especially the staff at the Central Oregon Agricultural Experiment Center, for facilitating our studies in central Oregon; Bruce MacBryde (U.S. Department of Agriculture, Washington, DC) for a comprehensive review of Agrostis literature; Joseph Wipff (Barenbrug USA, Albany, OR) for helpful discussions on Agrostis biology, and Leah Brilman (Seed Research of Oregon) for providing A. stolonifera for use as sentinel plants. This research was funded by the U.S. Environmental Protection Agency, Office of Research and Development, National Health and Environmental Effects Research Laboratory. This document has been cleared by the U.S. Environmental Protection Agency for publication.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GM, genetically modified; ha, hectare.
References
- 1.Ellstrand, N. C. (2003) Dangerous Liasions? When Cultivated Plants Mate With Wild Relatives (John Hopkins Univ. Press, Baltimore).
- 2.Klinger, T., Elam, D. R. & Ellstrand, N. C. (1991) Conserv. Biol. 5, 531-535. [Google Scholar]
- 3.Arias, D. M. & Rieseberg, L. H. (1994) Theor. Appl. Genet. 89, 665-660. [DOI] [PubMed] [Google Scholar]
- 4.Kirkpatrick, K. J. & Wilson, H. D. (1988) Am. J. Botany 75, 519-527. [Google Scholar]
- 5.Reiger, M. A., Lamond, M., Preston, C., Powles, S. B. & Roush, R. T. (2002) Science 296, 2386-2388. [DOI] [PubMed] [Google Scholar]
- 6.Department of Agriculture (2002) Oregon Administrative Rule 603-052-1240.
- 7.Duich, J. M. (1985) Weeds, Trees and Turf 24, 72-78. [Google Scholar]
- 8.Balasko, J. A., Evers, G. E. & Duell, R. W. (1995) in Forages: An Introduction to Grassland Agriculture, eds. Barnes, R. F., Miller, D. A. & Nelson, C. J. (Iowa State Univ. Press, Ames) 1, 357-373. [Google Scholar]
- 9.Smith, R. A. H. & Bradshaw, A. D. (1979) J. Appl. Ecol. 16, 595-612. [Google Scholar]
- 10.Hares, R. J. & Ward, N. I. (1999) Sci. Total Environ. 235, 169-178. [DOI] [PubMed] [Google Scholar]
- 11.Soreng, R. J. & Peterson, P. M. (2003) Contrib. U.S. Natl. Herb. 48, 42-89. [Google Scholar]
- 12.Kartesz, J. T. (2003) in Synthesis of the North American Flora, eds. Kartesz, J. T. & Meacham, C. A. [CD-ROM] (Biota of North America Program, University of North Carolina, Chapel Hill), Version 1.985.
- 13.Grime, J. P., Hodgson, J. G. & Hunt, R. (1988) Comparative Plant Ecology: A Functional Approach to Common British Species (Unwin Hyman, London), pp. 58-65.
- 14.Kik, C., Jongman, M. & van Andel, J. (1991) Plant Species Biol. 6, 47-54. [Google Scholar]
- 15.Hunt, R., Nicholls, A. O. & Pathy, S. A. (1987) Oikos 50, 53-59. [Google Scholar]
- 16.MacBryde, B. (2004) Perspective on Creeping Bentgrass, Agrostis stolonifera (U.S. Department of Agriculture/Animals and Plants Health Inspection Service/Biotechnology Regulatory Services, Washington, DC).
- 17.Davies, W. E. (1953) Br. Agric. Bull. 5, 313-315. [Google Scholar]
- 18.Belanger, F. C., Meagher, T. R., Day, P. R., Plumley, K. & Meyer, W. A. (2003) Crop Sci. 43, 240-246. [Google Scholar]
- 19.Jones, K. (1956) J. Genet. 54, 377-393. [Google Scholar]
- 20.Warnke, S. E., Douches, D. S. & Branham, B. E. (1998) Crop Sci. 38, 801-805. [Google Scholar]
- 21.Bonos, S. A., Plumley, K. A. & Meyer, W. A. (2002) Crop Sci. 42, 192-196. [DOI] [PubMed] [Google Scholar]
- 22.Edgar, E. & Forde, M. B. (1991) New Zealand J. Bot. 29, 139-161. [Google Scholar]
- 23.Sell, P. & Murrell, G. (1996) Flora of Great Britain and Ireland (Cambridge Univ. Press, Cambridge, U.K.), Vol. 5, pp. 186-191. [Google Scholar]
- 24.Edgar, E. & Connor, H. E. (2000) Flora of New Zealand (Manaaki Whenua, Lincoln, New Zealand), Vol. 5, pp. 225-242. [Google Scholar]
- 25.Wipff, J. K. & Fricker, C. (2001) Int. Turfgrass Soc. Res. J. 9, 224-242. [Google Scholar]
- 26.Christoffer, P. M. (2003) M.S. thesis (Washington State Univ., Pullman).
- 27.Meagher, T. R., Belanger, F. C. & Day, P. R. (2003) Philos. Trans. R. Soc. London B 358, 1157-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fei, S. & Nelson, E. (2003) Crop Sci. 43, 2177-2181. [Google Scholar]
- 29.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, N., Kotz, S. & Balakrishnan, N. (1944) Continuous Univariate Distributions (Wiley, New York), 2nd Ed., Vols. I and II.
- 31.Kolmogorov, A. N. (1933) G. Ist. Ital. degli Attuari 4, 83-91. [Google Scholar]
- 32.Smirnov, N. V. (1948) Ann. Math. Stat. 19, 279-281. [Google Scholar]
- 33.Smirnov, N. V. (1939) Bull. Moscow Univ., 2, 3-16. [Google Scholar]
- 34.Wand, M. P. & Jones, M. C. (1995) Kernel Smoothing: Monographs on Statistics and Applied Probability (Chapman & Hall, London), p. 7.
- 35.Conner, R. H., Glare, T. R. & Nap, J. P. (2003) Plant J. 33, 19-46. [DOI] [PubMed] [Google Scholar]
- 36.Butler, M., Gilmore, L. & Campbell, C. (2003) in 2002 Central Oregon Agricultural Research Center Annual Report (Oregon State Univ. Press, Corvallis).
- 37.Grime, J. P. (1977) Am. Nat. 111, 1169-1194. [Google Scholar]
- 38.Schippers, P., van Groenendael, J. M., Vleeshouwers, L. M. & Hunt, R. (2001) Oikos 95, 198-210. [Google Scholar]
- 39.Boedeltje, G., Bakker, J. P., Bekker, R. M., van Groenendael, J. M. & Soesbergen, M. (2003) J. Ecol. 91, 855-866. [Google Scholar]
- 40.National Research Council (2004) Biological Confinement of Genetically Engineered Organisms (Natl. Acad. Press, Washington, DC).
- 41.Haygood, R., Ives, A. R. & Andow, D. (2004) Ecol. Lett. 7, 213-220. [Google Scholar]