Abstract
Zea mays L. ssp. mays, or corn, one of the most important crops and a model for plant genetics, has a genome ≈80% the size of the human genome. To gain global insight into the organization of its genome, we have sequenced the ends of large insert clones, yielding a cumulative length of one-eighth of the genome with a DNA sequence read every 6.2 kb, thereby describing a large percentage of the genes and transposable elements of maize in an unbiased approach. Based on the accumulative 307 Mb of sequence, repeat sequences occupy 58% and genic regions occupy 7.5%. A conservative estimate predicts ≈59,000 genes, which is higher than in any other organism sequenced so far. Because the sequences are derived from bacterial artificial chromosome clones, which are ordered in overlapping bins, tagged genes are also ordered along continuous chromosomal segments. Based on this positional information, roughly one-third of the genes appear to consist of tandemly arrayed gene families. Although the ancestor of maize arose by tetraploidization, fewer than half of the genes appear to be present in two orthologous copies, indicating that the maize genome has undergone significant gene loss since the duplication event.
Keywords: maize genome, whole-genome sequence tags, map-based sequence, whole-genome duplication, gene families
Plant genomes differ from mammalian genomes by their enormous range in size. Whereas mammalian genomes range between 2 and 3 gigabases (Gb), the major crop plants vary from 0.4 Gb in rice to 16 Gb in wheat. The model plant Arabidopsis has an even smaller genome of 0.125 Gb and was, therefore, the first plant to be sequenced in its entirety (1). The only other plant genome that has been sequenced is from rice (http://rgp.dna.affrc.go.jp). Arabidopsis and rice belong to the two major divisions of the plant kingdom, the dicotyledonous and monocotyledonous plants, respectively. The grass family (Gramineae) is one of the largest monocotyledonous families; it arose 65 million years ago and is comprised of >10,000 species adapted to one-third of the arable land on earth (2). Cereals including rice, sorghum, sugarcane, oat, barley, wheat, and maize (corn) are examples of Gramineae species; they provide a major source of food and feed for humans.
A significant feature emerging from the genome sequences of Arabidopsis and rice is the large number of genes compared to mammalian genomes. Although the gene number in Arabidopsis is comparable to that in human and mouse, rice appears to have a much larger gene set, due largely to gene amplification (3). Does the gene number in plants at all correlate to their genome size (4)? Such a question arises, particularly in light of the fact that many crop plants have undergone whole-genome duplication. Maize is an interesting example of such a duplication event. In contrast to wheat, maize has not maintained homoeologous chromosomes, but rather has undergone reassortment of the homoeologous regions acquired from the two progenitor genomes. These regions were first demonstrated cytologically between nonhomologous chromosomes (5) and then later by genetic linkage mapping of nontandem gene duplicates (6). A comprehensive genetic analysis of homoeologous regions was performed with DNA markers (7) and by comparative mapping to close relatives of maize (8, 9).
Although genetic and cytogenetic analyses provided us with a global view of the organization of the maize genome, a more detailed analysis will come from its DNA sequence. In fact, the maize genome is most likely the next plant genome that will be sequenced after Arabidopsis and rice. However, because of its suspected greater sequence complexity than the human genome, maize is also thought to be the next technical challenge in genome sequencing. When large genomes are dissected into overlapping bacterial artificial chromosome (BAC) clones, these clones can be assembled into megabase (Mb)-sized chromosomal fragments through fingerprinting methods. After the fragments are assigned to their chromosomal locations, sequences from the BAC clones become positioned relative to the genetic map, thereby serving as anchors for other sequences.
Following this concept, we have generated ≈475,000 maize BAC end sequences (BESs) with a cumulative length of 307 Mb, providing an 8-fold coverage of the genome. BES reads averaged 647 bp with an average distribution of one end every 6.2 kb of the genome. Besides constituting a framework for sequencing the maize genome, the BES provided us with a comprehensive, quantitative data set, which allowed us to assess maize transposable element (TE) and gene content. Moreover, by examining the physical linkage of BESs, we determined that a large proportion of the maize gene set consists of tandemly arrayed gene families, and that a heavy loss of unlinked duplicated genes must have occurred during the transition from a tetraploid to a diploid species.
Materials and Methods
Processing of BAC Clones. Data from three large insert BAC libraries of maize inbred B73 were used to anchor the DNA sequence to the genetic map. The libraries had previously been constructed by partial digestion of genomic DNA with HindIII, EcoRI, and MboI (10, 11), providing a 30-fold physical coverage of the 2.365-Gb genome (12).
BAC clones were retrieved from 384-well storage plates and grown in 96 deep-well plates to saturation. DNAs were extracted with Whatman Unifilters by using a Tomtec liquid handling system. DNA precipitates were resuspended in 40 μl of buffer; samples of 10 μl were used for the forward and reverse sequencing of the BAC ends with universal primers; another 10-μl aliquot was used for DNA fingerprinting. About 75% of the clones were assembled into a physical map of contiguous, overlapping clones (www.genome.arizona.edu/fpc/maize and http://pgir.rutgers.edu) by using the program fingerprinted contigs (fpc) (13). More than 50% (297,961 BACs) of the fingerprinted clones were sequenced from the ends, 176,643 from both ends and 121,318 from one end only, yielding a total of 474,604 BESs with an average read length of 647 bases at Q16. DNA sequences were processed by using lucy software (14) and deposited in the genome survey sequence (GSS) section of the GenBank database.
Gene Prediction Parameters. Masked BESs (details in Results and Discussion) were analyzed for their coding potential by applying extrinsic (homology-based) and intrinsic (ab initio gene prediction) criteria and methods. Detection of genes by a single read (average, 647 bp) has similar limitations to EST-based approaches, but can be enhanced by using combinatorial evidence scores. Therefore, in our homology-based methods, gene content analysis of the BES data are predicated on the tentative consensus (TC) sequences (of the SPUTNIK database (15) containing structured ESTs from all major plant-derived EST collections (550,000 TCs; >2.3 × 106 ESTs).
Results and Discussion
Genome Coverage of BAC Libraries. Deep coverage of BAC libraries and even distribution of BAC clones across the genome are a critical presupposition to deduce representative data. However, because the BACs were constructed by partial digestion and restriction sites are unevenly distributed throughout the genome, biases in representation frequently can be observed. For example, ribosomal DNA sequences are not well represented in the HindIII and EcoRI libraries relative to the MboI library, because of the lack of HindIII sites and the presence of only one EcoRI site compared to numerous MboI sites in the ribosomal DNA cluster (10). To determine the specific features of BES data from the HindIII, EcoRI, and MboI BAC libraries, each library was analyzed separately. As a benchmark, the repeat composition within BESs from each BAC library (for details, see below) was compared with 50,876 reads derived from a library made from unfiltered, randomly sheared genomic DNA (16). Table 1 and Fig. 3, which is published as supporting information on the PNAS web site, show that the MboI library recovered high-copy-number genes very well, whereas HindIII did not. HindIII covered retrotransposons better than EcoRI and MboI. All three libraries have reduced representation of simple sequence repeats and Ty3/gypsy-type retrotransposable elements, which are typical for heterochromatic regions and centromeres. Except for these sequences, most differences between the three different libraries averaged out when they were combined. Therefore, we conclude that the 307 Mb of BESs represents a well distributed random sampling of about one-eighth of the corn genome with a slight bias toward the euchromatic regions.
Construction of a Database of Maize Repeat Elements. Besides sufficient coverage of the genetic map, an essential first step to study the content of the maize genome is a meaningful definition of repeat sequences. Accordingly, we first set out to determine a biologically relevant threshold of repeat identity for inclusion in our analyses. One challenge of distinguishing between coding and noncoding portions of the genome consists of filtering protein-encoding TEs from gene families. Because maize repeats are typically longer than a single sequence read, the BES data set was not used for de novo repeat discovery. Instead, the repeat database was built from completely sequenced maize BAC clones and other related genomic sequences already deposited in the GenBank database, along with a survey of GenBank entries screened for repeat sequences by using typical features like polyproteins of retroelements, LTRs, and other sequence repeat motifs by using wu-blast (17) Version 2.0 for repeat detection (Fig. 4, which is published as supporting information on the PNAS web site). After collapsing a total of 7,760 sequences (15.2 Mb), 74% (5,700 sequences) remained as nonredundant reference sequences.
To define a repeat identity threshold that would enable selective and sensitive repeat detection and classification, a blast-identity limit-for-repeat detection was determined for several data sets by plotting the degree of detection of repeat elements against the identity threshold applied (Fig. 5, which is published as supporting information on the PNAS web site). Four data sets of GSSs were analyzed. Because a large percentage of the maize genome consists of TEs, several attempts were made to use fractionation methods to increase the information content of genome sequence by constructing genomic subclone libraries that are depleted in TEs. Three such fractionation methods have been reported for the maize genome. The first (high C0t-derived, HC) is a library derived by reassociation kinetics of denatured genomic DNA of inbred B73 (16); the second (methyl-filtered, MF) is a library generated by in vivo filtration of methylated from nonmethylated DNA of inbred B73 (16, 18); and the third (RescueMu) is a library derived from junction sequences of genomic insertion sites of the maize transposable element Mutator from other inbred lines (19). Therefore, in addition to the BES collection, the analysis used these specialized whole-genome data sets that were designed to specifically reduce representation of repeated elements. Fig. 5, which is published as supporting information on the PNAS web site, shows a plot of the degree of sequence identity of hits versus the percentage of hits falling into each class. The BESs show a higher level of repeat content than the fractionated sequence representations (MF, HC, and RescueMu), but the overall shapes of the curves are similar. Below an identity limit of 55%, the curves reach a plateau. All four GSS collections show a steep drop in the percentage of repeat nucleotides above a 60% identity limit, with the curves from fractionated data becoming less steep above 65-70%, suggesting that a threshold in this range will be most suitable. Lower thresholds are likely to lead to unspecific or overmasking of GSS data. Based on these observations, subsequently, an identity threshold of 70% for repeat masking was applied for all data sets.
The TE/Repeat Elements of Maize. The BESs provided us with the most comprehensive data set of the repeat content of the maize genome to date. Previous reports projected the amount of repetitive DNA in maize to be in the order of 60-80% (20). Applying conservative parameters, the BES data set arrived at the lower end of previous calculations. As derived from the BES collection, the number of repeat elements present in maize may be close to 58% in terms of number of nucleotides, which might be a slight under-representation in comparison to the 63% detected within a random sheared library (16) (Table 1); this might be attributable to a suppression of centromeric sequences within the BES collection. Not unexpectedly, the largest class of repeat elements is TEs, which were first discovered in maize (21) and have been studied genetically for many years. In recent years, as more maize genomic sequences have become available, they have also been studied extensively at the molecular level (22). Therefore, a sequence similarity-based classification scheme can be used to determine the copy number of different TE families (Table 2). The class I elements (retroelements) dominate over the class II elements (DNA transposons) by a huge margin of 56% to 1% of the genome sequences, respectively. In general, most plant genomes are rich in LTR-retrotransposons and miniature inverted-repeat TEs (22). On the other hand, the number of non-LTR retroelements, like short and long interspersed nuclear elements, is very small (<0.2%), in contrast to the human genome with >25% (23).
Table 1. Distribution of repetitive DNA in rice and various GSSs of maize as percent of genome.
| Details | Total BES | HindIII BES | EcoRI BES | MboI BES | UF | MF | HC | RescueMu | Rice |
|---|---|---|---|---|---|---|---|---|---|
| Total no. of sequences | 474,604 | 309,560 | 78,313 | 86,731 | 50,876 | 30,000 | 30,000 | 30,000 | 179 |
| Total no. of base pairs | 307,169,410 | 206,221,247 | 46,673,217 | 54,274,946 | 37,621,118 | 21,649,324 | 21,637,862 | 10,074,088 | 35,800,000 |
| Percent of genome, % | 12.99 | 8.72 | 1.97 | 2.29 | 1.59 | 0.92 | 0.91 | 0.43 | 8.33 |
| Class I retroelements, % | 55.60 | 58.39 | 46.77 | 52.58 | 57.73 | 15.70 | 6.55 | 4.91 | 19.28 |
| Ty1/copia-like elements, % | 24.54 | 25.33 | 20.54 | 24.97 | 17.50 | 8.37 | 2.54 | 1.98 | 3.67 |
| Ty3/gypsy-like elements, % | 21.43 | 23.39 | 15.69 | 18.94 | 31.25 | 4.32 | 2.22 | 1.71 | 8.90 |
| LINES + SINES, % | 0.18 | 0.23 | 0.06 | 0.06 | 0.04 | 0.06 | 0.18 | 0.13 | 0.96 |
| Other retroelements, % | 9.45 | 9.44 | 10.48 | 8.61 | 8.94 | 2.95 | 1.62 | 1.09 | 5.74 |
| Class II DNA transposons, % | 0.98 | 0.94 | 1.16 | 0.98 | 0.92 | 1.14 | 1.58 | 4.84 | 10.35 |
| hAT superfamily, % | 0.03 | 0.02 | 0.04 | 0.03 | 0.04 | 0.04 | 0.09 | 0.11 | 0.37 |
| CACTA superfamily, % | 0.32 | 0.31 | 0.38 | 0.30 | 0.21 | 0.21 | 0.17 | 0.76 | 2.83 |
| Mutator, % | 0.01 | 0.01 | 0.02 | 0.02 | 0.01 | 0.05 | 0.05 | 0.36 | 0.51 |
| MITEs, % | 0.17 | 0.16 | 0.20 | 0.17 | 0.19 | 0.39 | 0.62 | 1.08 | 3.96 |
| Other DNA transposons, % | 0.45 | 0.42 | 0.54 | 0.48 | 0.48 | 0.44 | 0.65 | 2.53 | 2.67 |
| Simple repeats, % | 0.40 | 0.27 | 0.47 | 0.83 | 1.66 | 1.27 | 0.19 | 0.17 | 1.03 |
| High-copy-number genes, % | 0.82 | 0.12 | 1.13 | 3.17 | 1.95 | 0.17 | 0.19 | 0.15 | 0.06 |
| Other repeats, % | 0.12 | 0.11 | 0.12 | 0.16 | 0.30 | 0.09 | 0.06 | 0.05 | 0.43 |
| Total repeats, % | 57.91 | 59.82 | 49.65 | 57.72 | 62.55 | 18.38 | 8.57 | 10.13 | 31.14 |
Greater than 70% identity limit. UF, The Institute for Genomic Research (TIGR) unfiltered; MF, TIGR MF; HC, TIGR high C0t; RescueMu, MaizeDB RescueMu; MITE, miniature inverted-repeat TE; LINE, long interspersed nuclear element; SINE, short interspersed nuclear element. Data for Rice comprise 179 pseudo BACs, i.e., 200 kb cut equally from all 12 chromosomes.
Table 2. Occurrence and distribution of repetitive DNA in the maize BAC end sequences.
| Class, subclass, group, clade | No. of hits | Percentage of hits, % | No. of bases, bp | Percentage of masked bases, % | Percentage of genome,* % |
|---|---|---|---|---|---|
| Class I elements (retroelements) | 92.65 | 96.01 | 55.60 | ||
| LTR-retrotransposons | 90.91 | 95.65 | 55.38 | ||
| Ty1/copia-like elements | 133,860 | 39.74 | 42.37 | 24.54 | |
| Ji | 71,923 | 21.35 | 42,306,471 | 23.78 | 13.77 |
| Opie | 37,451 | 11.12 | 22,168,294 | 12.46 | 7.22 |
| Prem | 14,129 | 4.19 | 6,439,116 | 3.62 | 2.10 |
| Other copia | 10,357 | 3.07 | 4,455,318 | 2.50 | 1.45 |
| Ty3/gypsy-like elements | 116,375 | 34.55 | 37.02 | 21.43 | |
| Cinful | 40,706 | 12.08 | 24,989,484 | 14.05 | 8.14 |
| Grande | 16,752 | 4.97 | 9,772,902 | 5.49 | 3.18 |
| Huck | 16,892 | 5.01 | 9,107,710 | 5.12 | 2.97 |
| Tekay | 3,116 | 0.93 | 1,472,939 | 0.83 | 0.48 |
| Zeon | 30,305 | 9.00 | 17,452,971 | 9.81 | 5.68 |
| Other gypsy | 8,604 | 2.55 | 3,043,159 | 1.71 | 0.99 |
| Other LTR-retrotransposons | 55,941 | 16.61 | 28,912,679 | 16.25 | 9.41 |
| Retroposons without LTR | 1.75 | 0.36 | 0.21 | ||
| LINEs | 1,237 | 0.37 | 285,530 | 0.16 | 0.09 |
| SINEs | 3,802 | 1.13 | 256,122 | 0.14 | 0.08 |
| Other unclassified retroelements | 847 | 0.25 | 104,778 | 0.06 | 0.03 |
| Class II elements (DNA transposons) | 4.12 | 1.69 | 0.98 | ||
| hAT superfamily (hobo-Ac-Tam3) | 4,310 | 0.15 | 84,656 | 0.05 | 0.03 |
| CATA superfamily (En/Spm) | 3,140 | 0.93 | 981,562 | 0.55 | 0.32 |
| MuDR/Mu superfamily | 502 | 0.15 | 42,734 | 0.02 | 0.01 |
| MITEs | 1.28 | 523,368 | 0.29 | 0.17 | |
| Helitrons | 144 | 0.04 | 11,174 | 0.01 | 0.00 |
| Other unclassified DNA transposons | 5,274 | 1.57 | 1,366,349 | 0.77 | 0.44 |
| Simple repeats | 1.42 | 0.68 | 0.40 | ||
| Centromeric repeats | 2,348 | 0.70 | 598,640 | 0.34 | 0.19 |
| Telomeric repeats | 465 | 0.14 | 131,884 | 0.07 | 0.04 |
| VNTR (mini- and microsatellites) | 1,030 | 0.31 | 85,500 | 0.05 | 0.03 |
| Knob-region | 956 | 0.28 | 402,235 | 0.23 | 0.13 |
| High-copy-number genes (ribosomal) | 4,908 | 1.46 | 2,506,369 | 1.41 | 0.82 |
| Other undefined repeats | 1,179 | 0.35 | 364,729 | 0.21 | 0.12 |
| Total | 336,840 | 100 | 177,871,282 | 100 | 57.91 |
Greater than 70% identity limit. MITE, miniature inverted-repeat TE; LINE, long interspersed nuclear element; SINE, short interspersed nuclear element; VNTR, variable number terminal repeat.
Based on 2,365 Mb.
Availability of the rice genome sequence provided us with an opportunity for a side-by-side comparison between the two species in terms of TE content and repeat class distribution. The smaller genome of rice correlated well with its lower content of TEs compared to maize. This finding is consistent with the expected impact on plant genomes of TEs, which predominantly expand intergenic sequences and thus the physical length of the genome but do not increase the length of the genetic map (8). In addition to the overall TE content, the relative composition of class I and class II elements in rice and maize differ significantly. In rice, class I elements comprise 19% of the genome, and class II elements comprise 10% of the genome, in contrast to the 56-fold difference observed in maize. This difference could be due in part to the recent observation that rice may have lost nearly 40% of its LTR retrotransposons in the last 8 million years (24).
Features of TE. Different TEs have differential insertion specificities that are characteristic of each class. For example, in plants, the Ty1/copia elements were first identified as insertions near maize genes, whereas the highly repetitive Ty3/gypsy elements have a preference to insert into or near other repetitive elements (25). Moreover, in maize and other plant species, the class II TEs such as Ac/Ds, En/Spm, Mu and miniature inverted-repeat TEs are known to insert preferentially into genes and low-copy-number DNA, which are relatively hypomethylated. This finding explains the presence of class II elements in all three fractionated libraries (Table 1).
It is becoming clear from the analysis of many genomes that TEs are a ubiquitous feature in the organization of chromosomes. Surprisingly, even compact genomes, like those of pufferfish (0.4 Gb), were recently found to exhibit a richer diversity of retrotransposons than the human and mouse genomes, which are roughly 7 times larger (26). A very wide range of variation is observed in the structure of retroelements, ranging from nucleotide substitutions and small insertions/deletions to large rearrangements (27). Retroelements are therefore thought to evolve faster than the nonrepetitive portion of any genome (22). Because TEs comprise the single most abundant component (40-80%) of many large genomes, TE mining is an essential part of genome research that will enable us to determine their key role in genome evolution (28).
How Well Are TEs Fractionated from the Rest of the Genome? Comparison of the BESs to data sets of fractionated genome sequences revealed that the three fractionated libraries have reduced repeat sequence representation to a limited extent (3- to 6-fold, Table 1). To assess the effectiveness with which repeat elements were reduced in the fractionated data sets, the comparison was extended to the different classes of repeat elements. We observed biases in which types of repeat sequences were represented in each library type (Fig. 6, which is published as supporting information on the PNAS web site). In the MF and HC data sets, the dominant class of repeat elements is LTR retrotransposons. The RescueMu data set contains similar numbers of LTR retrotransposons and DNA transposons, indicating that Mu insertions occur in both classes of TEs, although at a low frequency. The dominance of class I TEs in the MF and HC data sets might reflect that LTR retrotransposons are frequently nested and/or fragmented and, therefore, recalcitrant to filtration by methylation or reassociation kinetics. Interestingly, the MF fraction has a significant proportion of centromere-specific repeats, indicating the possible importance of nonmethylation of chromatin structure in centromere function. This finding would also be consistent with the transcription of centromere-specific retrotransposons, as has been suggested for rice centromeres (29). Thus, different fractionation techniques tag interesting functional aspects of genomic sequences, but are unlikely to provide a simple separation of genomic DNA into genes and TEs.
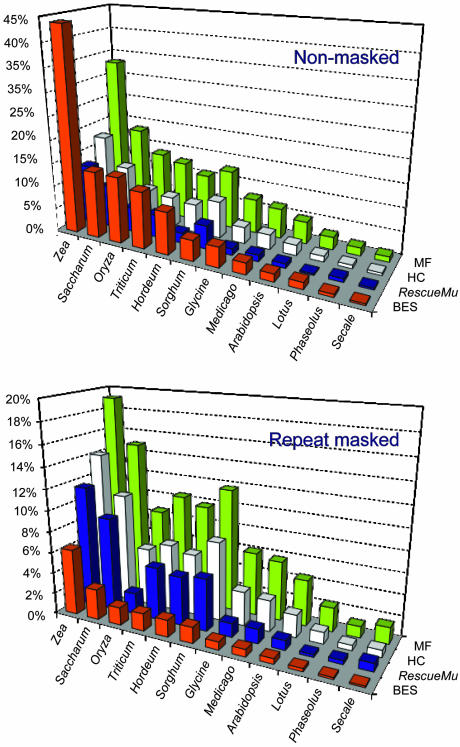
Significant Levels of Transcripts from Repeat Elements. To identify the transcribed portion of the genome, we first compared masked and unmasked BESs against large EST/TC data sets from different plant species. Such a comparison allowed us to determine the degree of sequence similarity between species and detect contaminations with sequences derived from nonmaize species. In addition, it gave us an estimate of the representation of repeat elements within EST databases. As a reference set for the BESs, we again used the three genomic libraries (HC, MF, and RescueMu) that have reduced repeat element representation. All data sets (both nonmasked and repeat masked) were screened in all six translated frames (tblastx using 10-35) against a collection of EST clusters and TCs from maize and six other members (sugarcane, rice, wheat, barley, Sorghum, and Secale) of the Gramineae family, along with four legumes (Leguminoseae), Medicago, Phaseolus, Glycine, and Lotus and also from Arabidopsis (Brassicaceae). The species specificities and the phylogenetic distances correlated, consistent with the lack of nonmaize sequences within the BES collection (Fig. 1). The difference in the hit percentages between nonmasked and repeat-masked is indicative of EST databases containing sequences from retroelements. In general, the differences in BES hit rates were caused by only a small portion of the respective ESTs/TCs. Even at high stringency (10-50), 1.5% of the TCs have a very high number of nonspecific hits. Because nonmasked BESs contain a large proportion of retrotransposon-related genes, the small proportion of TE-related ESTs becomes heavily emphasized. There is also an increase in the hit rate of nonmasked as compared to repeat-masked sequences in other plant species (sugarcane, rice, wheat, barley, Sorghum, and Glycine), indicating a conservation of transcribed TEs between species (30).
Fig. 1.
Analysis of coding potential of the GSSs by tblastx comparisons against complete collections of EST clusters from maize and 11 other plant species (E value = 10-35). The percentage of GSSs matching against the respective EST collection has been displayed. Data are given for both nonmasked as well as repeat-masked GSS populations. EST clusters have not been repeat-masked. MF, The Institute for Genomic Research (TIGR) MF enriched library; HC, TIGR high-C0t enriched library; RescueMu, MaizeDB RescueMu library.
Gene Content Analysis. After masking repeat element sequences, we used the BESs to estimate the gene content of the maize genome. Applying the most stringent criteria, i.e., either homology to known genes and/or gene prediction by at least two gene finders, 7.5% of the nucleotides within BESs appear to derive from transcribed regions. Based on an estimated size of 2,365 Gb for the maize genome (12), this extrapolates to 178 Mb for the transcribed portion of the genome, excluding TE-related transcripts. A preliminary analysis indicated that the size of known maize genes was comparable to that of the average known rice gene of ≈3 kb (31). This result suggests the presence of ≈59,000 genes in maize, significantly higher than the 45,000 estimated for rice (3), but with a lower average gene density of 1 per 40 kb. If the same analysis is applied to the fractionated GSS collection, gene coverage increases only by a factor of 2 (HC), 2.5 (RescueMu), and 3 (MF) (Fig. 7, which is published as supporting information on the PNAS web site).
Because most BESs cover only a small portion of a gene, we enhanced the functional analysis of these sequences by forming TCs from BES-tagged known transcripts (Materials and Methods) by applying a stringent threshold of E ≤ 10-20. In this way we could overcome the restriction of comparably short BESs with the longer TC-derived peptide sequences and arrive at a better representation of BESs assigned to functional categories (Table 3). About 9.1% have a match to plant-derived ESTs (Sputnik/TC, E ≤ 10-35). Among the TC hits, 7,628 were derived from maize, representing 22% of the total unique ESTs known from maize.
Table 3. Distribution of the functional classes of genes tagged to maize BESs and TCs against the Arabidopsis proteome.
| No. | Category | Arabidopsis proteome,* % | Assignment of BESs, % | Assignment of BES-tagged TCs, % |
|---|---|---|---|---|
| 1 | Metabolism | 12 | 11.30 | 11.86 |
| 2 | Transport facilitation, intracellular transport | 7 | 9.17 | 7.66 |
| 3 | Cellular communication, signal transduction | 5 | 7.87 | 5.32 |
| 4 | Cell rescue, defense, cell death, aging | 6 | 7.32 | 7.26 |
| 5 | Cell growth, cell division, DNA synthesis | 6 | 7.22 | 8.53 |
| 6 | Cellular organization | 5 | 6.90 | 7.77 |
| 7 | Cellular biogenesis | 3 | 6.90 | 7.77 |
| 8 | Transcription | 9 | 5.80 | 7.43 |
| 9 | Protein destination | 5 | 5.78 | 6.66 |
| 10 | Energy | 2 | 4.00 | 3.90 |
| 11 | Protein synthesis | 2 | 3.62 | 4.01 |
| 12 | Ionic homeostasis | 1 | 2.57 | 2.94 |
| 13 | Classification not yet clear-cut/unclassified proteins | 37 | 21.55 | 18.89 |
See ref. 1.
The TC collection was then screened against the Arabidopsis functional classification (Table 3) available through the MIPS Arabidopsis thaliana (MAtDB) and SPUTNIK databases (15, 32). In a general overview, the top three functional categories in maize are “metabolism” (11.3%), “transport facilitation, intracellular transport” (9.2%), and “cellular communication, signal transduction” (7.9%). Surprisingly, the category of transcription factors (5.8%) is lower than expected from previous analysis (Fig. 8, which is published as supporting information on the PNAS web site).
Genome Topology. What percentage of genes is tandemly arrayed and how many genes were derived from each of the two progenitors of maize (unlinked duplicate genes)? The latter question helps us to assess the degree of how much of whole-genome duplication is still recognizable at the gene level. However, for both questions, we needed to determine which BESs are physically linked. This became possible because the same clones have also been fingerprinted and assembled into contigs by using FPC (13). TCs based on BESs, as described above, with overlapping match characteristics were grouped into bins of homologous TCs or gene family signatures (GFSs). Because GFSs can be linked to fingerprinted contigs via individual BES names, relative positions of GFS can be determined and regional correlations can be analyzed. Therefore, the combination of the FPC information of BAC clones and their sequence information enabled us to address questions on the extent of local genic duplications (e.g., tandem arrays) and of global duplications.
The TCs were grouped into 9,129 distinct GFSs by using the BES-directed clustering strategy. After filtering for highly abundant GFSs, 9,038 were used for the subsequent analysis. By using this approach, we detected 3,064 individual tandem arrays. As expected from the distribution of BESs along the chromosome, the sizes ranged from 2 to 15 members with a maximum proportion (73%) of 2 members (Fig. 2). Of the total of 21,098 BES associated to GFSs, 7,427 fell into this category, which results in an estimate of one-third (35%) of the genes being organized in tandem arrays in maize. Because this number represents the lower limit in maize, it exceeds the degree of tandem gene duplications found in rice (25%) (31) and certainly in Arabidopsis (17%) (1).
Fig. 2.
Estimation of the number of singletons, gene families, and tandem repeats in maize. (Upper) Singletons and gene families in maize. The numbers of how many genes exist in which copy numbers. (Lower) Tandem repeats in maize. The number of tandemly arrayed genes according to their size classes.
We also tagged orthologous regions by applying the same experimental design used for the detection of tandem gene duplications. Using highly stringent criteria (E ≤ 10-20), we examined fpc-derived BAC contigs for BESs anchored within the respective contigs and associated them to GFSs. Of 1,802 fingerprinted contigs tested, 1,078 (60%) had at least two GFSs located on two corresponding contigs and 513 (28%) had at least three corresponding GFSs. Moreover, 34% (7,175 of 21,098) of individual GFSs anchored to BES fell into this category, implying exhaustive molecular traces of the ancient tetraploidization of the maize genome. Nevertheless, considering that we tagged ≈70% of the genes, this finding represents a very conservative estimate, and the degree of retained duplicates might be markedly higher.
Conclusions
A high-density sequence coverage of the genome of maize inbred B73 has given us the first comprehensive and quantitative overview of the DNA organization of the maize genome. Two global aspects of gene content are striking and appear contradictory. Based on whole-genome duplication from two progenitors, the duplicate gene number (two gene copies in unlinked positions) is surprisingly low (<50%). On the other side, tandem gene amplification appears to be unusually high. The reduction of duplicate genes could be explained by a recent study in which it was shown that, between orthologous intervals of the two homoeologous regions of the maize genome and the single homoeologous regions of the sorghum and rice genomes, a relatively small proportion of genes were conserved as duplicate factors in maize (also <50%) (33). If a composite is formed from both homoeologous regions of the maize genome, collinearity with sorghum and rice increases to ≈86%. This heavy loss of duplicate genes would be consistent with the change from a tetravalent genome of the progenitors of maize to today's diploid genome, which could be referred to as the diploidization process. Interestingly, a similar process seems to have occurred in yeast, although to a more extreme level with nearly 90% loss of duplicated genes (34). However, in contrast to yeast, the remaining gene number has increased dramatically, because of tandemly amplified gene families. One explanation could come from the phylogenetic analysis of the 41-member zein gene family, which indicated that tandem gene amplification occurred within the last 4.5 million years (35), whereas the two progenitors of maize arose ≈11.9 million years ago (mya) and hybridized to form maize between 11.9 and 4.8 mya (36).
Although we knew that the maize genome is rich in LTR-type retrotransposons, their density must be quite variable and their number may contribute to only slightly more than half of the genome size. The predicted gene sequences make up only 7.5% of the genome, and all repeat elements make up 58% of the genome, but what is located in the remaining 34.5%? Interestingly, two recent examples have shown that unique sequences potentially contain important regulatory features and can be separated from the coding regions by the insertion of retroelements in the range of 100 kb (37, 38). Therefore, the space between the known repeat elements and the identifiable coding region models will require more functional analysis.
Supplementary Material
Acknowledgments
We thank Drs. Chad Nusbaum and Bruce Birren of the Broad Institute (Massachusetts Institute of Technology) for critical reading; K. Ward (for laboratory supply management) and S. Young, S. Kavchok, G. Keizer, A. B. Nelson, V. Zohovetz, K. Rouzard, and R. Sugiyama (for DNA sequencing) at The Plant Genome Initiative at Rutgers; D. Stum, L. Scott, A. Lo, and K. Collura (for DNA sequencing) and C. Mueller and D. Kudrna (for clone management) at Arizona Genomics Institute, and J. Hatfield, K. Rao (data processing) at the Arizona Genomics Computational Laboratory. This project was supported by National Science Foundation Plant Genome Grant 0211851. Work at the Munich Information Center for Protein Sequences was supported in part by the Genomanalyse im Biologischen System Pflanze program of the German Ministry for Education and Research.
Abbreviations: Gb, gigabase(s); Mb, megabase(s); BAC, bacterial artificial chromosome; BES, BAC end sequence; TE, transposable element; GSS, genome survey sequence; TC, tentative consensus; HC, high C0t-derived; MF, methyl-filtered; GFS, gene family signature.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. can be found in Table 4, which is published as supporting information on the PNAS web site).
References
- 1.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 2.Kellogg, E. A. (2001) Plant Physiol. 125, 1198-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai, J. Dey, N., Kim, C.-S., Bharti, A. K., Rudd, S., Mayer, K. F. X., Larkins, B., Becraft, P. & Messing, J. (2004) Genome Res. 14, 1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messing, J. (2001) Trends Plant Sci. 6, 195-196. [DOI] [PubMed] [Google Scholar]
- 5.McClintock, B. (1930) Proc. Natl. Acad. Sci. USA 16, 791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhoades, M. M. (1951) Am. Nat. 85, 105-110. [Google Scholar]
- 7.Helentjaris, T., Weber, D. & Wright, S. (1988) Genetics 118, 353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn, S. & Tanksley, S. D. (1993) Proc. Natl. Acad. Sci. USA 90, 7980-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale, M. D. & Devos, K. M. (1998) Proc. Natl. Acad. Sci. USA 95, 1971-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim, Y. S., Davis, G. L., Duru, N. A., Musket, T. A., Linton, E. W., Messing, J., McMullen, M. D., Soderlund, C. A., Polacco, M. L., Gardiner, J. M., et al. (2002) Plant Physiol. 130, 1686-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomkins, J. P., Davis, G., Main, D., Yim, Y. S., Duru, N., Musket, T., Goicoechea, J. L., Frisch, D. A., Coe, E. H., Jr., & Wing, R. A. (2002) Crop Sci. 42, 928-933. [Google Scholar]
- 12.Bennett, M. D. & Laurie, D. A. (1995) Maydica 40, 199-204. [Google Scholar]
- 13.Soderlund, C., Humphrey, S. Dunhum, A. & French, L. (2002) Genome Res. 10, 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou, H. H. & Holmes, M. H. (2001) Bioinformatics 17, 1093-1104. [DOI] [PubMed] [Google Scholar]
- 15.Rudd, S., Mewes, H.-W. & Mayer, K. F. X. (2003) Nucleic Acids Res. 31, 128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitelaw, C. A, Barbazuk, W. B., Pertea, G., Chan, A. P., Cheung, F., Lee, Y., Zheng, L., van Heeringen, S., Karamycheva, S., Bennetzen, J. L., et al. (2003) Science 302, 2118-2120. [DOI] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 18.Palmer, L. E., Rabinowicz, P. D., O'Shaughnessy, A. L., Balija, V. S., Nascimento, L. U., Dike, S., de la Bastide, M., Martiennssen, R. A. & McCombie, W. R. (2003) Science 302, 2115-2157. [DOI] [PubMed] [Google Scholar]
- 19.Lunde, C. F., Morrow, D. J., Roy, L. M. & Walbot, V. (2003) Funct. Integr. Genomics 3, 25-32. [DOI] [PubMed] [Google Scholar]
- 20.Flavell, R. B., Bennett, M. D., Smith, J. B. & Smith, D. B. (1974) Biochem. Genet. 12, 257-269. [DOI] [PubMed] [Google Scholar]
- 21.McClintock, B. (1948) Carnegie Inst. Wash. Year Book 47, 155-169. [PubMed] [Google Scholar]
- 22.Feschotte, C., Jiang, N. & Wessler, S. R. (2002) Nat. Rev. Genet. 3, 329-341. [DOI] [PubMed] [Google Scholar]
- 23.International Human Genome Sequencing Consortium (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 24.Ma, J., Devos, K. M. & Bennetzen, J. L. (2004) Genome Res. 14, 860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennetzen, J. L. (1996) Trends Microbiol. 4, 347-353. [DOI] [PubMed] [Google Scholar]
- 26.Volff, J. N., Bouneau, L., Ozouf-Costaz, C. & Fischer, C. (2003) Trends Genet. 19, 674-678. [DOI] [PubMed] [Google Scholar]
- 27.Moore, G., Lucas, H., Batty, N. & Flavell, R. (1991) Genomics 10, 461-468. [DOI] [PubMed] [Google Scholar]
- 28.Kazazian, H. H., Jr. (2004) Science 303, 1626-1632. [DOI] [PubMed] [Google Scholar]
- 29.Wu, J., Yamagata, H., Hayashi-Tsugane, M., Hijishita, S., Fujisawa, M., Shibata, M., Ito, Y., Nakamura, M., Sakaguchi, M., Yoshihara, R., et al. (2004) Plant Cell 16, 967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicient, C. M., Kalendar, R. & Schulman, A. H. (2001) Genome Res. 11, 2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Rice Chromosome 10 Sequencing Consortium (2003) Science 300, 1566-1569. [DOI] [PubMed] [Google Scholar]
- 32.Schoof, H., Ernst, R., Nazarov, V., Pfeifer, L., Mewes, H. W. & Mayer, K. F. (2004) Nucleic Acids Res. 32, D373-D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, J., Ma, J., Swigonova, Z., Ramakrishna, W., Linton, E., Llaca, V., Tanyolac, B., Park, Y.-J., Jeong, O.-Y., Bennetzen, J. L., et al. (2004) Genome Res. 14, 1924-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellis, M., Birren, B. W. & Lander, E. S. (2004) Nature 428, 617-624. [DOI] [PubMed] [Google Scholar]
- 35.Song, R. & Messing, J. (2002) Plant Physiol. 130, 1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swigonova, Z., Lai, J., Ma, J., Ramakrishna, W., Llaca, V., Bennetzen, J. L. & Messing, J. (2004) Genome Res. 14, 1916-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stam, M., Belele, C., Dorweiler, J. E. & Chandler, V. L. (2002) Genes Dev. 16, 1906-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark, R. M., Linton, E., Messing, J. & Doebley, J. F. (2004) Proc. Natl. Acad. Sci. USA 101, 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.