Abstract
The plant mitochondrial thioredoxin (Trx) system has been described as containing an NADPH-dependent Trx reductase and Trx o. In addition to the mitochondrial isoform, Trx o, plants are known to contain several chloroplastic Trx isoforms and the cytosolic Trx h isoforms. We report here the presence in plant mitochondria of a Trx isoform (PtTrxh2) belonging to the Trx h group. Western blot analyses with mitochondrial proteins isolated from both poplar and GFP fusion constructs indicate that PtTrxh2 is targeted to plant mitochondria. The recombinant protein, PtTrxh2, has been shown to be reduced efficiently by the mitochondrial Trx reductase AtNTRA. PtTrxh2 is also able to reduce alternative oxidase homodimers and to allow its activation by pyruvate. In contrast, neither PtTrxh2 nor AtTrxo1 exhibits activity with several poplar glutathione peroxidases and especially a putative mitochondrial isoform. Incubation of PtTrxh2 with glutathione disulfide led to the formation of glutathionylated Trx, identified by mass spectrometry. The formation of a glutathione adduct increases the redox potential of PtTrxh2 from -290 to -225 mV. In addition to Trx o, this study shows that Trx h could also be present in mitochondria. This previously unrecognized complexity is not unexpected, considering the multiple redox-regulated processes found in plant mitochondria.
Keywords: poplar, glutathione peroxidase, glutathionylation, redox potential
Thioredoxins (Trxs) are small proteins involved in cellular redox regulation. These ubiquitous proteins, which are present in all organisms from prokaryotes to eukaryotes, display a particularly large diversity in photosynthetic organisms (1, 2). For example, at least 21 genes encoding Trxs have been detected in the fully sequenced genome of Arabidopsis thaliana (3). In plants, several Trxs are found: the chloroplastic Trxs f, m, x, y, and CDSP32, the mitochondrial Trxs o, and the Trxs h, which previously have been thought to be present exclusively in the cytosol. The chloroplastic Trxs, which are nuclear-encoded, are reduced by ferredoxin in a reaction catalyzed by a heterodimeric ferredoxin-Trx reductase (2). Recently, a mitochondrial system involving a NADPH-Trx reductase (NTR), NTRA, and at least one Trx o has been detected in A. thaliana (4).
The members of the third Trx group, called Trxs h, possess at least one specific characteristic that allows their identification: the presence of a conserved Trp residue (W16 in poplar Trx h1) that produces a unique characteristic shoulder at 290 nm in the UV-region absorbance spectrum (5). Based on primary structure analysis, Trxs h could be classified into three subgroups (3, 6). Members of subgroups I and II are reduced by NTR, whereas members of subgroup III are dependent on the glutathione/glutaredoxin system (7). In addition, all members of the second group contain N-terminal extensions (3). The significance of these extensions remains unclear, given that prediction programs, such as targetp (www.cbs.dtu.dk/services/TargetP) or psort (http://psort.ims.u-tokyo.ac.jp), suggest that a majority of these proteins should be cytosolic (3).
A poplar Trx, called PtTrxh2, belonging to subgroup II, was characterized recently (8). The recombinant protein has been produced in Escherichia coli. In this case, the N-terminal extension was cleaved in the E. coli cells, with the first 19 amino acids missing, suggesting the presence of a putative cleavage site in the N-terminal extension of the protein.
We report here that PtTrxh2 is associated with mitochondria. The redox properties of PtTrxh2 were investigated by examining its interaction with two potential mitochondrial Trx targets, alternative oxidase (AOX) and glutathione peroxidase (Gpx). In addition, we report that this Trx displays a potential site for glutathionylation and that formation of the glutathione adduct alters its redox potential.
Materials and Methods
GFP Experiments. For in vivo intracellular localization, the cDNA sequences of the putative transit peptide and the first putative α-helix of PtTrxh2 were cloned between the NcoI and BamHI sites of plasmid pCK-GFP3A, which allows the expression of protein-enhanced-GFP fusions under the control of a 35S promoter. The following oligonucleotides were used to obtain a chimeric protein containing the first 65 amino acids of PtTrxh2 connected to GFP: forward oligonucleotide, 5′-CCCCCATGGCTAATTTTGATCACTCCAATGGA-3′; reverse oligonucleotide, 5′-CCCCGGATCCGCCGTGAATTCAATCAC-3′. The resultant plasmid was transfected in tobacco leaves by bombardment, and images were obtained with a Zeiss LSM510 confocal microscope. The pCK-mRFP plasmid, which harbors the DsRED1 gene (Clontech), which codes for the red fluorescent protein fused to the mitochondrial yeast COXIV presequence, was used as a mitochondrial marker. It was cotransfected with the tested construction.
Mitochondria Purification. Mitochondria, isolated from 13-day-old soybean (Glycine max) cotyledons, were purified as described in ref. 9. For poplar (Populus tremula × Populus alba, clone 717-1-B4, Institut National de la Recherche Agronomique Orléans, Olivet Cedex, France), growing leaves from 6-month-old cuttings were used. The leaves (60 g) were washed with cold distilled water, cut, and placed in 300 ml of extraction medium [30 mM Mops buffer, pH 7.4/0.4 M mannitol/1% (wt/vol) BSA/3 mM cysteine/4 mM DTT/1.25 mM EGTA/2.5 mM MgCl2/0.4% (wt/vol) polyvinylpyrrolidone/0.4% (wt/vol) phenol/chloroform/polyvinylpolypyrrolidone]. After two rounds of homogenization at a low speed for 2 s with a Waring blender, the homogenate was rapidly filtered through two layers of Miracloth (100 μm). The leaf fragments retained by the net were homogenized again (three times for 2 s) with 200 ml of the extraction medium and filtered as described above. The two filtrates were pooled and subjected to differential centrifugations as described in ref. 9. Mitochondria were resuspended in washing medium [0.4 mM mannitol/10 mM Mops buffer, pH 7.2/0.5% BSA (wt/vol)] and purified as described in ref. 10. Mitochondria were washed twice in 30 ml of washing medium devoid of BSA. The final pellet, suspended to ≈1.5 μg of protein per μl of washing medium, was stored at -80°C.
Western Blot Analysis. Polyclonal antibodies (IgGs) against poplar PtTrxh2 and poplar chloroplastic methionine sulfoxide reductase A were raised in rabbits and purified from the serum by affinity chromatography involving covalently linking proteins to Sepharose columns by using a technique similar to the one described for type II peroxiredoxin in ref. 11. Antibodies against glycine cleavage system subunits P and T were gifts from J. Bourguignon (Centre Nationale de la Recherche Scientifique, Grenoble, France) (12).
Protein extraction from leaves, stems, and roots was carried out as described in ref. 11. Western blots were performed by using polyvinylidene difluoride membranes (Millipore) and the Immun-Star Goat Anti-Rabbit Detection kit (Bio-Rad).
Cloning of Processed PtTrxh2. Because the full-length recombinant protein is processed in E. coli (8), the ORF of poplar Trxh2, in which the nucleotide sequence encoding the first 19 amino acids was deleted, was cloned by PCR, by using the previously described plasmid pET-popTrxh-2L (8) as a template, into the expression plasmid pET-3d between the two restriction sites NcoI and BamHI with the following oligonucleotides: forward, 5′-CCCCCATGGCTAATTTTGATCACTCCAATGGA-3′; reverse, 5′-CCCCCGGATCCTTATATCATCCCATGCTTCTCAAT-3′. In this construction, the recombinant protein starts with the sequence MANFDH.
Expression and Purification of the Recombinant Proteins. For Pt-Trxh2 production, E. coli strain BL21(DE3) was cotransformed with the helper plasmid pSBET and the recombinant plasmid (13). Production and purification of the recombinant protein were performed as described in ref. 8.
The procedures for the expression and purification of AtNTRA, AtNTRB, AtTrxo1, PtTrxh1, PtTrxh3, and PtPrxQ are described in refs. 4 and 14-17.
Trx Activity. The reduction of Trx by NADPH and the recombinant A. thaliana NTRA or -B was followed at 412 nm by using 5,5′-dithiobis(2-nitrobenzoic acid) as a substrate (4).
AOX Reduction and Activity. The reduction of soybean AOX by PtTrxh2 was performed as follows. PtTrxh2 was reduced by incubation with 10 mM DTT for 30 min and then dialyzed to remove DTT. The absence of detectable DTT in the protein preparation was checked by using 5,5′-dithiobis(2-nitrobenzoic acid). Purified frozen/thawed soybean mitochondria samples containing 2.5, 5, or 10 μg of total protein were incubated for 30 min with 150 μM PtTrxh2 or 40 mM DTT before being subjected to nonreductive SDS/PAGE. AOX immunodetection also was performed by using monoclonal antibodies [a generous gift from T. E. Elthon (University of Nebraska, Lincoln) to Y.J.] raised against the Sauromatum guttatum AOX (18).
To test the AOX activity, mitochondrial oxygen consumption was measured with a Clark-type O2-electrode (Rank Brothers, Cambridge, U.K.). Purified soybean mitochondria were oxidized with 10 mM diamide, washed, and frozen, allowing membranes to break down. The mitochondria then were suspended in 1 ml of reaction medium containing 0.3 M sucrose, 5 mM KH2PO4, 10 mM N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (pH 7.0), 10 mM NaCl, 2 mM MgSO4, and 0.1% (wt/vol) BSA at 25°C. PtTrxh2 (500 μM) was reduced in 30 mM Tris·HCl (pH 8)/1 mM EDTA buffer by using the NADPH/AtNTRB system described in ref. 8. Subsequent additions were made as described in Results.
Gpx Activity. The Gpx activity was measured by following H2O2 reduction. The reaction medium had the following composition: 50 mM Tris·HCl, pH 8.0/500 μM DTT/500 μM H2O2 with varying amounts of PtTrxs and PtGpxs. After 1 min of incubation at ambient temperature, the reaction was started by adding H2O2. For each time point, 50 μl of the reaction mixture was added to 950 μl of FOX2 reagent as described in ref. 17. The absorbance at 560 nm was read after a 1-h incubation.
Glutathionylation Experiments. The reaction mixture (50 μl) containing 30 mM Tris·HCl (pH 8), 1 mM DTT, and 50 μg of poplar Trx h1, h2, or h3 (concentration of ≈80 μM) was incubated 10 min before adding 5 mM oxidized glutathione (GSSG). The excess of GSSG was removed by extensive dialysis (1/25,000), and the resultant glutathionylated protein was concentrated by ultrafiltration.
Redox Potential Determination. Titration of Trx h2 redox potential was determined by using the monobromobimane method described in ref. 19. PtTrxh2 redox potential also was determined by using atNTRB-catalyzed poising by direct reduction by NADP+/NADPH (20). PtTrxh2 and GSSG-treated PtTrxh2 (10-40 μM) were mixed with 108 μM NADPH in 500 μl. Then, 85 nM AtNTRB was added, and the NADPH oxidized was determined at 340 nm against an identical blank without NADPH. To determine the redox potential, the experiment was continued by adding 1 mM NADP+ to reverse the equilibrium of the reaction. The concentration of the NADP+ stock solution was checked by reading the A at 260 nm, assuming that ε = 15.3 mM-1·cm-1. From the equilibrium concentrations, redox potentials were calculated by using the Nernst equation (20).
Electrospray MS. A Micromass Q-Tof Ultima (Waters) hybrid tandem mass spectrometer was used for the acquisition of the electrospray ionization mass spectra. This instrument is equipped with a nanoflow-electrospray source. The samples were infused into the mass spectrometer by using nanoflow capillaries (Proxeon Biosystems, Odense, Denmark). The needle voltage was ≈1,800 V, and the collision energy was 10 eV for the MS analyses. Samples for flow injection analyses were diluted 1:20 with a solution of 50:50 acetonitrile and 0.1% formic acid. Data analysis was accomplished with a MassLynx data system and transform deconvolution software supplied by the manufacturer (Waters).
Results
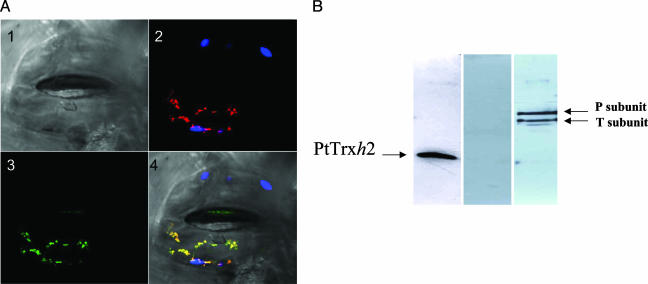
Mitochondrial Localization of PtTrxh2. Two sets of experiments aimed at investigating the subcellular localization of PtTrxh2 were undertaken. First, a transient expression experiment was carried out by using a chimeric protein obtained by fusing the putative transit peptide of PtTrxh2 to GFP. After bombardment of tobacco leaf epidermis, the protein-GFP fusion was found only in mitochondria (Fig. 1A).
Fig. 1.
Mitochondrial localization of PtTrxh2. (A) Mitochondrial localization of the PtTrxh2 fused to enhanced GFP in N. tabacum stomata. (Image 1) Image of stomata from Nicotiana benthamiana under visible light. Only the lower guard cell was transfected. (Image 2) Autofluorescence of chlorophyll (blue) and fluorescence of the mitochondrial marker (red). (Image 3) Fluorescence of the fusion protein (green). (Image 4) Merged images. (B) Immunodetection of PtTrxh2 (Left), methionine sulfoxide reductase A (Center), and glycine decarboxylase complex subunits P and T (Right) in poplar purified mitochondria.
The mitochondrial localization was further demonstrated by using specific antibodies together with mitochondria purified from poplar leaves (Fig. 1B). The quality of the mitochondrial preparation was checked by using antibodies recognizing both chloroplastic and cytosolic methionine sulfoxide reductases (N.R., unpublished data). In this case, no signal could be detected by using the mitochondrial preparation (Fig. 1B Center), whereas a strong signal could be detected in whole poplar leaf extracts (data not shown). The PtTrxh2 signal intensity (Fig. 1B Left) was similar to that obtained by using antibodies raised against glycine decarboxylase complex subunits (Fig. 1B Right) used as mitochondrial markers.
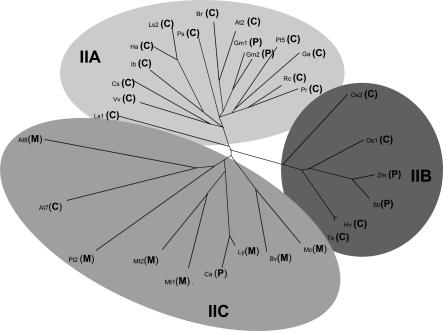
PtTrxh2 Belongs to a Specific Subgroup of Trxs h. Trx h have been classified into three different subgroups based on primary structure analysis (3). Subgroup II includes Trxs exhibiting an N-terminal extension (6). A phylogenic tree has been constructed by using various Trxs h belonging to subgroup II (Fig. 2). These Trxs could be further divided into three subclasses: one group containing proteins related to AtTrxh2 called IIA; one group containing proteins from cereals, which cluster separately (IIB); and a third group (IIC) that contains proteins related to AtTrxh8 and also includes PtTrxh2. Prediction of subcellular localization of each protein was performed by using a combination of two programs, targetp and ipsort. With the exception of AtTrxh7, PtTrxh2-related members all are predicted to possess a peptide signal. In contrast, AtTrxh2-related sequences are mainly predicted to be cytosolic. However, it should be pointed out that soybean Trxs, which have been shown to be anchored to plasma membranes (21), are predicted to be chloroplastic proteins by these programs. These analyses suggest that mitochondrial Trxs h could constitute a quite homogeneous subclass in Trx h subgroup II.
Fig. 2.
Phylogenetic tree of various Trxs h belonging to the subgroup II. The clustalw software was used to generate the phylogenetic tree of the following Trxs (GenBank accession numbers in parentheses): At2, A. thaliana Trxh2 (S58123); At7, A. thaliana Trxh7 (AAD39316); At8, A. thaliana Trxh8 (AAG52561); Bv, Beta vulgaris (BQ489103); Br, Brassica rapa (AF352030); Cs, Citrus sinensis (CF837405); Ca, Capsicum annuum (CA523711); Ga, Gossypium arboretum (BG440056); Gm1, G. max (AW620807); Gm2, G. max (BM188930); Ha, Helianthus annus (TC13569); Hv, Hordeum vulgare (BI960260); Ib, Ipomoea batatas (AY344228); Mc, Mesembryanthemum crystallinum (BE033809); Mt1, Medicago truncatula (AW560796); Mt2, M. truncatula (AW686237); Ls1, Lactucca sativa (BU008396); Ls2, L. sativa (TC9551); Ly, Lycopersicon esculentum (BG126499); Pr, Prunus armeniaca (CB818939); Os1; Oryza sativa (AK062383); Os2, O. sativa (CB681257); Ps, Pisum sativum (AY170651); Pt2, Populus trichocarpa Trxh2 (AF483266); Pt5, P. trichocarpa Trxh5 (BU869308); Rc, Rosa chinensis (BI978567); Sb, Sorghum bicolor (AW747151); Ts, Triticum aestivum (CD914485); Vv, Vitis vinifera (CF513794); Zm, Zea mays (AY104013). The subcellular prediction (C, cytosol; M, mitochondria; P, plastid) was performed by using ipsort (http://hypothesiscreator.net/iPSORT).
Expression of PtTrxh2. The expression of PtTrxh2 in plant organs was investigated by recording the number of ESTs coding for this protein among all of the poplar ESTs present in the GenBank database. Around 80 ESTs are present of a total 210 detected for the whole Trx h group. For comparison, only six ESTs encoding Trx o have been found in the database. The Pttrxh2 gene thus appears to be transcribed at high levels in leaves (≈69 hits) and at low levels in roots (5 hits) and petioles (5 hits). Trx h2 also was followed at the protein level in roots, stems, and leaves. The protein is present in similar detectable amounts in all parts of the plant (data not shown).
atNTRA and atNTRB. The presence of NADPH Trx reductase in A. thaliana mitochondria is demonstrated in ref. 4. The activity of PtTrxh2 has been monitored in the presence of both cytosolic (AtNTRB) and mitochondrial (AtNTRA) NADPH Trx A. thaliana reductases (data not shown). The observed activity when using 5,5′-dithiobis(2-nitrobenzoic acid) as an electron acceptor was similar regardless of which NTR was used, suggesting that PtTrxh2 could be reduced by NTR in mitochondria.
In additional experiments, SDS/PAGE performed without reductant addition showed that PtTrxh2 is present mainly as homodimers, which could be reduced by DTT (data not shown). The thiol content of the preparation was determined by using the 5,5′-dithiobis(2-nitrobenzoic acid) method. As expected, nearly three thiols are titrated for the reduced protein and nearly no SH group (0.03 SH per protein) is present in the oxidized protein, suggesting that Cys-29 is involved in a disulfide bridge, possibly linking two monomers together.
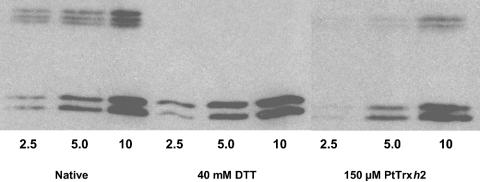
AOX Reduction. AOX, a potential mitochondrial Trx target, has been shown to be redox-regulated (22). Western blot experiments allowing the detection of oxidized AOX dimers as well as reduced AOX monomers were performed by using PtTrxh2-treated or untreated soybean mitochondria (Fig. 3). As a control, similar experiments were carried out by using DTT as reductant. A significant reduction of inactive dimers to monomers by PtTrxh2 was observed, suggesting that Trx could regulate AOX dimerization and possibly its activity in mitochondria.
Fig. 3.
AOX reduction. Purified and frozen/thawed soybean mitochondria (2.5, 5, and 10 μg of proteins) were mixed with reduced 150 μM PtTrxh2or40 mM DTT and incubated for 30 min before being subjected to SDS/PAGE. AOX dimers and monomers were immunodetected.
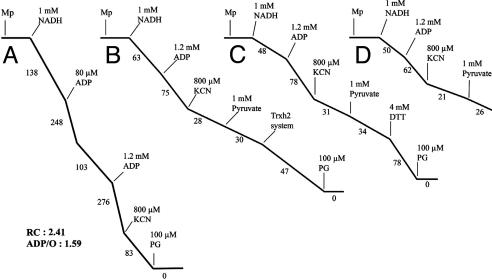
To confirm this hypothesis, the activity of AOX also was monitored in the presence or absence of NTR-reduced PtTrxh2 by using purified soybean mitochondria (Fig. 4). Purified mitochondria presented a high phosphorylative efficiency and a cyanide resistance estimated at 30% of the state-3 rate when NADH was used as substrate (Fig. 4A). The cyanide-insensitive respiration was fully inhibited by propylgalate, a specific inhibitor of AOX. To study the effect of PtTrxh2, mitochondria were oxidized by using diamide (3 mM), and oxygen uptake was recorded after a 2-min incubation in the electrode medium devoid of sucrose. Under these conditions, the subsequent addition of pyruvate poorly stimulated the cyanide-insensitive respiration (Fig. 4B), confirming that the AOX protein needs to be reduced to be activated by this effector. The addition of NTR-reduced PtTrxh2 enhanced the recorded AOX activity (Fig. 4B). A similar effect was obtained with DTT (Fig. 4C). Moreover, the addition of the reductant system of Trxs, depleted of Trxh2, poorly modified the oxygen uptake (Fig. 4D). Similar experiments performed with AtTrxo1 and the cytosolic PtTrxh1 indicated that these isoforms also could activate AOX (data not shown).
Fig. 4.
AOX activation. NADH oxidation by mitochondria isolated from soybean cotyledons. (A) Purified mitochondria in respiration medium. (B-D) Purified soybean mitochondria were incubated for 30 min with 10 mM diamide and washed two times, and the oxygen uptake was measured in sucrose-depleted respiration medium. Where indicated, the following additions were made to the oxygen electrode chamber: 35 μM NTR reduced PtTrxh2 (B). PtTrxh2 (875 μM) was previously reduced during 15 min in the presence of atNTRB (12.5 μM) and NADPH (2.5 mM). As control, either 4 mM DTT (C) or only NADPH/NTR (D) was added. Numbers on the traces represent nmol O2 per min per mg of protein.
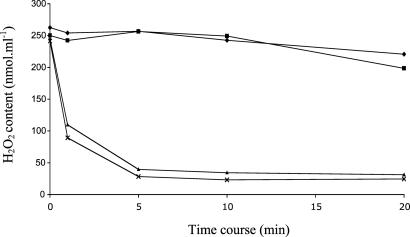
Gpx Activity. Despite their classification, plant Gpxs use Trxs for peroxide reduction (23). The ability of PtTrxh2 to reduce poplar Gpxs in vitro has been investigated. A representative experiment involving the putative mitochondrial PtGpx3 is shown in Fig. 5. Surprisingly, PtTrxh2 remained completely inactive in this test, in contrast to the cytosolic PtTrxh1 and PtTrxh3. Similar results were obtained with the other tested Gpx isoforms, PtGpx1 and PtGpx5 (data not shown). The mitochondrial AtTrxo1 was also unable to reduce the different isoforms of PtGpx (data not shown).
Fig. 5.
Gpx activity. The reduction of H2O2 by poplar Gpx 3 (PtGpx3) was measured by following the disappearance of 500 μM H2O2 with 500 μM DTT or 20 μM of different Trx isoforms and 2 μM Gpx3 in a typical ferrous oxidation-xylenol orange (FOX) assay. ♦, minus Trx; ▪, 20 μM PtTrxh2; ▴, 20 μM PtTrxh1; X, 20 μM PtTrxh3.
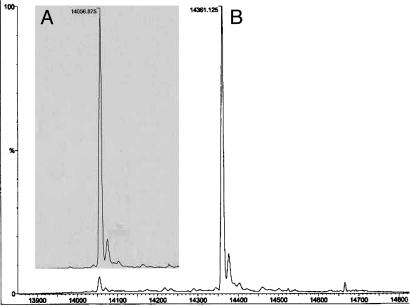
Glutathionylation of PtTrxh2. To investigate the susceptibility of PtTrxh2 to glutathionylation, the protein was reduced by DTT, incubated with excess GSSG, and analyzed by quadrupole time-of-flight. Reduced recombinant PtTrxh2 had the expected molecular mass of 14,056.875 Da. Quadrupole time-of-flight analysis of PtTrxh2 that had been treated with GSSG showed the formation of a peak with a mass of 14,361.125 Da, compatible with the addition of one glutathione molecule (305.5 Da). A representative analysis of untreated PtTrxh2 and GSSG-treated PtTrxh2 is shown in Fig. 6. When the GSSG-treated PtTrxh2 was rereduced by using 10 mM DTT, a peak at 14,059 was obtained (data not shown), as expected for reductive cleavage of the adduct. PtTrxh1 and PtTrxh3, two other poplar Trxs h, also were tested. In both cases, no glutathione addition was detected on these Trxs in these experimental conditions.
Fig. 6.
Glutathionylation of PtTrxh2. The deconvoluted spectra of native PtTrxh2 (A) and PtTrxh2 incubated with 5 mM GSSG (B) are shown.
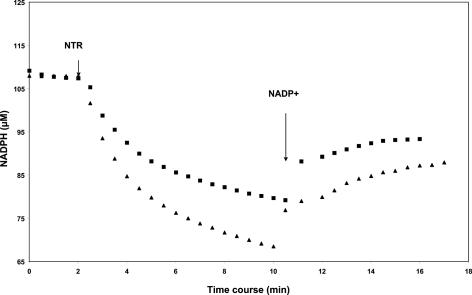
To investigate the effect of glutathionylation on the enzymatic activity of PtTrxh2, the time courses of reduction and reoxidation of disulfides in GSSG-treated PtTrxh2 and PtTrxh2 in the presence of 85 nM AtNTRB are compared in Fig. 7. For PtTrxh2, the amount of oxidized NADPH determined in the first part of the curves indicates that the three cysteines are reduced (≈1.5 oxidized NADPH per protein). In contrast, the amount of oxidized NADPH corresponds to only one disulfide bridge in GSSG-treated PtTrxh2 (≈1 oxidized NADPH per protein). From equilibrium constants, redox potentials of -287 ± 15 mV for PtTrxh2 and -225 ± 15 mV for GSSG-treated PtTrxh2 were calculated by using a value of -320 mV for the couple NADP+/NADPH at pH 7.0 and 20°C. The oxidation-reduction midpoint potential (Em) of PtTrxh2 also was measured by using the monobromobimane titration method. The Em value is approximately -292 mV ± 10 mV at pH 7.0, a value similar to that found with the NTR method and for other Trxs h (24, 25).
Fig. 7.
Determination of redox potential of PtTrxh2 and glutathionylated PtTrxh2. Reduction of disulfides in 26.6 μM PtTrxh2 (▴) and 26 μM GSSG-treated PtTrxh2 (▪) was started by the addition of 85-nm Trx reductase B from A. thaliana. When the reaction was stopped, NADP+ was added to a final concentration of 1.0 mM. The formation of NADPH was followed from the increase at 340 nm.
Discussion
In poplar, as well as in other plants, the Trx h group can be divided into three distinct subgroups. The main characteristic of subgroup II members is the presence of an amino acid N-terminal extension. Nevertheless, it had been thought that these isoforms were localized in the cytosol (2, 3). In this study, we demonstrate that at least one poplar Trx h (PtTrxh2) belonging to this subgroup is associated with mitochondria. This mitochondrial localization is in agreement with previous evidence for the presence of Trxs and particularly Trxs h in plant mitochondria (26, 27). This mitochondrial localization of PtTrxh2 raises the possibility of subcellular organelle localization for other Trxs h with N-terminal extensions. The phylogenic tree presented in this study raises the possibility that mitochondrial Trxhs could constitute a distinct subdivision of the Trxh subgroup II. However, the physiological significance of the N-terminal extensions found in other proteins of this subgroup remains to be elucidated.
The mitochondrial localization of PtTrxh2 increases the complexity of the Trx system in this organelle, because it was recently shown that Trx o and NTRA are also localized in mitochondria (4). In poplar, EST database analysis indicated the presence of at least one mitochondrial Trx o (7). The presence of at least two distinct isoforms of Trx in mitochondria raises the question of the specificity and also the function of these Trxs in this organelle, both isoforms being reduced by NTR. EST database analysis suggested that PtTrxh2 may be expressed to a significantly larger extent than PtTrxo, suggesting a major role for this isoform in poplar.
The functions of Trx in plant mitochondria remain largely unknown. Nevertheless, recent data collected by using proteomic approaches suggest the presence of at least 50 potential Trx-linked proteins involved in different fundamental processes (28). Among these potential targets, Gpx has been identified. Interestingly, PtTrxh2 and atTrxo1 are not able to serve as electron donors for different isoforms of poplar Gpx, whereas these Gpxs are reduced by either PtTrxh1 or PtTrxh3, two putative cytosolic isoforms. Among the tested PtGpxs, PtGpx3 is predicted to be targeted to mitochondria and could be classified as part of a mitochondrial Gpx subgroup including AtGpx6 (29). The mitochondrial potato homolog of AtGpx6 has been found to be a potential target of Trx (28). Because the Em value of PtTrxh2 is quite similar to those of other Trxs (24) and that PtTrxh2 is able to interact with nonmitochondrial proteins (8, 17), it is likely that structural features, rather than thermodynamic factors, are responsible for the absence of interactions between PtTrxh2 and PtGpxs. The details of this Trx/target interaction specificity thus require further investigation. Taken together, these data suggest that additional Trxs could exist in mitochondria and that the Trx system of this organelle may be as complex as that found in chloroplasts. It is also possible that other unidentified reductants may be present in mitochondria.
Another potential target of mitochondrial Trxs is AOX. Besides the cytochrome c pathway, plant mitochondria have an alternative respiratory pathway involving the homodimeric AOX. AOX plays a role in decreasing the formation of mitochondrial reactive oxygen species (30-32). A redox regulation of AOX has been identified. When the subunits are linked by a disulfide bond, the enzyme is inactive (22). In contrast, when the disulfide is reduced, monomers could be activated by α-keto acids (33, 34). The reductive activation of AOX dimers by PtTrxh2 and AtTrxo1 described in this study suggests that Trxs could play a major role in the activation of AOX and also in reactive oxygen species detoxification.
Another characteristic of PtTrxh2 is its ability to be glutathionylated. This property may be unique for PtTrxh2, because two other poplar Trxs, PtTrxh1 and PtTrxh3, are not glutathionylated. Data on glutathionylation processes in plants are scarce. In contrast, this phenomenon is well known in mammalian systems. For instance, glutathionylation of human Trx has been shown to reduce its enzymatic activity (35). In this study, we have shown that glutathionylation significantly alters the PtTrxh2 redox potential, suggesting that glutathione adduct could also modulate its enzymatic activity. The physiological significance of this posttranslational modification of PtTrxh2 remains to be elucidated and deserves further investigation.
Abbreviations: AOX, alternative oxidase; Gpx, glutathione peroxidase; GSSG, oxidized glutathione; Trx, thioredoxin; NTR, NADPH-Trx reductase.
References
- 1.Buchanan, B. B., Schürmann, P., Wolosiuk, R. A. & Jacquot, J. P. (2002) Photosynth. Res. 73, 215-222. [DOI] [PubMed] [Google Scholar]
- 2.Schürmann, P. & Jacquot, J. P. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371-400. [DOI] [PubMed] [Google Scholar]
- 3.Meyer, Y., Vignols, F. & Reichheld, J. P. (2002) Methods Enzymol. 347, 394-402. [DOI] [PubMed] [Google Scholar]
- 4.Laloi, C., Rayapuram, N., Chartier, Y., Grienenberger, J. M., Bonnard, G. & Meyer, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14144-14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaire, S. D., Richardson, J. M., Goyer, A., Keryer, E., Lancelin, J. M., Makhatadze, G. I. & Jacquot, J. P. (2000) Biochim. Biophys. Acta 1476, 311-323. [DOI] [PubMed] [Google Scholar]
- 6.Gelhaye, E., Rouhier, N. & Jacquot, J. P. (2004) Plant Physiol. Biochem. 42, 265-271. [DOI] [PubMed] [Google Scholar]
- 7.Gelhaye, E., Rouhier, N. & Jacquot, J. P. (2003) FEBS Lett. 555, 443-448. [DOI] [PubMed] [Google Scholar]
- 8.Gelhaye, E., Rouhier, N., Laurent, P., Sautière, P. E., Martin, F. & Jacquot, J. P. (2002) Physiol. Plant. 114, 165-171. [DOI] [PubMed] [Google Scholar]
- 9.Gerard, J. & Dizengremel, P. (1988) Plant Sci. 56, 167-171. [Google Scholar]
- 10.Kelly, B. M. & Wiskich, J. T. (1988) Plant Physiol. 87, 705-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouhier, N., Gelhaye, E., Sautiere, P. E., Brun, A., Laurent, P., Tagu, D., Gerard, J., de Fay, E., Meyer, Y. & Jacquot, J. P. (2001) Plant Physiol. 127, 1299-1309. [PMC free article] [PubMed] [Google Scholar]
- 12.Vauclare, P., Diallo, N., Bourguignon, J., Macherel, D. & Douce, R. (1996) Plant Physiol. 112, 1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schenk, P. M., Baumann, S., Mattes, R. & Steinbiss, H. H. (1995) BioTechniques 19, 196-198. [PubMed] [Google Scholar]
- 14.Jacquot, J. P., Rivera-Madrid, R., Marinho, P., Kollarova, M., Le Marechal, P., Miginiac-Maslow, M. & Meyer, Y. (1994) J. Mol. Biol. 235, 1357-1363. [DOI] [PubMed] [Google Scholar]
- 15.Behm, M. & Jacquot, J. P. (2000) Plant Physiol. Biochem. 39, 363-369. [Google Scholar]
- 16.Gelhaye, E., Rouhier, N., Vlamis-Gardikas, A., Girardet, J. M., Sautière, P. E., Sayzet, M. & Jacquot, J. P. (2003) Plant Physiol. Biochem. 41, 629-635. [Google Scholar]
- 17.Rouhier, N., Gelhaye, E., Gualberto, J. M., Jordy, M. N., De Fay, E., Hirasawa, M., Duplessis, S., Lemaire, S. D., Frey, P., Martin, F., et al. (2004) Plant Physiol. 134, 1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elthon, T. E., Nickels, R. L. & McIntosh, L. (1989) Plant Physiol. 89, 1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krimm, I., Lemaire, S., Ruelland, E., Miginiac-Maslow, M., Jacquot, J. P., Hirasawa, M., Knaff, D. B. & Lancelin, J. M. (1998) Eur. J. Biochem. 255, 185-195. [DOI] [PubMed] [Google Scholar]
- 20.Krause, G., Lundström, J., Barea, J. L., Pueyo de la Cuesta, C. & Holmgren, A. (1991) J. Biol. Chem. 266, 9494-9500. [PubMed] [Google Scholar]
- 21.Shi, J. & Bhattacharyya, M. K. (1996) Plant Mol. Biol. 32, 653-662. [DOI] [PubMed] [Google Scholar]
- 22.Umbach, A. L. & Siedow, J. N. (1993) Plant Physiol. 103, 845-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung, B. G., Lee, K. O., Lee S. S., Chi, Y. H., Jang, H. H., Kang, S. S., Lee, K., Lim, D., Yoon, S. C., Yun, D. J., et al. (2002) J. Biol. Chem. 277, 12572-12578. [DOI] [PubMed] [Google Scholar]
- 24.Bréhelin, C., Laloi, C., Setterdahl, A. T., Knaff, D. B. & Meyer, Y. (2004) Photosynth. Res. 79, 233-248. [DOI] [PubMed] [Google Scholar]
- 25.Wangensteen, O. S., Chueca, A., Hirasawa, M., Sahrawy, M., Knaff, D. B. & Lopez Gorge, J. (2001) Biochim. Biophys. Acta 1547, 156-166. [DOI] [PubMed] [Google Scholar]
- 26.Marcus, F., Chamberlain, S. H., Chu, C., Masiarz, F. R., Shin, S., Yee, B. C. & Buchanan, B. B. (1991) Arch. Biochem. Biophys. 287, 195-198. [DOI] [PubMed] [Google Scholar]
- 27.Konrad, A., Banze, M. & Follmann, F. (1996) J. Plant Physiol. 149, 317-321. [Google Scholar]
- 28.Balmer, Y., Vensel, W. H., Tanaka, C. K., Hurkman, W. J., Gelhaye, E., Rouhier, N., Jacquot, J. P., Manieri, W., Schürmann, P., Droux, M. & Buchanan, B. B. (2004) Proc. Natl. Acad. Sci. USA 101, 2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez Milla, M. A., Maurer, A., Rodriguez Huete, A. & Gustafson, J. P. (2003) Plant J. 36, 602-615. [DOI] [PubMed] [Google Scholar]
- 30.Purvis, A. C. (1997) Physiol. Plant. 100, 165-170. [Google Scholar]
- 31.Maxwell, D. P., Wang, Y. & McIntosh, L. (1999) Proc. Natl. Acad. Sci. USA 96, 8271-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yip, J. Y. H. & Vanlerberghe, G. C. (2001) Physiol. Plant. 112, 327-337. [DOI] [PubMed] [Google Scholar]
- 33.Millar, A. H., Wiskich, J. T., Whelan, J. & Dat, A. D. (1993) FEBS Lett. 329, 259-262. [DOI] [PubMed] [Google Scholar]
- 34.Pastore, D., Trono, D., Laus, M. N., Di Fonzo, N. & Passarella, S. (2001) Plant Cell Physiol. 42, 1373-1382. [DOI] [PubMed] [Google Scholar]
- 35.Casagrande, S., Bonetto, V., Fratelli, M., Gianazza, E., Eberini, I., Massignan, T., Salmona, M., Chang, G., Holmgren, A. & Ghezzi, P. (2002) Proc. Natl. Acad. Sci. USA 99, 9745-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]