Abstract
Omp85-like proteins represent a family of proteins involved in protein translocation, and they are present in all domains of life, except archaea. In eukaryotes, Omp85-like proteins have been demonstrated to form tetrameric pore-forming complexes that interact directly with their substrate proteins. Studies performed with bacterial Omp85-like proteins have demonstrated pore-forming activity but no evidence of multimerization. In this article, we characterize the Haemophilus influenzae HMW1B protein, an Omp85-like protein that has been demonstrated to be critical for secretion of the H. influenzae HMW1 adhesin. Analysis of purified protein by biochemical and electron microscopic techniques revealed that HMW1B forms a tetramer. Examination using liposome-swelling assays demonstrated that HMW1B has pore-forming activity, with a pore size of ≈2.7 nm. Far-Western blot analysis established that HMW1B interacts with the N terminus of HMW1. These results provide evidence that a bacterial Omp85-like protein forms a tetramer and interacts directly with a substrate protein, suggesting that the architecture and physical properties of Omp85-like proteins have been conserved throughout evolution.
Omp85-like proteins represent a large family defined by phylogenetic relatedness, secondary-structure predictions, and a role in protein translocation (1-5). Members of this family are present in diverse organisms across the evolutionary spectrum, including bacteria, fungi, plants, and animals (1, 4). The widespread nature of Omp85-like proteins underscores their critical role in cellular processes, highlighted by the fact that many are required for cell viability (3, 4, 6-9). Although all Omp85-like proteins share sequence homology, they can be grouped into two classes based on the specific role that they play in protein translocation. One class is typified by Saccharomyces cerevisiae Sam50 and Neurospora crassa Tob55, which are chiefly involved in insertion of β-barrel proteins into the outer membrane (OM) of mitochondria (6, 9). The second class is epitomized by the chloroplast protein Toc75, which is involved in secretion of proteins across a membrane (10, 11). Interestingly, both classes of Omp85-like proteins are present in Gram-negative bacteria. Neisseria meningitidis Omp85, the namesake of this family of proteins, has been demonstrated recently to be critical for the insertion of proteins into the bacterial OM (7). Other Gram-negative bacterial Omp85-like proteins, including Serratia marcescens ShlB and Bordetella pertussis FhaC [and other members of the so-called two-partner secretion (TPS) pathway], have been demonstrated to function in protein export across the OM (12).
The high level of amino acid homology among Omp85-like proteins from bacteria to humans raises the possibility that members of this family have similar structures. Although no crystal structures exist so far for Omp85-like proteins, several of these proteins have been subjected to biochemical studies of overall architecture. In eukaryotes, both classes of Omp85-like proteins have been demonstrated to form tetramers and to have pore activity (6, 11, 13). Additional analysis of eukaryotic Omp85-like proteins has revealed that protein import is facilitated by a direct interaction with the precursor substrate protein (6, 14). In bacteria, the only two family members that have been studied in detail are ShlB and FhaC, both of which have been demonstrated to have pore activity. However, no convincing evidence for multimer formation has been obtained for either ShlB or FhaC, thus contrasting with eukaryotic Omp85-like proteins (15, 16). These findings have led others to suggest that the tetrameric Omp85 protein translocation system in eukaryotes may have evolved from more primitive, monomeric bacterial Omp85 homologs (3, 5).
In this study, we examined Haemophilus influenzae HMW1B, an OM protein that has been demonstrated to be critical for the secretion of the H. influenzae HMW1 adhesin across the OM and is considered to be a member of the TPS pathway, like ShlB and FhaC (12, 17). We found that HMW1B exhibits heat-modifiable mobility, a hallmark of β-barrel proteins. Examination by size-exclusion chromatography, Blue Native PAGE, and electron microscopy indicated that detergent-extracted HMW1B forms a tetramer. Liposome-swelling assays revealed that HMW1B has pore-forming activity, with a pore size of ≈2.7 nm. Additional analysis established that HMW1B interacts with the HMW1 N terminus, which serves as a targeting signal and facilitates translocation of the substrate protein. Together, these observations suggest that Omp85-like proteins have maintained a common structural architecture and conserved intrinsic properties throughout evolution, highlighting the general importance of tetrameric pore-forming proteins in protein translocation.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions. Escherichia coli DH5α (Invitrogen) and E. coli BL21(DE3) (18) are laboratory strains that have been described. E. coli BL21(DE3)omp8 is a porin-deficient strain that lacks OM proteins OmpF, OmpC, OmpA, and LamB (19) and was a generous gift from V. Braun (University of Tubingen, Tubingen, Germany). Plasmids used in this study are described in Supporting Materials and Methods and Table 1, which are published as supporting information on the PNAS web site. E. coli strains were grown on LB agar or in LB broth and were stored at -80°C in LB broth with 50% glycerol. Antibiotic concentrations used to select for plasmids included 100 μg·ml-1 ampicillin and 30 μg·ml-1 chloramphenicol.
Recombinant DNA Methods. DNA ligations, restriction endonuclease digestions, gel electrophoresis, and PCR were performed according to standard techniques (20). Plasmids were introduced into E. coli by electroporation (21).
Cell Fractionation and Protein Analysis. Whole-cell sonicates were prepared by resuspending bacterial pellets in 10 mM Hepes (pH 7.4) and sonicating to clarity. OM proteins were recovered on the basis of Sarkosyl insolubility, as described by Carlone et al. (22). Western blotting was performed with a rabbit polyclonal antiserum raised against HMW1B or a rabbit polyclonal antiserum raised against the histidine-affinity tag (HAT) epitope (Clontech). An anti-rabbit IgG antiserum conjugated to horseradish peroxidase (Sigma) was used as the secondary antibody, and detection of antibody reactivity was accomplished by incubation of the membrane in a chemiluminescent substrate solution (Pierce) and exposure to film.
For heat-modifiability assays, samples were resuspended in an equal volume of 0.4% SDS sample buffer (0.4% SDS/125 mM Tris, pH 6.8/40% glycerol/0.01% bromophenol blue). The samples were incubated for 5 min at either 25°C or 100°C before loading on a 10% polyacrylamide gel. Electrophoresis was performed at 4°C using precooled running buffer (0.1% SDS/25 mM Tris base/192 mM glycine).
Culture supernatants were precipitated by adding trichloroacetic acid to a final concentration of 10% (vol/vol), incubating for 10 min at 4°C, and then centrifuging at 15,600 × g at 4°C for 10 min.
Protein Purification. To purify HMW1B, overnight cultures of E. coli BL21(DE3)omp8 harboring pHAT::HMW1B were harvested and resuspended in 10 mM Hepes (pH 7.4) supplemented with Complete Mini protease inhibitor mixture tablets (Roche). Bacteria were disrupted with a French press (SLM Instruments, Rochester, NY), and OM proteins were recovered on the basis of Sarkosyl insolubility (22). OMs were solubilized with 1% Elugent (Calbiochem)/20 mM Hepes, pH 8.0/150 mM NaCl by rocking for 1 h at 25°C. The insoluble OM fraction was pelleted by centrifugation at 40,000 × g for 45 min at 4°C. The soluble OM fraction, containing the majority of HMW1B, was loaded onto a column containing Talon beads (Clontech) and equilibrated with 0.1% n-dodecyl-β-d-maltoside (DDM)/20 mM Hepes, pH 8.0/150 mM NaCl. By using an Akta Purifier (Amersham Biosciences), HMW1B was purified by using a gradient of 0-300 mM imidazole. The fractions containing HMW1B as assessed by immunoblotting were pooled and concentrated by using an Amicon Ultra 10 concentrator (Millipore) according to the manufacturer's instructions. The concentrated fraction was then further purified by size-exclusion chromatography by using a HiPrep 16/60 Sephacryl S-200 HR column (Amersham Biosciences) that had been equilibrated previously with 0.1% DDM/20 mM Hepes, pH 8.0/150 mM NaCl. The HMW1B-containing fractions were concentrated by using an Amicon Ultra 10 concentrator (Millipore). The N-terminal β-barrel domain of OmpA was purified in a similar manner, starting with a culture of BL21(DE3) harboring pHAT::P-49/OmpA21-191.
To purify HMW11-441 and its derivatives, strains were grown in M9 media to an OD600 of 0.6. Isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 0.2 mM, and the culture was grown for an additional 90 min at 37°C. Bacteria were harvested; resuspended in 20 mM Hepes, pH 8.0/100 mM NaCl; and disrupted by sonication. The induced protein formed inclusion bodies that were solubilized in 20 mM Hepes, pH 8.0/100 mM NaCl/8 M urea. The insoluble fraction was pelleted by centrifugation at 19,000 × g for 20 min at 4°C. The soluble fraction was incubated with Talon beads (Clontech) for 30 min at 25°C. HMW11-441, HMW11-441/IAIGI, HMW11-441/ITIG, and HMW11-441/IAIGI/ITIG were eluted from the beads by using 0.2 M imidazole/20 mM Hepes, pH 8.0/100 mM NaCl/8 M urea.
CD. CD spectra were obtained on a Jasco 600 CD spectrophotometer (Jasco, Easton, MD) by using a quartz cell with a 0.1-cm path length. HMW1B was purified as described above and used at a final concentration of 0.25 mg/ml. Five scans were performed at room temperature, and the contribution of buffer alone was subtracted. Estimation of secondary-structure content was performed by using the cdsstr algorithm (available on the Dichroweb web site, www.cryst.bbk.ac.uk/cdweb) (23, 24). Mean residue ellipticities were calculated by considering a mean residue mass of 111.55 Da.
Blue Native PAGE. Blue Native PAGE was performed as described (25). Coomassie blue G was added to all samples before electrophoresis, achieving a final concentration of 0.05%. Samples were electrophoresed on a 9% polyacrylamide gel with 0.01% Coomassie blue G in the cathode buffer. Native gel protein standards were obtained from Amersham Biosciences.
Electron Microscopy. A 5-μl droplet of purified HMW1B was pipetted onto a glow-discharged 300-mesh copper grid covered with a thin layer of continuous carbon film. After a 1-min incubation, excess solution on the grid was blotted with a piece of filter paper. The grid was then washed with two drops of deionized water, stained in 2% uranyl acetate aqueous solution for 30 sec, blotted, and left to air dry. Electron microscopy was performed by using a JEOL 1200EX microscope, and images were recorded on a Gatan 791 charge-coupled device camera (Warrendale, PA). The single-particle image-analysis software package eman was used for particle selection, classification, and class averaging on an SGI Fuel workstation (26).
Liposome-Swelling Assays. Swelling assays were performed as described by Nikaido et al. (27). Briefly, multilammelar liposomes were prepared from 2.4 μmol of egg phosphatidylcholine (Avanti Polar Lipids) and 0.2 μmol of dicetyl phosphate (Sigma). After addition of HMW1B, BSA, or OmpA21-191 and generation of proteoliposomes, samples were resuspended in 10 mM Tris·HCl (pH 8.0) containing 15% (wt/vol) Dextran T-40 (Amersham Biosciences). Control proteoliposomes were prepared by using 0.5 μg of total OM proteins recovered from E. coli BL21(DE3). Swelling of the proteoliposomes was recorded on a DU-640 spectrophotometer (Beckman Coulter).
Far Western Blot Analysis. Proteins were separated by SDS/PAGE using a 10% polyacrylamide gel and were transferred to a nitrocellulose membrane, as described by Sambrook and Russell (20). After incubation of the membrane in blocking reagent (Roche) for 1 h at room temperature, the membrane was incubated with either 10 μg/ml HMW1B or 10 μg/ml OmpA21-191 for 1 h at room temperature. After extensive washing, Western blot analysis using antiserum against the HAT epitope was performed.
Results
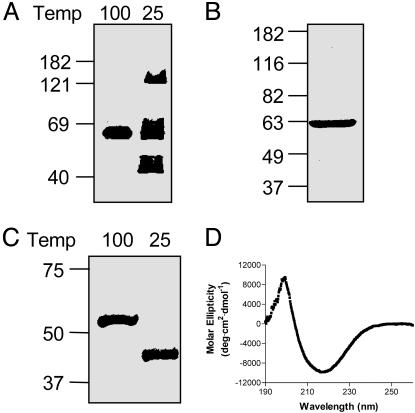
HMW1B Likely Forms a β-Barrel. Given that all bacterial OM proteins for which crystal structures exist have been found to form β-barrels (28), we hypothesized that HMW1B forms a β-barrel. To test this hypothesis, we isolated OM fractions from DH5α expressing HMW1B, incubated these fractions in sample buffer for 5 min at either 25°C or 100°C, and performed SDS/PAGE and Western blot analysis (Fig. 1A). When heated at 100°C, HMW1B migrated at ≈60 kDa, consistent with its predicted molecular mass. However, when incubated at 25°C, a significant amount of HMW1B migrated at ≈45 kDa, thus exhibiting heat-modifiable mobility, a hallmark of β-barrel structures (29). Note that at 25°C some of the HMW1B migrated as a high-molecular species, with a band at ≈125 kDa, suggesting that HMW1B may form a multimer in the OM.
Fig. 1.
HMW1B likely forms a β-barrel. (A) Immunoblot analysis of OM proteins isolated from E. coli DH5α expressing HMW1B. Samples were incubated at either 100°C or 25°C for 5 min before loading on the gel, and the membrane was probed with a polyclonal antiserum against HMW1B. (B) Coomassie blue-stained gel of purified HMW1B. Purified HMW1B was boiled for 5 min and then resolved on an SDS/PAGE gel and stained with Coomassie blue. (C) Heat-modifiability of purified HMW1B. Samples were incubated at either 100°C or 25°C for 5 min and were then resolved on an SDS/PAGE gel and stained with Coomassie blue. (D) Circular dichroism spectrum of HMW1B. Purified HMW1B in 0.1% DDM/20 mM Hepes/150 mM NaCl was measured at room temperature, and the average of five buffer-corrected scans is shown.
To aid in purification and further characterization of the structure and function of HMW1B, we generated an HMW1B derivative with a HAT epitope at the immediate N terminus. The resulting protein was capable of fully complementing a HMW1B-deficient strain (data not shown), indicating function in vivo. This protein was over-expressed in BL21(DE3)omp8, a strain that is deficient in the major porins OmpF, OmpC, OmpA, and LamB. OM proteins were extracted by using Elugent, and HMW1B was purified by metal-affinity chromatography to apparent homogeneity (Fig. 1B). Similar to HMW1B in OM fractions, purified HMW1B exhibited heat modifiability, suggesting preservation of conformation during the purification process (Fig. 1C). CD analysis of purified HMW1B revealed a high degree of β-strand structure, with 34% β-strand, 18% α-helix, 22% turns, and 27% random coil (Fig. 1D). These data agree well with secondary-structure predictions, which predict 22 transmembrane strands representing 38% of the HMW1B mature sequence (17). Together, these data suggest that HMW1B likely forms a β-barrel in the OM.
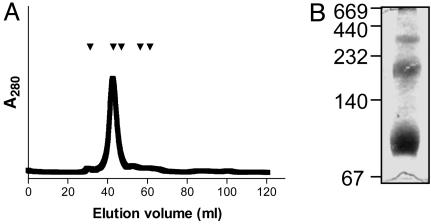
HMW1B Forms a Multimer. To investigate more rigorously the prospect that HMW1B forms a multimer (Fig. 1 A), we examined purified HMW1B by size-exclusion chromatography. As shown in Fig. 2A, HMW1B eluted with a retention time consistent with a molecular mass of ≈237 kDa, suggesting a tetrameric structure. However, to prevent precipitation of HMW1B, the size-exclusion chromatography was performed with 0.1% DDM in the buffer, raising the possibility that the apparent molecular mass might have been influenced by detergent bound to HMW1B. To avoid this concern, we examined HMW1B by Blue Native PAGE, which allows membrane proteins to be electrophoresed in the absence of detergent by using Coomassie blue G dye to maintain protein solubility (25). Although Coomassie blue G binds to proteins, the contribution of Coomassie blue G to electrophoretic mobility can be eliminated by dividing the apparent molecular mass by 1.8 (30). As shown in Fig. 2B, Blue Native PAGE of purified HMW1B demonstrated three distinct species with apparent masses of ≈400, ≈220, and ≈100 kDa. Accounting for the contribution of Coomassie blue G dye to the molecular mass, these molecular masses correspond approximately to a tetramer, a dimer, and a monomer (220, 122, and 55 kDa). Additionally, analysis of purified HMW1B by dynamic light scattering provided results consistent with a tetramer (data not shown).
Fig. 2.
Analysis of the oligomeric state of HMW1B. (A) Size-exclusion chromatography of HMW1B. Arrowheads indicate elution volumes of standards. From left to right, standards included thyroglobulin (Mr, 669 kDa), catalase (Mr, 232 kDa), aldolase (Mr, 158 kDa), albumin (Mr, 67 kDa), and ovalbumin (Mr, 43 kDa). Monomeric HMW1B has a molecular mass of 61.4 kDa. (B) Blue native PAGE gel of purified HMW1B.
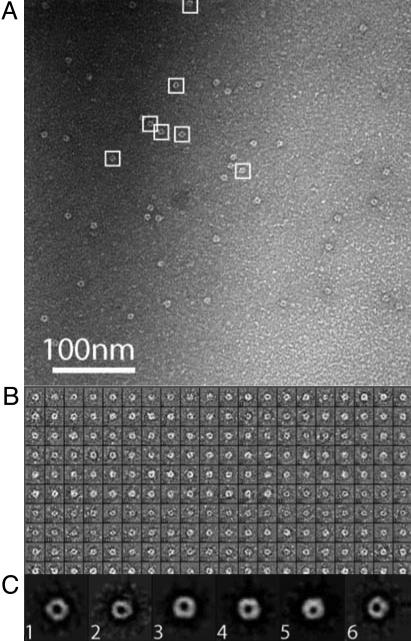
To gain additional information about the structure of HMW1B, we performed electron-microscopic analysis of negatively stained HMW1B. As shown in Fig. 3A, HMW1B exhibited well defined ring-like morphology. Further image analysis was performed to determine the symmetry of the particles. A total of 200 particles with good contrast were randomly selected from raw electron images (Fig. 3B). Each individual particle image was properly scaled and centered, and the multivariate statistic analysis method was used to separate them into six classes (26). Particles belonging to the same class were aligned translationally and rotationally and were then averaged, and results are shown in Fig. 3C. These class averages have a much higher signal-to-noise ratio than the raw images shown in Fig. 3B. The tetragonal architecture of the particles is the most clear in classes 1, 3, and 4, which are top-on views. Classes 2, 5, and 6 are projections of tetragonal particles with significant out-of-plane tilt. Based on measurements of classes 1, 3, and 4, the particles were estimated to have an outer diameter of ≈8 nm and an apparent central channel of ≈2.5 nm.
Fig. 3.
Single-particle analysis of negatively stained HMW1B. (A) A raw electron micrograph of HMW1B. A few particles of tetragonal shape are indicated by white boxes. (B) Gallery of 200 raw particles. (C) Class averages of the HMW1B particles.
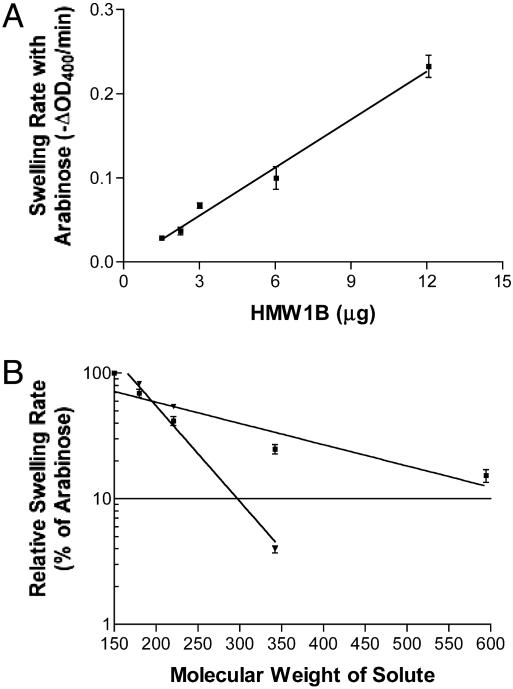
HMW1B Has Pore Activity. We have suggested (17) that HMW1 is translocated to the bacterial surface by HMW1B. To test whether HMW1B forms a conduit in the OM to facilitate translocation, we examined HMW1B by using the liposome-swelling assay, which has been used previously to study the pore-forming activities of bacterial porins and several protein-specific translocators (16, 27, 31-34). As shown in Fig. 4A, porin activity was directly proportional to the amount of HMW1B added to the liposomes. The N-terminal β-barrel domain of OmpA (purified from the OM by using the same method used to purify HMW1B) and BSA served as negative controls and demonstrated no pore-forming activity (data not shown). The specific activity of the HMW1B proteoliposome for arabinose was ≈0.019 ΔA400 per min/μg, which is considerably lower than that of OmpF (35). This low specific activity raises the possibility that HMW1B in the proteoliposome is intermittently in a closed or otherwise nonfunctional conformation (16, 34).
Fig. 4.
Liposome-swelling assay. (A) Swelling rates of HMW1B proteoliposomes with l-arabinose as the solute. A representative experiment is shown. Each data point represents the mean ± SE of at least three readings. (B) Swelling rates of proteoliposomes reconstituted with 0.6-12 μg of HMW1B (▪) or 0.5 μg of E. coli BL21(DE3) OM proteins (▾) in the presence of solutes of various molecular weights. Swelling rates are plotted as the percentage of the swelling rate obtained with arabinose. Solutes included l-arabinose (Mr, 150), d-galactose (Mr, 180), N-acetyl-d-glucosamine (Mr, 221), sucrose (Mr, 342), and d-raffinose (Mr, 504). The plot represents the mean ± SE of three experiments.
To estimate the pore size of HMW1B, rates of swelling were obtained for sugars of various molecular weights and expressed relative to swelling rates obtained with arabinose (Fig. 4B). As a positive control, liposomes reconstituted with OM fractions prepared from E. coli BL21(DE3) were examined in a similar manner. The molecular weight of a solute that results in 10% of the swelling activity of arabinose [Mr(0.1 Ara)] is a parameter that has been used extensively to calculate the diameter of channels in OM proteins (27). The Mr(0.1 Ara) for the proteoliposomes reconstituted with the OM fraction from E. coli BL21(DE3) was calculated to be 297, which is close to the published value for OmpF (280) (27). The best-fit regression line for HMW1B proteoliposomes indicates that the Mr(0.1 Ara) is equal to 680. Comparing this value with the Mr(0.1 Ara) for OmpF and considering the fact that OmpF has a pore with a diameter of 1.1 nm (36), the pore size of HMW1B is on the order of 2.7 nm.
HMW1B Interacts with the N-Terminal Fragment of HMW1. Having demonstrated that HMW1B forms a multimeric pore, we questioned how HMW1B recognizes HMW1 to facilitate secretion of this substrate across the OM. In earlier work (37), site-directed mutagenesis established that NPNGI and NTNG amino acid motifs contained within the N-terminal fragment of HMW1 are critical for secretion. Similar work performed with ShlA and filamentous hemagglutinin (FHA), along with C-terminal truncations of ShlA, HpmA, and FHA, have led to the designation of a “secretion domain” required for translocation of TPS pathway exoproteins (designated TpsA) (12, 38, 39). Although these experiments indicated the necessity of the secretion domain, they failed to establish whether this domain is sufficient for secretion or interacts directly with the translocator protein.
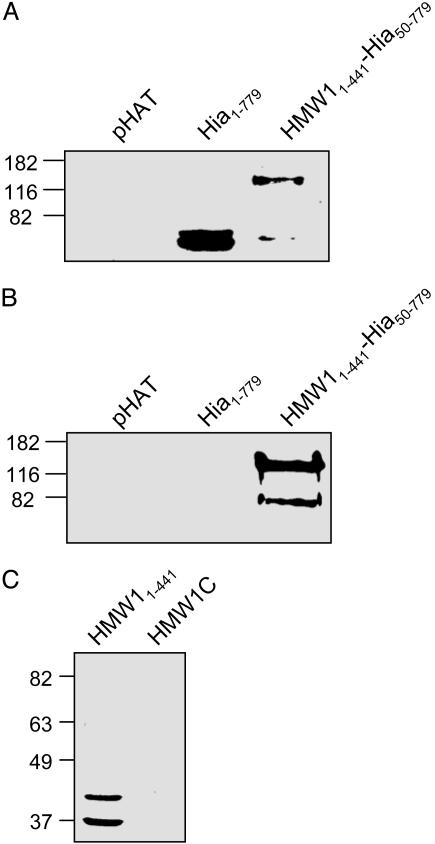
To investigate the question of sufficiency, we fused the N-terminal fragment of HMW1 to a portion of the H. influenzae Hia adhesin, generating an HMW11-441-Hia50-779 chimeric protein. Hia is an autotransporter that mediates adherence to cultured epithelial cells via two distinct binding domains contained within residues 50-252 and 580-714, and it has a translocator domain contained within residues 1,023-1,098 (40-43). Whole-cell sonicates and trichloroacetic acid-precipitated supernatant proteins generated from DH5α expressing either Hia1-779 or the HMW1-Hia chimeric protein in the presence of HMW1B were examined by immunoblot analysis. We hypothesized that if the N-terminal fragment of HMW1 is necessary and sufficient for translocation through HMW1B, only the chimeric protein should be present in the supernatant. Given that the chimera has no membrane anchor, it should be freely secreted into the supernatant. As shown in Fig. 5A, both Hia1-779 and the HMW1-Hia chimeric protein were detectable in whole-cell sonicates. However, only the chimera was present in the supernatant, indicating that HMW11-441 is sufficient for secretion through HMW1B (Fig. 5B). It is of note that the chimera was not present in the supernatant when coexpressed with the translocator domain of Hia (data not shown), suggesting specificity between HMW11-441 and its cognate translocator HMW1B.
Fig. 5.
Role of HMW11-441 in HMW1B-mediated translocation. (A and B) Immunoblot analysis using polyclonal antiserum against the HAT epitope of whole-cell sonicates (A) or trichloroacetic acid-precipitated culture supernatants (B) generated from E. coli BL21(DE3) expressing the indicated construct in the presence of HMW1B. (C) Far-Western blot analysis of purified HMW11-441 and HMW1C, which were separated by SDS/PAGE and overlaid with purified HMW1B. HMW1C has a molecular mass of 74 kDa.
To extend these results, we questioned the mechanism by which HMW1B recognizes the secretion domain, hypothesizing that HMW1B must interact directly with HMW11-441. Accordingly, we examined purified HMW11-441 in a far-Western blot analysis, overlaying with purified HAT-tagged HMW1B and probing with an antibody against the HAT epitope. As shown in Fig. 5C, HMW11-441 interacts with HMW1B. In control far-Western blots, we found no evidence of interaction between HMW1C and HMW1B (Fig. 5C) or between HMW11-441 and purified HAT-tagged OmpA21-191 (data not shown), indicating that the interaction between the HMW1 N-terminal fragment and HMW1B is specific. To investigate whether the NPNGI and NTNG motifs contained within HMW11-441 have any role in mediating the interaction between this fragment of HMW1 and HMW1B, we performed similar far-Western blot analyses by using purified HMW11-441 where the conserved NPNGI and NTNG motifs had been mutated to IAIGI and ITIG, respectively. These mutant N-terminal fragments were still able to bind HMW1B, indicating that these motifs are not involved in interacting with HMW1B (data not shown).
Discussion
In this study, we purified HMW1B to gain insight into its architecture and function. By using biochemical and electron microscopy techniques, we established that HMW1B exists as a tetramer with an external diameter of ≈8 nm and has pore activity with a pore size of ≈2.7 nm. In addition, we demonstrated that purified HMW1B interacts with the N-terminal fragment of HMW1, ultimately leading to secretion of native HMW1.
Based on phylogenetic analysis and secondary-structure predictions, HMW1B is believed to be a member of the Omp85 family (1). Other investigators have suggested that eukaryotic Omp85 family members are related evolutionarily to prokaryotic Omp85 proteins, raising the possibility that these two groups of proteins have similar structural and functional characteristics (1, 3, 5). However, previous studies performed with bacterial Omp85-like proteins have not supported this notion. Although several bacterial family members have been demonstrated to have pore activity, none had been found to be multimeric (7, 15, 16). Immunoblot analysis revealed that N. meningitidis Omp85 exists in a high-molecular weight complex (7); however, earlier work by Manning et al. demonstrated that this protein interacts with several heterologous proteins in the OM (44), thus precluding any conclusion regarding the structural organization of N. meningitidis Omp85. The current study provides a clear demonstration that a bacterial Omp85-like protein forms a multimer, suggesting that the architecture of Omp85-like proteins is conserved among bacteria and eukaryotes.
Consistent with a role for HMW1B in protein secretion, liposome-swelling assays revealed pore activity, with a pore size of ≈2.7 nm. This pore size is in agreement with the reported dimensions of other bacterial and eukaryotic Omp85-like proteins, and it is large enough to allow passage of an unfolded or partially unfolded substrate protein (6, 13, 15, 16, 34). Both the relatively low specific activity of HMW1B for arabinose and the biphasic nature of the relative swelling data may be due to differences between the nature of HMW1B, which translocates a specific protein substrate, and bacterial porins, which are generally nonspecific diffusion channels (16, 33, 34).
Analysis of high-resolution images of HMW1B supported the biochemical analyses and liposome-swelling results, demonstrating a tetrameric complex with an apparent ≈2.5-nm central channel. At this point, it is unclear whether the apparent central channel is the protein-secreting pore. The faster-migrating ≈40 kDa species observed in heat-modifiability assays suggests that each monomer of HMW1B adopts a folded β-barrel structure, implying that the apparent central channel is ringed by the hydrophobic exterior of multiple β-barrels and arguing against a protein-secreting central channel (45). PapC, an OM protein required for assembly of P pili in uropathogenic E. coli, has a faster-migrating monomer form in heat-modifiability assays and has recently been observed to be a dimer, with each monomer forming a channel (H.L., L. Qian, Z. Chen, D. Thahbot, G. Liu, T. Liu, and D.G.T., unpublished data) (34). In this regard, it is noteworthy that the translocon of the outer envelope of chloroplasts (the Toc complex, comprising Toc159, Toc34, and the Omp85-like protein Toc75) forms four independent pores (13). With this information in mind, it is intriguing to speculate that each monomer of HMW1B forms a secretion-competent pore, leading to four pores per HMW1B complex.
Mutational analysis and deletion studies performed with several different TpsA proteins (TPS pathway exoproteins) has suggested that these proteins contain an N-terminal domain that targets them to their cognate translocator (generally designated as TpsB) (12, 37, 38). Several investigators have speculated that this secretion domain interacts with TpsB proteins sometime before translocation across the OM (12, 37, 39). However, neither of these points has been demonstrated experimentally. By generating an HMW1-Hia chimeric protein, we established that the N-terminal fragment of HMW1 is sufficient for translocation across the OM in an HMW1B-specific manner. Moreover, far-Western blot analysis using purified proteins demonstrated that HMW1B interacts with HMW11-441. The finding of a direct interaction between HMW1B and its substrate protein is similar to observations with eukaryotic Omp85-like proteins (6, 7, 9, 14). The recently solved crystal structure of the secretion domain from B. pertussis FHA (which shares significant homology with the HMW1 secretion domain) demonstrated that the conserved asparagine-containing motifs form type I β-turns, leading the authors to speculate that these residues play a critical role in overall protein stability. Our results with the asparagine mutants in HMW11-441 are consistent with this speculation, demonstrating no defect in interaction with HMW1B. The crystal structure of the FHA secretion domain should provide a platform to begin to identify the residues involved in HMW1 targeting to and interaction with HMW1B.
The TPS pathway is characterized by an OM TpsB translocator protein and a secreted TpsA protein, and it has been classified as a subfamily of the autotransporter family found in Gram-negative bacteria (46). However, in contrast to TPS family members, autotransporter proteins consist of a single polypeptide with a C-terminal β-barrel domain that allows for translocation of the internal “passenger” domain to the surface. Examination of the translocator domains of monomeric, “classic” autotransporters and the recently described trimeric class of autotransporters reveals no significant structural resemblance to TPS translocator proteins (40, 47, 48). Given the lack of sequence, phylogenetic, and structural relatedness between TPS proteins and autotransporters, we suggest that TPS proteins should be classified as a subfamily of the Omp85 family rather than as variant autotransporters.
We have examined the H. influenzae HMW1B protein, a member of the TPS family, and we have observed striking structural and functional similarities to Omp85-like proteins. Our findings demonstrate that the Omp85 family has maintained a similar structural architecture and conserved physical properties throughout evolution.
Supplementary Material
Acknowledgments
We thank Jim Bann for assistance with interpretation of CD spectra. This work was supported by U.S. Department of Health and Human Services Public Health Service Grants R01AI44167 and R01DC02873 (to J.W.S.), R01GM62987 (to D.G.T.), and F32AI10555 and T32AI07172 (to G.G.H.). H.L. was supported by Laboratory Directed Research and Development Project Number 03-096 at Brookhaven National Laboratory. N.S. was supported by U.S. Department of Health and Public Services Public Health Service Training Grant T32AI07172 and is a member of the Medical Scientist Training Program at the Washington University School of Medicine.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DDM, n-dodecyl-β-d-maltoside; OM, outer membrane; HAT, histidine-affinity tag; TPS, two-partner secretion; FHA, filamentous hemagglutinin.
References
- 1.Yen, M. R., Peabody, C. R., Partovi, S. M., Zhai, Y., Tseng, Y. H. & Saier, M. H. (2002) Biochim. Biophys. Acta 1562, 6-31. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Pulido, L., Devos, D., Genevrois, S., Vicente, M. & Valencia, A. (2003) Trends Biochem. Sci. 28, 523-526. [DOI] [PubMed] [Google Scholar]
- 3.Reumann, S., Davila-Aponte, J. & Keegstra, K. (1999) Proc. Natl. Acad. Sci. USA 96, 784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentle, I., Gabriel, K., Beech, P., Waller, R. & Lithgow, T. (2004) J. Cell Biol. 164, 19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolter, B., Soll, J., Schulz, A., Hinnah, S. & Wagner, R. (1998) Proc. Natl. Acad. Sci. USA 95, 15831-15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paschen, S. A., Waizenegger, T., Stan, T., Preuss, M., Cyrklaff, M., Hell, K., Rapaport, D. & Neupert, W. (2003) Nature 426, 862-866. [DOI] [PubMed] [Google Scholar]
- 7.Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. (2003) Science 299, 262-265. [DOI] [PubMed] [Google Scholar]
- 8.Genevrois, S., Steeghs, L., Roholl, P., Letesson, J. J. & van der Ley, P. (2003) EMBO J. 22, 1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozjak, V., Wiedemann, N., Milenkovic, D., Lohaus, C., Meyer, H. E., Guiard, B., Meisinger, C. & Pfanner, N. (2003) J. Biol. Chem. 278, 48520-48523. [DOI] [PubMed] [Google Scholar]
- 10.Schnell, D. J., Kessler, F. & Blobel, G. (1994) Science 266, 1007-1012. [DOI] [PubMed] [Google Scholar]
- 11.Hinnah, S. C., Hill, K., Wagner, R., Schlicher, T. & Soll, J. (1997) EMBO J. 16, 7351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob-Dubuisson, F., Locht, C. & Antoine, R. (2001) Mol. Microbiol. 40, 306-313. [DOI] [PubMed] [Google Scholar]
- 13.Schleiff, E., Soll, J., Kuchler, M., Kuhlbrandt, W. & Harrer, R. (2003) J. Cell Biol. 160, 541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Y., Kouranov, A., LaSala, S. E. & Schnell, D. J. (1996) J. Cell Biol. 134, 315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konninger, U. W., Hobbie, S. & Braun, V. (1999) Mol. Microbiol. 32, 1212-1225. [DOI] [PubMed] [Google Scholar]
- 16.Jacob-Dubuisson, F., El-Hamel, C., Saint, N., Guedin, S., Willery, E., Molle, G. & Locht, C. (1999) J. Biol. Chem. 274, 37731-37735. [DOI] [PubMed] [Google Scholar]
- 17.St. Geme, J. W., III, & Grass, S. (1998) Mol. Microbiol. 27, 617-630. [DOI] [PubMed] [Google Scholar]
- 18.Studier, F. W. & Moffatt, B. A. (1986) J. Mol. Biol. 189, 113-130. [DOI] [PubMed] [Google Scholar]
- 19.Prilipov, A., Phale, P. S., Van Gelder, P., Rosenbusch, J. P. & Koebnik, R. (1998) FEMS Microbiol. Lett. 163, 65-72. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J. & Russel, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 21.Dower, W. J., Miller, J. F. & Ragsdale, C. W. (1988) Nucleic Acids Res. 16, 6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlone, G. M., Thomas, M. L., Rumschlag, H. S. & Sottnek, F. O. (1986) J. Clin. Microbiol. 24, 330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobley, A., Whitmore, L. & Wallace, B. A. (2002) Bioinformatics 18, 211-212. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama, N. & Woody, R. W. (2000) Anal. Biochem. 287, 252-260. [DOI] [PubMed] [Google Scholar]
- 25.Schagger, H. & von Jagow, G. (1991) Anal. Biochem. 199, 223-231. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke, S. J., Baldwin, P. R. & Chiu, W. (1999) J. Struct. Biol. 128, 82-97. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido, H., Nikaido, K. & Harayama, S. (1991) J. Biol. Chem. 266, 770-779. [PubMed] [Google Scholar]
- 28.Koebnik, R., Locher, K. P. & Van Gelder, P. (2000) Mol. Microbiol. 37, 239-253. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, K. & Mizushima, S. (1976) J. Biochem. (Tokyo) 80, 1411-1422. [DOI] [PubMed] [Google Scholar]
- 30.Heuberger, E. H., Veenhoff, L. M., Duurkens, R. H., Friesen, R. H. & Poolman, B. (2002) J. Mol. Biol. 317, 591-600. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura, F., Zalman, L. S. & Nikaido, H. (1983) J. Biol. Chem. 258, 2308-2314. [PubMed] [Google Scholar]
- 32.Lee, H. W. & Byun, S. M. (2003) Biochem. Biophys. Res. Commun. 307, 820-825. [DOI] [PubMed] [Google Scholar]
- 33.Veiga, E., Sugawara, E., Nikaido, H., de Lorenzo, V. & Fernandez, L. A. (2002) EMBO J. 21, 2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., Saulino, E. T., Lombardo, M. J., Roth, R., Heuser, J. & Hultgren, S. J. (1998) Proc. Natl. Acad. Sci. USA 95, 3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugawara, E. & Nikaido, H. (1992) J. Biol. Chem. 267, 2507-2511. [PubMed] [Google Scholar]
- 36.Cowan, S. W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R. A., Jansonius, J. N. & Rosenbusch, J. P. (1992) Nature 358, 727-733. [DOI] [PubMed] [Google Scholar]
- 37.Grass, S. & St. Geme, J. W., III. (2000) Mol. Microbiol. 36, 55-67. [DOI] [PubMed] [Google Scholar]
- 38.Schonherr, R., Tsolis, R., Focareta, T. & Braun, V. (1993) Mol. Microbiol. 9, 1229-1237. [DOI] [PubMed] [Google Scholar]
- 39.Clantin, B., Hodak, H., Willery, E., Locht, C., Jacob-Dubuisson, F. & Villeret, V. (2004) Proc. Natl. Acad. Sci. USA 101, 6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surana, N. K., Cutter, D., Barenkamp, S. J. & St. Geme, J. W., III. (2004) J. Biol. Chem. 279, 14679-14685. [DOI] [PubMed] [Google Scholar]
- 41.St. Geme, J. W., III, & Cutter, D. (2000) J. Bacteriol. 182, 6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo, H. J., Cotter, S. E., Laarmann, S., Juehne, T., St. Geme, J. W., III, & Waksman, G. (2004) EMBO J. 23, 1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laarmann, S., Cutter, D., Juehne, T., Barenkamp, S. J. & St. Geme, J. W., III (2002) Mol. Microbiol. 46, 731-743. [DOI] [PubMed] [Google Scholar]
- 44.Manning, D. S., Reschke, D. K. & Judd, R. C. (1998) Microb. Pathog. 25, 11-21. [DOI] [PubMed] [Google Scholar]
- 45.Schulz, G. E. (2000) Curr. Opin. Struct. Biol. 10, 443-447. [DOI] [PubMed] [Google Scholar]
- 46.Henderson, I. R., Cappello, R. & Nataro, J. P. (2000) Trends Microbiol. 8, 529-532. [DOI] [PubMed] [Google Scholar]
- 47.Oomen, C. J., Van Ulsen, P., Van Gelder, P., Feijen, M., Tommassen, J. & Gros, P. (2004) EMBO J. 23, 1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roggenkamp, A., Ackermann, N., Jacobi, C. A., Truelzsch, K., Hoffmann, H. & Heesemann, J. (2003) J. Bacteriol. 185, 3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.