Abstract
Toward the end of gestation in mammals, the fetal lung undergoes a process of differentiation that is required for transition to air breathing at birth. Respiratory epithelial cells synthesize the surfactant proteins and lipids that together form the pulmonary surfactant complex necessary for lung function. Failure of this process causes respiratory distress syndrome, a leading cause of perinatal death and morbidity in newborn infants. Here we demonstrate that expression of the forkhead gene Foxa2 in respiratory epithelial cells of the peripheral lung controls pulmonary maturation at birth. Newborn mice lacking Foxa2 expression in the lung develop severe pulmonary disease on the first day of life, with all of the morphological, molecular, and biochemical features of respiratory distress syndrome in preterm infants, including atelectasis, hyaline membranes, and the lack of pulmonary surfactant lipids and proteins. RNA microarray analysis at embryonic day 18.5 demonstrated that Foxa2-regulated expression of a group of genes mediating surfactant protein and lipid synthesis, host defense, and antioxidant production. Foxa2 regulates a complex pulmonary program of epithelial cell maturation required for transition to air breathing at birth.
Keywords: forkhead, transcription factor, pulmonary epithelium, surfactant
In vertebrates, adaptation to air breathing required for terrestrial life was achieved by formation of the lung, providing an extensive surface area mediating transfer of oxygen and carbon dioxide. Whereas gas exchange in the fetus is provided solely by the placenta, respiration depends entirely on the lung after birth. Structural and functional maturation of the lung is required for neonatal adaptation to breathing, a process that occurs in late gestation in mammals. Unfortunately, when pregnancy is complicated by premature birth, the peripheral lung often has not undergone adequate maturation and lacks pulmonary surfactant. Surfactant deficiency results in respiratory distress within hours after birth (1).
Lung tubules undergo branching morphogenesis to form the conducting airways and peripheral lung tubules that will form the gas exchange region required for postnatal respiration. During late stages of fetal lung morphogenesis, peripheral regions of the lung saccules dilate, the mesenchyme thins, and an extensive pulmonary vascular bed grows into close apposition to respiratory epithelial cells. During this time, the cuboidal pretype II epithelial cells of the peripheral lung tubules differentiate to form cuboidal type II and squamous type I cells (2). Type I cells come into close contact with endothelial cells in alveolar capillaries, forming an efficient gas exchange area that is required for respiration after birth. Perinatal maturation of type II cells is associated with marked ultrastructural and biochemical changes that include reduced glycogen content, enhanced surfactant protein and lipid synthesis, increased numbers of lamellar bodies, surfactant secretion, and maturation of the microvilli at the apical surface of type II cells (3). Surfactant proteins and phospholipids reduce surface tension at the air-liquid interface in the alveolus. Both phospholipid and protein components of the surfactant complex play critical roles in surfactant function and pulmonary homeostasis at birth. Deficiency or dysfunction of surfactant proteins and phospholipids causes respiratory distress syndrome (RDS), a common disorder affecting preterm infants (1, 4).
Lung morphogenesis is mediated by numerous transcription factors, growth factors, and other signaling molecules that coordinate cell proliferation and differentiation. Foxa2 is a winged-helix transcription factor known to play critical roles in the formation of the primitive streak and the endoderm (5). In mice, Foxa2 is first expressed in the primitive streak shortly after the onset of gastrulation (6). Later in embryonic development, Foxa2 is expressed in endodermal-derived tissues, including liver, lung, pancreas, and intestine (5, 7). Germ-line deletion of Foxa2 causes failure of anterior-posterior patterning and foregut endoderm development, demonstrating its critical role in the formation of the early embryo (5). In the lung, Foxa2 expression is restricted to subsets of respiratory epithelial cells. Foxa2 is coexpressed with a number of proteins that mark epithelial cell differentiation, including thyroid transcription factor 1 (TTF-1), Clara cell secretary protein (CCSP), and surfactant proteins SP-A, SP-B, SP-C, and SP-D. The surfactant proteins are components of the surfactant and host defense systems required for postnatal adaptation to air breathing (8, 9). Previous in vitro studies demonstrated that Foxa2-enhanced expression of SP-B (10), TTF-1 (11), and CCSP (12, 13), supporting its potential role in the regulation of gene expression and/or differentiation of type II cells in the lung. Although the transcription factors and signaling events mediating early lung formation have been subject to active study, those coordinating the complex morphological, biochemical, and gene-expression changes that occur in the respiratory epithelium during perinatal lung maturation are unknown.
Because deletion of Foxa2 in the mouse causes embryonic death before formation of the lung, its potential roles in postnatal pulmonary lung adaptation have not been determined. To identify the potential roles of Foxa2 in postnatal pulmonary formation and function, we deleted the Foxa2 gene in subsets of respiratory epithelial cells by using a conditional Cre/LoxP recombination system (14). Deletion of Foxa2 caused emphysema, inflammation, and goblet cell hyperplasia in mice surviving to postnatal days (P) 16-30 (14). About 20% of Foxa2Δ/Δ mice survived until P30; however, the timing and cause of death at earlier times was not discerned. In the present study, we sought to determine the causes and mechanisms underlying postnatal death observed after deletion of Foxa2Δ/Δ in the lung.
Methods
Animals and Transgene Genotype. Foxa2loxP/loxP mice were generated at the University of Pennsylvania (15). Homologous recombination between loxP sites was accomplished by using (tetO)7CMV-Cre mice, kindly provided by Corrinne Lobe (University of Toronto, Toronto) (16). For lung-specific, doxycycline-induced recombination, the SP-C-rtTA-/tg transgenic line was used (14, 17). Triple-transgenic mice were generated by crossing (tetO)7Cre-/tg/Foxa2loxP/loxP and SP-C-rtTA-/tg/Foxa2loxP/loxP. Littermates of all other genotypes served as controls. Transgenic mice were identified by PCR with genomic DNA from the tails of fetal and postnatal mice as described in ref. 14.
Animal Husbandry and Doxycycline Administration. Animals were maintained in pathogen-free conditions according to protocols approved by Institutional Animal Care and Use Committee of the Cincinnati Children's Hospital Research Foundation. All animals were housed in humidity- and temperature-controlled rooms on a 12-h/12-h light-dark cycle and were allowed food and water ad libitum. There was no serologic evidence of pulmonary pathogens or bacterial infections in sentinel mice maintained within the colony. No serological or histological evidence of viral or bacterial infection was detected in representative mice. Gestation was dated by detection of the vaginal plug. Dams bearing double- and triple-transgenic pups were maintained on doxycycline in food (25 mg/g; Harlan Teklad, Madison, WI) since embryonic day (E) 0. The mice were killed by injection of anesthetic and exsanguinated.
Morphological Analysis. Tissues from fetal and neonatal lungs were prepared as described in ref. 18. Five-micrometer tissue sections were immunostained by Foxa2 antibody (1:800, sheep immunoaffinity purified IgG, Upstate Biotechnology, Lake Placid, NY). Electron microscopy was performed on lung tissue obtained from E18.5 Foxa2Δ/Δ mice and littermate controls after fixation in glutaraldehyde as described in ref. 19. The platelet endothelial cellular adhesion molecule was detected with antiserum from Santa Cruz Biotechnology. All experiments shown are representative of findings from at least two independent dams, generating at least four triple-transgenic offspring that were compared with littermates.
RNA Isolation and Analysis. RNA was prepared by using TRIzol reagent (Life Technologies, Rockville, MD) according to the manufacturer's specifications. SP-A, SP-B, SP-C, CCSP, and SP-D mRNAs were quantified by S1 nuclease protection assay or with ribosomal protein L32 as an internal control (20). RNA was treated with DNase at 37°C for 30 min. DNase was removed by DNase inactivation reagent (Ambion, Austin, TX) before cDNA synthesis. DNase-treated total-lung RNA (1 μg) was reverse transcribed and analyzed by real-time PCR with the Smart Cycler System (Cepheid, Sunnyvale, CA). The relative concentration of each mRNA was normalized to the concentration of GAPDH mRNA in each sample. GAPDH mRNA was quantitated by using 5′-CTT CAC CAC CAT GGA GGA GGC and 3′-GGC ATG GAC TGT GGT CAT GAG primers. Other primers are listed in Table 2, which is published as supporting information on the PNAS web site. Differences were assessed by two-tailed Student's t test.
RNA Microarray Analysis. Methods for RNA isolation, amplification, and data analysis are essentially as described in ref. 21. Lung cRNA was hybridized to the murine genome MOE430A (consists of ≈22,000 gene entries) chips (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol. Affymetrix microarray suite 5.0 was used to scan and quantitate the gene chips under default scan settings. Intensity data were collected from each chip and scaled to a target intensity of 1,500, and the results were analyzed by using genespring 6.0 (Silicon Genetics, Redwood City, CA). Eight chips from four pairwise experiments were used. Two-step normalization was used with each chip normalizing to the median of all genes on the chip, and each RNA from Foxa2Δ/Δ mouse was normalized to its specific control (age-matched littermates). Data were further transformed into log ratio for analysis and symmetry of distribution and analyzed as described in ref. 21. Promoter sequences of differentially expressed genes (-900 to +100) were searched for Foxa2 binding sites by using matinspector (Genomatrix, Munich), a weight matrix-based tool with a large library of Genomatix precompiled position weight matrix (22). The consensus Foxa2 site included A/G y aAAC/TA (23). The cutoffs were set up so that the core similarity was 1 and the matrix similarity was >0.9. Genes with potential Foxa2-binding sites in a 1-kb promoter region were displayed in a feature map (refs. 24 and 25 and data not shown).
Surfactant Analysis. Phospholipids were isolated from chloroform/methanol extracts of lungs from two mice for each assay. Phospholipids were separated by 2D thin-layered chromatography for compositional analyses (26). For surfactant protein analysis, extracted lipids [0.44 nmol of saturated phosphatidylcholine (PC)] were subjected to SDS/PAGE and transferred to nitrocellulose. Western blotting was performed with antiserum against mature SP-B peptide by using AB3426 (Chemicon) (26).
Transcription of the Lyz-M, Lyz-P, and Scgb1a1 Promoters in Vitro. The lysozyme P and lysozyme M promoter-luciferase constructs were kindly provided by H. Akinbi (Cincinnati Children's Hospital Medical Center). These promoters consisted of 1 kb of the 5′ regulatory region of the mouse genes. The Scgb1a1-luciferase construct consisted of 2.3 kb of the 5′ regulatory region encoding the rat Scgb1a1 gene. Promoter-luciferase constructs were cotransfected with a Foxa2 expression plasmid into HeLa cells. FuGENE 6 (Roche Molecular Biochemicals) was used for transfection. Forty-eight hours after transfection, luciferase activity was assessed and normalized for cotransfection by β-galactosidase activity.
Statistical Analysis. ANOVA or Student's t test was used to determine the levels of difference between groups, and P values for significance were set to 0.05. Values for all measurements were expressed as the mean ± SE.
Results and Discussion
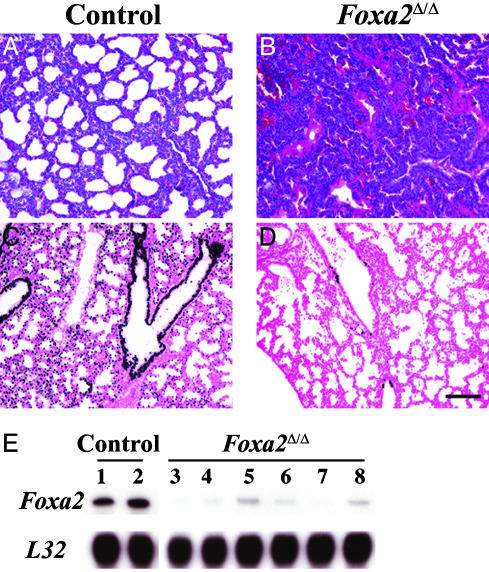
Foxa2 was deleted by expression of Cre recombinase under conditional control of the reverse tetracycline transactivator expressed in respiratory epithelial cells of the fetal lung under control of the surfactant protein C promoter (see Methods), producing Foxa2Δ/Δ pups. When SP-C-rtTA-/tg, (tetO)7Cre-/tg, Foxa2loxP/loxP dams (n = 6 litters) were maintained on doxycycline from E0 and genotyped between P1 and P5, only 46% of the Foxa2Δ/Δ pups survived and most died on P1. The extent of Foxa2 deletion was assessed the day before birth (E18.5), demonstrating extensive reduction of Foxa2 by immunohistochemistry (Fig. 1 C and D) and RNA analysis (Fig. 1E). Lung weights were similar, and there was no evidence of cell injury or inflammation before birth. Although the extent of Foxa2 reduction was variable, nuclear staining of Foxa2 and the levels of Foxa2 mRNA were markedly reduced in the terminal lung saccules and in epithelial cells of intrapulmonary conducting airways of the Foxa2Δ/Δ mice (Fig. 1D). In contrast, nuclear Foxa2 staining was detected in alveolar type II cells and in conducting airway epithelial cells in control mice (Fig. 1C). Pulmonary vascular structures, as indicated by platelet endothelial cellular adhesion molecule staining, were unchanged after deletion of Foxa2 (Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 1.
Lung atelectasis and hyaline membrane formation in Foxa2Δ/Δ mice. Lung sections from control (A) and Foxa2Δ/Δ (B) mice were prepared on P1 and stained with hematoxylin/eosin. Severe atelectasis and hyaline membranes (B) were observed in a Foxa2Δ/Δ with respiratory distress on P1. Normal lung inflation and histology are observed in age-matched controls (A). Lung sections were prepared on E18.5 for immunohistochemistry by using anti-Foxa2 antibody. Nuclear Foxa2 staining was observed in epithelial cells of conducting and peripheral airways and alveoli (C) of control littermates and was absent or decreased in Foxa2Δ/Δ mice (D). No morphological changes were observed in Foxa2Δ/Δ at the light microscopy level at E15.5 (data not shown) or E18.5 (D). (Scale bar, 100 μm.) RNase protection assay was used for estimation of Foxa2 mRNA in lung RNA from control and Foxa2 littermates at E18.5 and compared with L32 mRNA (E). Lung RNA from these pups was used for RNA microarray analysis. Observations are representative of n = 5-10 pups.
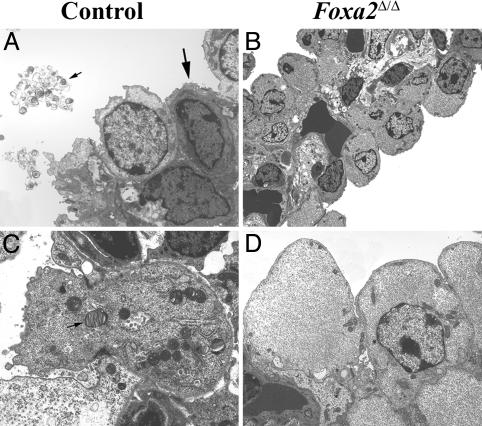
When observed during delivery, most Foxa2Δ/Δ mice became oxygenated after birth. About one-half of the Foxa2Δ/Δ mice developed severe respiratory distress and were found gasping, cyanotic, or dead within the first 2 days after birth. The precise timing of death was frequently complicated by maternal cannibalism. When observed during normal delivery (E19.5-E20), ≈50% of the Foxa2Δ/Δ mice developed respiratory distress within 2-3 h after birth. Non-Foxa2Δ/Δ littermates survived normally. Histologic examination of the pups found in distress or moribund revealed atelectasis, with formation of hyaline membranes lining terminal airways, typical of findings in preterm infants with RDS (Fig. 1B). Pathology was severe, but variable, with focal or extensive atelectasis, hyaline membrane formation, and pulmonary congestion. On examination by electron microcopy, ultrastructural features of lungs from E18.5 Foxa2Δ/Δ mice were immature compared with their littermates (Fig. 2). In controls, cuboidal type II cells contained numerous lamellar bodies, apical microvilli, and highly organized rosette glycogen (Fig. 2 A and C). Lamellar bodies were observed in the lumen of peripheral airspaces. Alveolar walls were thin, well vascularized, and lined both by squamous type I cells and cuboidal type II cells, indicating structural maturation of the peripheral lung saccules. In Foxa2Δ/Δ pups, alveolar walls were thickened and lined by immature cuboidal type II cells. Squamous type I cells were not observed in Foxa2Δ/Δ mice (Fig. 2 B and D). The integrity of the pulmonary vascular system was carefully assessed. Although the microvessels were imbedded within relatively thickened alveolar walls, there was no evidence of capillary leakage or endothelial cell abnormalities. Cytoplasmic glycogen was dispersed, and apical microvilli were lacking. Lamellar bodies, the intracellular storage form of pulmonary surfactant, were absent in alveolar type II cells, and secreted surfactant was not detected in the air spaces at E18.5. Taken together, the ultrastructure of the peripheral lung saccules of the Foxa2Δ/Δ mice was typical of that normally seen much earlier in gestation (E16-E17) (27).
Fig. 2.
Lung ultrastructure in Foxa2Δ/Δ mice. Electron microscopy was performed on lungs from E18.5 Foxa2Δ/Δ (B and D) and littermates (A and C). Squamous type I cells (thick arrow) and cuboidal type II cells (thin arrow in C) containing lamellar bodies, apical microvilli, and highly organized rosette glycogen were observed in lungs of control mice (A and C). The densely stained cell below the type I cell is a capillary endothelial cell creating the gas-exchange area (A). Type I cells were not observed in the lungs of Foxa2Δ/Δ mice. The pulmonary vascular bed in Foxa2Δ/Δ mice was intact, but in the absence of type I cells, the thin-walled alveolar-capillary interface seen in the control mice was not formed. Type II cells were immature, lamellar bodies were absent in Foxa2Δ/Δ mice, apical microvilli were sparse, and particulate glycogen was absent in Foxa2Δ/Δ mice. Thin arrow (A) shows secreted surfactant. The micro-graphs are representative of two Foxa2Δ/Δ and littermate controls.
Lung PC and saturated PC, critical components of pulmonary surfactant, normally increase before birth (27). Decreased PC- or lecithin-to-sphingomyelin ratios in amniotic fluid or lung secretions are used clinically to predict the risk of RDS in preterm infants (27, 28). At E18.5, fractional content of PC and saturated PC were significantly decreased, whereas that of phosphatidylethanolamine (PE), sphingomyelin (SM), and phosphatidylserine (PS) was increased in the lungs of Foxa2Δ/Δ mice (Table 1).
Table 1. Phospholipid composition after deletion of Foxa2.
| Control | Foxa2Δ/Δ | |
|---|---|---|
| n | 6 | 6 |
| Lung weight, g | 0.033 ± 0.004 | 0.029 ± 0.006* |
| Body weight, g | 1.24 ± 0.074 | 1.30 ± 0.079 |
| Total lipid, μg per g of body weight | 434 ± 48 | 327 ± 49 |
| SatPC, nmol per g of body weight | 77 ± 4 | 37 ± 3* |
| Phospholipid composition,† % | ||
| PC | 63.1 ± 1.4 | 48.4 ± 3.5* |
| Phosphatidylethanolamine | 3.2 ± 0.3 | 7.1 ± 0.6* |
| Phosphatidylglycerol | 18.2 ± 2.0 | 22.3 ± 1.0 |
| Phosphatidylinositol | 1.3 ± 0.2 | 1.3 ± 0.2 |
| Sphigomyelin | 8.7 ± 0.6 | 11.1 ± 0.3* |
| Lyso-PC | 2.2 ± 0.2 | 2.4 ± 0.4 |
| Phosphatidylserine | 3.3 ± 0.3 | 6.4 ± 0.8* |
Values are mean ± SE.
p < 0.05 vs. control littermates.
Three samples consisting of the pooled lung homogenate from two mice were evaluated for the relative abundance of phospholipids. PC was significantly decreased in Foxa2Δ/Δ mice, whereas phosphatidylethanolamine, sphingomyelin, and phosphatidylserine were significantly increased.
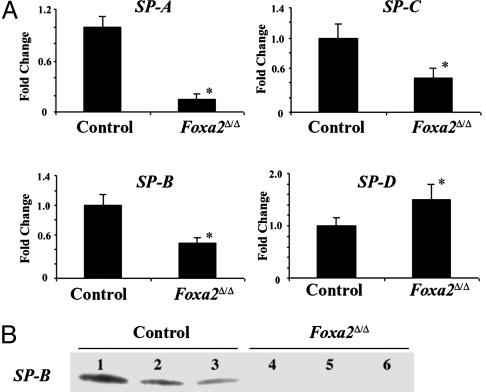
Because surfactant proteins A-D play critical roles in surfactant function and homeostasis (29), their expression was assessed in Foxa2Δ/Δ mice at E18.5 (Fig. 3A). SP-A, SP-B, and SP-C mRNAs were significantly decreased in the Foxa2Δ/Δ mice. The active SP-B peptide, produced by proteolytic processing of proSP-B, was markedly decreased in lung homogenates made from the Foxa2Δ/Δ mice before birth (Fig. 3B). Because SP-B is required for the formation of lamellar bodies, tubular myelin, and neonatal lung function in mice and humans (30, 31), decreased production and processing of SP-B alone likely contributes to respiratory distress in newborn Foxa2Δ/Δ mice. However, because lung maturation and expression of surfactant proteins A, C, and D are not altered in gene-targeted SP-B mice (30), the decrease in SP-B does not account for the structural and biochemical abnormalities seen in the lungs of Foxa2Δ/Δ mice.
Fig. 3.
Surfactant protein expression. (A) Decreased surfactant protein mRNAs in Foxa2Δ/Δ mice. Total lung RNA was prepared from fetal mouse lung at E18.5. S1 nuclease protection analysis for SP-A, SP-B, SP-C, SP-D, and ribosomal protein L32 mRNAs was performed with 3 μg of total RNA. Protected fragments were quantified by phosphorimaging, and mRNAs were normalized relative to L32 as an internal control. The mean of the mRNA content in control littermates was set to 1. Relative values of SP-A, SP-B, and SP-C mRNAs in Foxa2Δ/Δ mice are shown as mean ± SE and were compared with controls with the two-tailed t test (n = 4-8 animals per group; *, P < 0.05 vs. control). (B) Decreased SP-B protein in lung homogenates from Foxa2Δ/Δ mice. Lung homogenates were prepared from Foxa2Δ/Δ (n = 6) and littermate controls (n = 6) at E18.5. A representative immunoblot of mature SP-B from control (lanes 1-3) and Foxa2Δ/Δ (lanes 4-6) mice is shown.
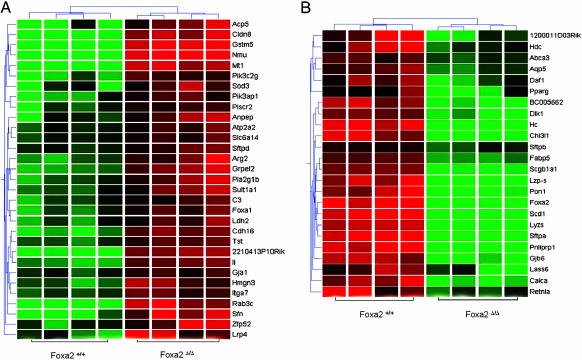
To identify the genes regulated by Foxa2 in the lung, RNA from Foxa2Δ/Δ and control lungs were compared at E18.5 by using Affymetrix murine genome MOE430 gene chips. Lung tissue from Foxa2Δ/Δ mice, in which Foxa2 mRNA was nearly absent, were chosen for microarray analysis (Fig. 1E). Lung Foxa2 mRNA was decreased 11-fold in the Foxa2Δ/Δ mice, supporting the RNase protection data. Microarray analysis identified 24 mRNAs that were significantly decreased and 33 mRNAs that were significantly increased by 1.5-fold or more in the Foxa2Δ/Δ mice (Fig. 4 and Table 3, which is published as supporting information on the PNAS web site). Expression of genes encoding surfactant proteins A-D and other genes regulating lipid homeostasis, host defense, and antioxidant production were influenced by deletion of Foxa2. Expression of genes regulating lipid homeostasis, including Scd-1, Abca3, Dlk1, Pparg, Retnla, Pnliprp1, Pon1, and Fabp5, was observed. Of these, Abca3 is of critical importance to lung function in the Foxa2Δ/Δ mice, being expressed in the limiting membrane of type II epithelial cells in the lung (32). Recently, mutations in the human ABCA3 transporter gene were associated with lethal neonatal respiratory distress in term infants and with ultrastructural abnormalities in lamellar bodies (33). Genes involved in lung innate host defense also were decreased, including Sftpa, Scgb1a1, Hc, Lyz-M, Lyz-P, Marco, Chi3l1, Defb35, and Daf1, and, therefore, represent a group of genes that are likely critical for protection of the lung from infection after birth. Increased expression of genes regulating antioxidant production, complement, and antigen presentation was observed, including Sod3, Gstm5, Mt1, Li, C3, and H2-DMb2. Changes in the relative expression of a number of these genes were verified by RT-PCR supporting the validity of the microarray data (Table 2). Of interest, Foxa1 mRNA was increased ≈2-fold in the Foxa2Δ/Δ mice. Because Foxa1 and Foxa2 are closely related forkhead family members, share similar structures, and are coexpressed in respiratory epithelial cells, increased expression of Foxa1 may represent a compensatory response to the deletion of Foxa2.
Many of the mRNAs that were decreased in Foxa2Δ/Δ mice are expressed in epithelial cells, rather than mesenchymal cells, in the lung and, therefore, may represent direct transcriptional targets of Foxa2. Potential Foxa2 binding sites located within 1 kb (-900 to +100) of their transcription start sites were identified in many of these genes. Genes whose expression was altered in the microarray analyses of the Foxa2Δ/Δ mice and contained potential Foxa2 elements (Fig. 6, which is published as supporting information on the PNAS web site). Among those genes, Scd-1, Abca3, Pparg, Retnal, Sftpa, Sftpb, Sftpd, Scgb1a1, Defb35, Lyz-P, and Lyz-M are coexpressed with Foxa2 in lung epithelial cells. However, the Foxa2 consensus binding site is variable and found with high frequency in the genome. To assess whether Foxa2 regulated several of the potential target genes, transfection assays were performed with reporter constructs made with the 5′ flanking regions of potential target genes after cotransfection with a Foxa2 expression vector in HeLa cells. Transfection with Foxa2 activated promoter elements from the Lyz-P, Lyz-M, and Scgb1a1 genes, demonstrating that these genes are likely direct transcriptional targets of Foxa2 (Fig. 7, which is published as supporting information on the PNAS web site). Foxa2 elements were previously identified in the 5′ regions of SP-B and CCSP promoters, and site-specific mutations in the Foxa2 binding sites in the SP-B and CCSP promoters were shown to inhibit promoter activity in vitro (10, 34).
Direct observation of pups born at E19-E19.5 demonstrated that both Foxa2Δ/Δ and control littermates initiated respiration and became oxygenated at birth; however, Foxa2Δ/Δ mice died within hours after birth, and nearly one-half were dead by P5. Many Foxa2Δ/Δ pups were found moribund on P1. Because dead pups are eaten by their dam, the precise timing of death of the Foxa2Δ/Δ mice was often unclear. All Foxa2Δ/Δ mice succumbed from severe lung disease by P60. Because the efficiency of Foxa2 deletion is variable in this model, differences in survival of Foxa2Δ/Δ mice may be influenced, at least in part, to the extent of Foxa2 recombination. Decreased SP-A, SP-B, and increased SP-D were observed after deletion of Foxa2 in respiratory epithelial cells. Foxa2 and the surfactant proteins are colocalized in respiratory epithelial cells of both conducting and peripheral airways. Although Foxa2 binding sites were identified within proximal regions of the Sftpa and Sftpc genes, direct stimulation of their transcription was not observed in transfection assays in vitro (ref. 35 and data not shown), indicating that Foxa2 may cooperate with other transcription factors or indirectly regulate expression of target genes. It is also possible that Foxa2 influences gene expression by interacting with elements not included in the regulatory regions of the gene constructs tested. Previous studies identified the function of Foxa2 regulatory elements in the Sftpb gene (10).
Fig. 4.
Identification of mRNAs influenced by deletion of Foxa2 in the lung. Two-dimensional hierarchical clustering shows 55 genes that were significantly altered in response to Foxa2Δ/Δ. The intensity in the red and green color ranges indicates the up- and down-regulation of RNAs, respectively. Each row represents a single gene, and each column represents a biological replicate lung mRNA isolated for Foxa2+/[supi]+ or Foxa2Δ/Δ pups at E18.5. Each box represents a normalized gene expression value. Clustering was performed by Unweighted Pair Group Method with Arithmetic Mean (UPGMA), and similarity was measured by standard correlation (see Methods).
Respiratory distress syndrome occurs primarily in premature infants and is caused by pulmonary immaturity and the lack of surfactant. Both SP-B and ABCA3 are required for successful transition to air breathing at birth, and mutations in both genes cause lethal respiratory distress and severe acute or chronic lung disease in humans (31, 36) and mice (30). Targeted disruption of Sftpb in mice and mutations in ABCA3 in humans perturbed formation of lamellar bodies, causing respiratory failure shortly after birth (30, 33). In amniotic fluid, SP-B and phospholipid concentrations increase in parallel during late gestation (37). Reduction of SP-B is associated with surfactant dysfunction and respiratory failure in the perinatal and postnatal periods (30, 38).
Phospholipid composition of the lungs of Foxa2Δ/Δ mice was consistent with delayed pulmonary maturation as seen in preterm infants at risk for RDS. In the present study, deletion of Foxa2 inhibited expression of a number of genes involved in lipid metabolism including several of the genes previously identified as Foxa2 targets during preadipocyte differentiation (39, 40) (Fig. 6). Scd-1 mRNA increases in late gestation and is expressed in respiratory epithelial cells (41). In a recent study from this laboratory, both surfactant proteins and Scd-1 were decreased in the lungs of mice bearing mutations in phosphorylation sites in TTF-1 (42). Cis-acting sites for both TTF-1 and Foxa2 are found in the promoter regions of many of the transcriptional target genes influenced by microarray in the Foxa2Δ/Δ mice, supporting the concept that Foxa2 and TTF-1 may cooperate in regulation in modulating surfactant homeostasis and host defense.
In the present studies, deletion of Foxa2 was associated with a 2-fold increase in Foxa1 mRNA, raising the possibility that Foxa1 may partially compensate for loss of Foxa2 in respiratory epithelial cells. Foxa1 and Foxa2 are expressed in similar temporal-spatial patterns in respiratory epithelial cells of the conducting and peripheral airways (43). They share 93% amino acid homology in the winged-helix DNA binding domain but are conserved to a lesser extent in transcription activator or repressor domains. In previous in vitro studies, both distinct and similar effects of Foxa1 and Foxa2 have been reported (10, 12, 13, 34, 35, 44).
Although mutation and deletion of a number of transcription factors have been associated with pulmonary abnormalities that cause neonatal respiratory failure, most of these genes cause lethal lung malformations rather than RDS (reviewed in ref. 43). Of clinical importance, disruption of corticosterone releasing factor (CRH) or the glucocorticoid receptor (GR) disrupts lung morphogenesis, retards prenatal lung maturation, and causes respiratory failure at birth (45, 46). However, effects of glucocorticoids are mediated primarily by their actions on lung fibroblasts and are not accompanied by changes in surfactant protein expression.
In summary, Foxa2 regulates groups of genes mediating surfactant homeostasis and host defense that are required for postnatal adaptation to air breathing. Deletion of Foxa2 in the epithelial cells of the developing mouse lung inhibited the differentiation of respiratory epithelial cells and caused neonatal respiratory failure with the characteristics of RDS in preterm infants. Thus, expression of a single gene in epithelial cells of the lung regulates a genetic program critical for respiration after birth.
Supplementary Material
Acknowledgments
We thank Ms. Ann Maher for secretarial support. This work was supported by National Institutes of Health Grants HL56387 (to J.A.W.), HL75770, and HL38859.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CCSP, Clara cell secretary protein; En, embryonic day n; PC, phosphatidylcholine; Pn, postnatal day n; RDS, respiratory distress syndrome; TTF-1, thyroid transcription factor-1.
References
- 1.Avery, M. E. & Mead, J. (1959) AMA J. Dis. Child. 97, 517-523. [DOI] [PubMed] [Google Scholar]
- 2.Burri, P. H. (1984) Annu. Rev. Physiol. 46, 617-628. [DOI] [PubMed] [Google Scholar]
- 3.Williams, M. C. & Mason, R. J. (1977) Am. Rev. Respir. Dis. 115, 37-47. [DOI] [PubMed] [Google Scholar]
- 4.Hallman, M., Haataja, R. & Marttila, R. (2002) Semin. Perinatol. 26, 450-460. [DOI] [PubMed] [Google Scholar]
- 5.Ang, S. L., Wierda, A., Wong, D., Stevens, K. A., Cascio, S., Rossant, J. & Zaret, K. S. (1993) Development (Cambridge, U.K.) 119, 1301-1315. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki, H. & Hogan, B. L. (1993) Development (Cambridge, U.K.) 118, 47-59. [DOI] [PubMed] [Google Scholar]
- 7.Monaghan, A. P., Kaestner, K. H., Grau, E. & Schutz, G. (1993) Development (Cambridge, U.K.) 119, 567-578. [DOI] [PubMed] [Google Scholar]
- 8.Zhou, L., Lim, L., Costa, R. H. & Whitsett, J. A. (1996) J. Histochem. Cytochem. 44, 1183-1193. [DOI] [PubMed] [Google Scholar]
- 9.Stahlman, M. T., Gray, M. E. & Whitsett, J. A. (1998) J. Histochem. Cytochem. 46, 955-962. [DOI] [PubMed] [Google Scholar]
- 10.Bohinski, R. J., Di Lauro, R. & Whitsett, J. A. (1994) Mol. Cell. Biol. 14, 5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda, K., Shaw-White, J. R., Wert, S. E. & Whitsett, J. A. (1996) Mol. Cell. Biol. 16, 3626-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bingle, C. D. & Gitlin, J. D. (1993) Biochem. J. 295, 227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bingle, C. D., Hackett, B. P., Moxley, M., Longmore, W. & Gitlin, J. D. (1995) Biochem. J. 308, 197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan, H., Kaestner, K. H., Ang, S. L., Ikegami, M., Finkelman, F. D., Stahlman, M. T., Fulkerson, P. C., Rothenberg, M. E. & Whitsett, J. A. (2004) Development (Cambridge, U.K.) 131, 953-964. [DOI] [PubMed] [Google Scholar]
- 15.Sund, N. J., Ang, S. L., Sackett, S. D., Shen, W., Daigle, N., Magnuson, M. A. & Kaestner, K. H. (2000) Mol. Cell. Biol. 20, 5175-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer, B. (1998) Methods 14, 381-392. [DOI] [PubMed] [Google Scholar]
- 17.Perl, A. K., Tichelaar, J. W. & Whitsett, J. A. (2002) Transgenic Res. 11, 21-29. [DOI] [PubMed] [Google Scholar]
- 18.Wert, S. E., Yoshida, M., LeVine, A. M., Ikegami, M., Jones, T., Ross, G. F., Fisher, J. H., Korfhagen, T. R. & Whitsett, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5972-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark, J. C., Tichelaar, J. W., Wert, S. E., Itoh, N., Perl, A. K., Stahlman, M. T. & Whitsett, J. A. (2001) Am. J. Physiol. 280, L705-L715. [DOI] [PubMed] [Google Scholar]
- 20.Dranoff, G., Crawford, A. D., Sadelain, M., Ream, B., Rashid, A., Bronson, R. T., Dickersin, G. R., Bachurski, C. J., Mark, E. L., Whitsett, J. A., et al. (1994) Science 264, 713-716. [DOI] [PubMed] [Google Scholar]
- 21.Xu, Y., Clark, J. C., Aronow, B. J., Dey, C. R., Liu, C., Wooldridge, J. L. & Whitsett, J. A. (2003) J. Biol. Chem. 278, 7674-7682. [DOI] [PubMed] [Google Scholar]
- 22.Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995) Nucleic Acids Res. 23, 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overdier, D. G., Porcella, A. & Costa, R. H. (1994) Mol. Cell. Biol. 14, 2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulzink, R. J., Weerdesteyn, H., Croes, A. F., Gerats, T., van Herpen, M. M. & van Helden, J. (2003) Plant Physiol. 132, 75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Helden, J. (2003) Nucleic Acids Res. 31, 3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikegami, M., Dhami, R. & Schuchman, E. H. (2003) Am. J. Physiol. 284, L518-L525. [DOI] [PubMed] [Google Scholar]
- 27.Randell, S. H. & Young, S. L. (2004) in Fetal and Neonatal Physiology, eds. Polin, R. A., Fox, W. W. & Abman, S. H. (Saunders, Philadelphia), pp. 1034-1040.
- 28.Gluck, L., Kulovich, M. V., Borer, R. C., Jr., Brenner, P. H., Anderson, G. G. & Spellacy, W. N. (1971) Am. J. Obstet. Gynecol. 109, 440-445. [DOI] [PubMed] [Google Scholar]
- 29.Whitsett, J. A. & Weaver, T. E. (2002) N. Engl. J. Med. 347, 2141-2148. [DOI] [PubMed] [Google Scholar]
- 30.Clark, J. C., Wert, S. E., Bachurski, C. J., Stahlman, M. T., Stripp, B. R., Weaver, T. E. & Whitsett, J. A. (1995) Proc. Natl. Acad. Sci. USA 92, 7794-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogee, L. M., Garnier, G., Dietz, H. C., Singer, L., Murphy, A. M., deMello, D. E. & Colten, H. R. (1994) J. Clin. Invest. 93, 1860-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamano, G., Funahashi, H., Kawanami, O., Zhao, L. X., Ban, N., Uchida, Y., Morohoshi, T., Ogawa, J., Shioda, S. & Inagaki, N. (2001) FEBS Lett. 508, 221-225. [DOI] [PubMed] [Google Scholar]
- 33.Shulenin, S., Nogee, L. M., Annilo, T., Wert, S. E., Whitsett, J. A. & Dean, M. (2004) N. Engl. J. Med. 350, 1296-1303. [DOI] [PubMed] [Google Scholar]
- 34.Sawaya, P. L. & Luse, D. S. (1993) J. Biol. Chem. 269, 22211-22216. [PubMed] [Google Scholar]
- 35.He, Y., Crouch, E. C., Rust, K., Spaite, E. & Broody, S. L. (2000) J. Biol. Chem. 275, 31051-31060. [DOI] [PubMed] [Google Scholar]
- 36.Nogee, L. M., Wert, S. E., Profitt, S. A., Hull, W. M. & Whitsett, J. A. (2000) Am. J. Respir. Crit. Care Med. 161, 973-981. [DOI] [PubMed] [Google Scholar]
- 37.Pryhuber, G. S., Hull, W. M., Fink, I., McMahan, M. J. & Whitsett, J. A. (1991) Pediatr. Res. 30, 597-605. [DOI] [PubMed] [Google Scholar]
- 38.Gregory, T. J., Longmore, W. J., Moxley, M. A., Whitsett, J. A., Reed, C. R., Fowler, A. A., III, Hudson, L. D., Maunder, R. J., Crim, C. & Hyers, T. M. (1991) J. Clin. Invest. 88, 1976-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfrum, C., Shih, D. Q., Kuwajima, S., Norris, A. W., Kahn, C. R. & Stoffel, M. (2003) J. Clin. Invest. 112, 345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villena, J. A., Kim, K. H. & Sul, H. S. (2002) Horm. Metab. Res. 34, 664-670. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, F., Pan, T., Nielsen, L. D. & Mason, R. J. (2004) Am. J. Respir. Cell Mol. Biol. 30, 174-183. [DOI] [PubMed] [Google Scholar]
- 42.deFelice, M., Silberschmidt, D., DiLauro, R., Xu, Y., Wert, S. E., Weaver, T. E., Bachurski, C. J., Clark, J. C. & Whitsett, J. A. (2003) J. Biol. Chem. 278, 35574-35583. [DOI] [PubMed] [Google Scholar]
- 43.Costa, R. H., Kalinichenko, V. V. & Lim, L. (2001) Am. J. Physiol. 280, L823-L838. [DOI] [PubMed] [Google Scholar]
- 44.Kaestner, K. H., Hiemisch, H., Luckow, B. & Schutz, G. (1994) Genomics 20, 377-385. [DOI] [PubMed] [Google Scholar]
- 45.Muglia, L. J., Bae, D. S., Brown, T. T., Vogt, S. K., Alvarez, J. G., Sunday, M. E. & Majzoub, J. A. (1999) Am. J. Respir. Cell Mol. Biol. 20, 181-188. [DOI] [PubMed] [Google Scholar]
- 46.Cole, T. J., Blendy, J. A., Monaghan, A. P., Krieglstein, K., Schmid, W., Aguzzi, A., Fantuzzi, G., Hummler, E., Unsicker, K. & Schutz, G. (1995) Genes Dev. 9, 1608-1621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.