Abstract
Respiratory Complex I from E. coli may exist in two interconverting forms: resting (R) and active (A). The R/A transition of purified, solubilized Complex I occurring upon turnover was studied employing two different fluorescent probes, Annine 6+, and NDB-acetogenin. NADH-induced fluorescent changes of both dyes bound to solubilized Complex I from E. coli were characterized as a function of the protein:dye ratio, temperature, ubiquinone redox state and the enzyme activity. Analysis of this data combined with time-resolved optical measurements of Complex I activity and spectral changes indicated two ubiquinone-binding sites; a possibility of reduction of the tightly-bound quinone in the resting state and reduction of the loosely-bound quinone in the active state is discussed. The results also indicate that upon the activation Complex I undergoes conformational changes which can be mapped to the junction of the hydrophilic and membrane domains in the region of the assumed acetogenin-binding site.
Keywords: Biological Sciences, Biochemistry, Biophysics
1. Introduction
NADH:ubiquinone oxidoreductase (Complex I) is the electron input enzyme in the respiratory chain of mitochondria and many bacteria. It catalyzes the electron transfer from NADH to ubiquinone along the intraprotein redox chain localized in the hydrophilic domain coupled with proton translocation across the membrane performed by antiporter-like membrane subunits. These two processes, electron and proton transfers, are widely separated in space [1, 2, 3] and time [4, 5, 6] in contrast to other known redox-driven proton pumps. There is a consensus that the electron transfer from NADH to the last FeS cluster in the intraprotein redox chain, N2, is not coupled with energy conversion (see for reviews [7, 8, 9]). Also, it has been found that the redox potential of the bound ubiquinone is lower than −300 mV (comparable with the redox potential of −320 mV of the donor NADH/NAD) [4, 10, 11]; therefore, its reduction by N2 is not accompanied by a significant energy drop. Such unusually low ubiquinone Em indicates restrictions in Q2− stabilization by bulk H+ due to particular properties of quinone-binding site. Thus, the key event in Complex I molecular mechanism, energy transduction, should occur beyond the primary reduction of the ubiquinone: upon its protonation and release [9, 10, 11]. Two possibilities are under consideration: the tightly-bound quinone can be released from the protein after its reduction and protonation [2, 12, 13] or it has only limited mobility and shuttles inside the long channel between its reduction site close to N2 and the second ubiquinone-binding site where it can reduce a quinone in the membrane pool [7, 9, 14].
The question is whether ubiquinone protonation and/or movement are accompanied by conformational changes that transduce energy to the membrane fragment upon the enzyme functioning. The experimental data on the redox-induced conformational changes within Complex I is rather limited. An analysis by FTIR (Fourier transform infrared spectroscopy) revealed the structural reorganization in Complex I from E. coli [15, 16] and mitochondria [17] in the hydrophilic domain around FeS clusters, particularly N2, upon the enzyme reduction. The analysis of cross-linking data of nearby subunits revealed redox-dependent conformational changes in the hydrophilic domain that are possibly extended into the membrane domain at least to the area of the interface of these domains [18]. This was confirmed by the high-resolution structural studies of the oxidized and reduced hydrophilic domain of Complex I from Thermus thermophilus: the enzyme reduction induced an adjustment in the nucleotide-binding pocket and a small shift of several α-helices at the interface with the membrane domain [19].
Site-directed spin labeling of Complex I from E. coli did not indicate redox-dependent changes in the enzyme [20], however, the limitation of the engineering positions exclusively to the surface of the hydrophilic domain and the loop of the hydrophobic M subunit could be a reason for the observed lack of the changes. The dependence of photoaffinity labeling by Complex I inhibitors, such as fenperoxymate and quinazoline, on NADH enabled Miyoshi and coauthors to conclude that redox-linked conformational changes occur at the interface of the hydrophilic and membrane domains of bovine heart mitochondrial Complex I [21, 22]. It should be stressed that mitochondrial Complex I may exist in two states, active (A) and resting or de-active (D) [23, 24, 25, 26]; upon A/D transition the enzyme undergoes conformational changes where three subunits at the junction of the hydrophilic and hydrophobic fragment are involved [27, 28, 29]. Therefore, the data mentioned above may not be indicative of the conformational changes upon turnover, but they could rather reflect particular events that occur in the enzyme upon its conversion to the functional state.
Recently we have shown that oxidized Complex I from E. coli also exists in de-active, resting (R), state and it converts to the active (A) state [30]. To test whether Complex I from E. coli undergoes conformational changes upon its reduction, during the activation phase, two hydrophobic fluorescent dyes, NBD-acetogenin (NBD-Acg) and Annine 6+ that respond to changes in the fluorophore surrounding were chosen in this study.
Studying of NADH-induced fluorescent responses of these probes bound to Complex I let us make a conclusion that these fluorescence changes reflect two events occurring upon the enzyme activation: conformational changes and bound ubiquinone reduction. The obtained data allowed us to suggest that i) the conformational changes occurring upon R/A conversion of Complex I can be mapped to the junction of the hydrophilic and membrane domain; ii) Complex I contains two quinone binding sites; one, tightly-bound, ubiquinone molecule is reduced in R form, and the other, bound loosely, is reduced and exchanged with the pool quinone only in the A form.
2. Materials and methods
2.1. Synthesis of NBD-Acg
NBD-Acg, compound 2, was prepared in 7 steps using the previously described synthetic intermediate 4, earlier prepared en route the synthesis of asimicin and bullatacin stereoisomers [31, 32] and the commercially available NBD-Cl (4-Chloro-7-nitrobenzofurazan), as outlined in Fig. 1. NBD-Cl and chemical reagents used in NBD-Acg synthesis are commercially available and were purchased from Sigma-Aldrich.
Fig. 1.
Synthesis of NBD-Acg, 2. Abbreviations: CH3CN, Acetonitrile; ClTi(OPr)3, Chlorotrisopropxytitanium (IV); CH2Cl2, Methylene chloride; DMF, Dimethyl formamide; DMSO, Dimethyl sulfoxide; Et2O, diethyl ether; EtOAc, Ethyl acetate; HF, Hydrogen fluoride; iPr2EtN, N,N-diisopropylethylamine; MeOH, Methanol; NaN3, Sodium azide; NaHCO3, Sodium hydrogen carbonate; Pd-C, Palladium on Carbon; RT, Room temperature.
2.2. Bacterial growth and purification of Complex I
The E. coli MWC215 (SmR ndh:CmR) and mutant, NuoM E144A [33] and NuoCD R274A [34] strains were grown in LB medium at 37 °C in a 25 L fermenter and harvested at the late exponential growth phase. The membranes for Complex I purification were prepared by passing the cells through an APV Gaulin homogenizer. Then Complex I was purified by two consecutive chromatography steps using DEAE-Trisacryl M (Bio-Sepra) anion exchanger columns and gel filtration on Superdex 200 prep grade (GE Healthcare), respectively, as described [35]. DQ, decylubiquinone (Santa Cruz Biotechnology), and HAR, hexaammineruthenium (III) chloride, reductase activities of Complex I were measured as described previously [36].
2.3. Kinetic measurements
Monitoring optical changes upon Complex I reduction by dithionite and NADH were carried out at room temperature under anaerobic conditions using a high-resolution CCD-array spectrometer (HR2000+, Ocean Optics) combined with a DH-2000-BAL light source with filtering technology that produces a smooth spectrum across the entire range (Ocean Optics). Standard quartz optic cuvette, 10 × 10 mm was used. Sample volume was 1 ml. Acquired time-resolved optical changes were combined to form the data surface. Obtained data analysis was carried out using the MATLAB software (the Mathworks, Inc.). Decomposition of the kinetic data surfaces was achieved by global fitting run under a MATLAB interface using the Rakowsky algorithm as described in [37, 38].
Fast measurements of Complex I activity by following NADH oxidation were performed by fast mixing of equal volumes of solubilized Complex I and buffer containing NADH (Unisoku Stopped-flow RSP-2000 apparatus) using a high-resolution CCD-array spectrometer (HR2000+, Ocean Optics) combined with a Xenon lamp. NADH oxidation track was fitted by exponential curve with time constant 1.2 s, the fluorescent response track was fitted by a logistic curve with a breakpoint at 2.1 s.
Fast measurements of fluorescence responses upon mixing Annine 6+ bound to Complex I with NADH-containing buffer (1:1 volume) were followed using the homemade setup comprised BioLogic SFM-300 stopped-flow apparatus, high energy blue LED (460 nm) as a light source and Hamamatsu PMT R2949 for the signal detection. Microvolume cuvette was used. For absorption measurements at 340 nm the same stopped-flow apparatus was used but with a Xenon lamp as the light source and imaging spectrograph CP140 (Horiba Scientific) coupled with fast linear scan camera (spL2048–140 km, Basler Inc.) as the signal detector as previously described [30].
The electric potential (Δψ) generation by Complex I reconstituted into liposomes was performed by monitoring absorbance changes (ΔA588-625) of Δψ-sensitive probe, Oxonol VI, as described in [36].
Stationary measurements of fluorescence were performed using Hitachi F-7000 fluorescence spectrophotometer, λex = 460 nm, λem = 550 nm for Annine 6+ and 540 nm for NDB-Acg at 25–30 °C except temperature dependence study where the temperature is specified. Anaerobic conditions were achieved by the initial purge of the solutions with argon, upon kinetic measurements the anaerobiosis was supported by blowing argon through the cuvette.
The assay buffer comprised 40 mM HEPES-KOH, pH 7.0, and 0.5 mM MgSO4 except Fig. 6 where it is specified. Complex I was added at a concentration of 0.5 mg/ml for spectral measurements, 15–30 μg/ml for Annine 6+ assay and 50–60 μg/ml for NBD-Acg assay except for stopped-flow experiments where protein concentration was doubled. Concentrations of fluorescent dyes were constant in all experiments: 0.6 μM Annine 6+ and 0.1 μM NBD-Ac. It should be noted that upon the concentration of purified enzyme the detergent, dodecyl β-D-maltopyranoside (DDM) (Glycon-Biochemicals GmbH), content could be also significantly concentrated. DDM micelles efficiently bound Annine 6+ and NBD-Acg and prevent binding of the dyes to the protein; therefore DDM was partially removed from some preparations. When DQ was used, the protein sample was incubated with it for at least 10 minutes to approach stationary ubiquinone distribution between water and micelles phases and, therefore, to avoid the rate limitation due to slow DQ equilibration between these phases. All experiments were reproduced at least three times.
Fig. 6.
Fluorescence of Annine6+ bound to proteoliposomes does not respond to Δψ, but it reflects redox state of ubiquinone. A. NADH-induced fluorescent responses of 0. 6 μM Annine 6+ bound to proteoliposomes with reconstituted Complex I. B. Monitoring NADH-induced Δψ generation by Complex I across the liposome membrane by means of 3 μM Oxonol VI. Medium: 100 mM Hepes-KOH, pH 7.0, 1 mM MgSO4, DQ 100 μM. Additions: NADH 150 μM, valinomycin 1 μM, quinol oxidase bo3 20 nM.
Chemical reagents used for purification, reconstitution into liposomes and study of purified Complex I from E. coli are commercially available and were purchased from Sigma-Aldrich except specified otherwise. Quinol oxidase, bo3, was purified from E. coli as described in [39].
3. Results
3.1. Characterization of fluorescent probes used in the study
NBD-acetogenin (NBD-Acg) (Fig. 2, Fig. 3) is an acetogenin analog in that the universal butenolide function was modified with an NBD fluorophore. Acetogenins are specific inhibitors of ubiquinone reductase activity of Complex I [40]. Like the parent acetogenin (Fig. 2, 1) [31]. NBD-Acg inhibits Complex I function potently (Fig. 3). The acetogenin moiety was reported to bind Complex I between the hydrophobic and hydrophilic domains in the tight vicinity of the ubiquinone channel outlet [41, 42, 43, 44]; fluorescence of NBD is highly sensitive to the hydrophobicity of its environment, thus it should indicate structural re-arrangements in this area.
Fig. 2.
Structure of a non-natural acetogenin stereoisomer (1), a related NBD (nitrobenzoxadiazole) analog (NBD-Acg, 2) and the Annine 6+ dye (3).
Fig. 3.
Titration of NADH:DQ oxidoreductase activity of solubilized Complex I by rolliniastatin (I1/2 = 11 nM), blue, and NBD-Acg (I1/2 = 50 nM), red.
Annine 6+ (Dr. Hinner & Dr. Hübener. Sensitive Farbstoffe GbR, Germany) [45] (Fig. 2, 3), belongs to the class of hemicyanine dyes with annelated benzene rings containing an intramolecular dipole [46] and therefore capable to respond to transmembrane electric potential [45, 46, 47] or, like a styrylpyridinium dye RH241, to an intraprotein charge movement [48] or a change in local dipole potential [49]. Annine 6+ is relatively soluble in water but, nevertheless, it displays strong binding to lipid membranes [45] and its fluorescence is utterly dependent on the environment polarity.
3.2. Fluorescent responses of NDB-Acg and Annine 6+ bound to Complex I
Both dyes have almost no fluorescence in water solution; their fluorescence is strongly increased upon binding solubilized Complex I. Both dyes bound to Complex I respond similarly to the enzyme reduction by NADH: their fluorescence increases. The response is reversed when NADH is oxidized (Fig. 4A). The amplitude of the response is not dependent on NADH concentration in the range of 10–100 μM. The fluorescent responses on NADH without added ubiquinone are strongly dependent on the dye:protein ratio what is discussed in detail below. An addition of decylubiquinone, DQ, results in strong quenching. The following NADH addition increases the fluorescence so long until NADH is not consumed (Fig. 4B). The nature of these responses was studied.
Fig. 4.
Fluorescence responses of Annine 6+ (ΔF550 nm, blue curves) and NBD-acetogenin (ΔF540 nm, red curves) bound to the solubilized Complex I. A. NADH addition is the absence of DQ. B. DQ addition results in the fluorescence quenching reversed by NADH. Fluorescent probes concentration: 0.6 μM Annine 6+ and 0.1 μM NBD-Ac. Complex I concentration: 20 μg/ml for Annine 6+ assay and 50 μg/ml for NBD-Acg assay.
3.3. Quenching of NDB-acetogenin and Annine 6+ fluorescence by ubiquinone depends on its redox state
Since the ubiquinone in contrast to ubiquinol was found to be a strong quencher for a number of lipophilic fluorescent probes [50, 51, 52, 53, 54] we suggested that the amplitude of fluorescent changes in the presence of the added ubiquinone reflects the level of ubiquinone reduction in the vicinity of the protein. This suggestion was proved for both dyes, Annine 6+ and NBD-acetogenin, using detergent micelles, non-functional Complex I and proteoliposomes with either DQ or ubiquinone 1, Q1.
3.3.1. Annine 6+ in a non-protein system.
Fluorescence of Annine 6+ was strongly increased in the presence of DDM at the detergent concentration higher than CMC and it was quenched by ubiquinone addition (Fig. 5A). Under anaerobic conditions in the absence of DQ an addition of the strong reductant dithionite has no effect on the fluorescence of Annine 6+, while dithionite restored the fluorescence quenched by DQ due to the quinone reduction.
Fig. 5.
Discrimination of ubiquinone redox states by Annine 6+ fluorescence in non-protein system (A) and bound to non-functional Complex I (B). A. 0.6 μM Annine 6+ was bound to DDM (0.1%) micelles. Under anaerobic conditions 60 μM DQ was added in the presence (dotted line) or in the absence of dithionite (solid line), indicated by DT. Addition of 0.5 mM dithionite to the oxidized DQ resulted in the fluorescence recovery while in the absence of DQ or when it was already reduced dithionite has no effect on Annine 6+ fluorescence. B. 0.6 μM Annine 6+ was bound to 20 μg/ml non-functional Complex I. The fluorescence was quenched by 60 μM Q1 (red curves) or DQ (blue curves) and restored in the presence of 2 mM dithiothreitol (dotted lines) in case of Q1 but not DQ.
3.3.2. Annine 6+ bound to non-functional, oxidized, Complex I
Another way to show the discrimination of ubiquinone redox states was to use the dye bound to Complex I when the enzyme is not active. The addition of DQ and Q1 results in fluorescence decrease, which can be restored in Q1 case if the medium is supplemented with dithiothreitol (DTT) as a discriminative quinone reductant. DTT is capable of reduction of Q1 but not of more hydrophobic DQ. It is important that DTT does not reduce Complex I or produce any changes in Complex I activity and optical properties. When Annine 6+ was bound to non-functional, oxidized, Complex I the ubiquinones, DQ and more hydrophilic Q1, both, caused quenching, but the fluorescence could be restored by DTT in the latter case (Fig. 5B).
3.3.3. Annine 6+ bound to proteoliposomes
It should be noted that Annine 6+ was used for the detection of the transmembrane electric potential based on the Stark effect [45, 47, 55]; however, this dye did not show Δψ-dependent response on proteoliposomes containing Complex I upon proton pumping. If there was such an effect, it was negligible and completely covered by the strong influence of ubiquinone redox transition. The NADH-induced increase of Annine 6+ fluorescence was insensitive to compounds that dissipate Δψ such as valinomycin. On the other hand, this increase was greatly diminished or completely abolished by re-oxidation of ubiquinone by oxygen as catalyzed by added quinol oxidase, bo3 (Fig. 6A ). NADH-induced Δψ generation by reconstituted Complex I was verified using Δψ–sensitive probe Oxonol VI (Fig. 6B).
3.3.4. NBD-Acg bound to detergent micelles and non-active Complex I
Unfortunately, the experiments under anaerobic conditions with dithionite cannot be reproduced to show the same phenomenon on NBD-Acg because the NDB fluorophore interacts with dithionite. However, similar effects could be observed with NBD-Acg bound to DDM micelles or non-active Complex I (Fig. 7A,B) although in less extent. DTT reducing Q1 but not DQ restored fluorescence quenched by quinone only in case of Q1.
Fig. 7.
Discrimination of ubiquinone redox states by fluorescence of 0.1 μM NBD-Acg bound to DDM micelles (A) or non-functional, oxidized Complex I (B). Fluorescence of NBD-Acg bound to DDM micelles was quenched by 60 μM Q1, restored upon its reduction by 4 mM dithiothreitol and quenched again in the presence of 20 nM ubiquinol oxidase bo3 (A). The same effect was observed with 60 μM Q1 (B, blue line) when NBD-Acg was bound with non-functional Complex I (10 μg/ml), while dithiothreitol did not affect quenching by 60 μM DQ (B, black line).
Summing up, in the presence of added quinone NADH-induced fluorescence changes of both probes are difficult to unequivocally interpret since they reflect the quinone reductase activity of Complex I as well as intraprotein events. Nonetheless, in the absence of added quinone these changes reflect the reduction of the bound quinone(s) and/or conformational changes in Complex I.
3.4. Dependence of Annine 6+ and NBD-Acg fluorescence on protein concentration
The dependence of fluorescence on the dye:protein ratio is a significant characteristic of the dye binding. Total fluorescence of used dyes simply increases with growing concentration of Complex I. However, the amplitude of specific responses of the bound dye to NADH addition to Complex I can be saturated. Titration of NBD-Acg fluorescence by Complex I showed that the amplitude of the response to NADH in the absence of ubiquinone is saturated at approximately equimolar concentrations of NBD-Acg and Complex I (Fig. 8), which indicates a specific binding of a single dye molecule to the site undergoing changes upon Complex I reduction. The saturation of NADH-induced response of Annine6+ fluorescence occurred at higher dye:protein ratio suggesting that 2–3 molecules of the dye bound to the enzyme are responsible for fluorescence changes upon Complex I reduction. The limited amount of sites accommodating molecules of Annine6+, which are susceptible to the enzyme reduction, also indicates a specific binding.
Fig. 8.
Dependence of NBD-Acg and Annine 6+ responses to 10 μM NADH addition on the concentration of solubilized Complex I in the absence of added quinone. NBD-Acg concentration was 0.1 μM, Annine 6+ 0.6 μM.
3.5. Temperature dependence of Annine 6+ and NBD-Acg fluorescence responses
The temperature dependence of the observed fluorescence changes may characterize the nature of events occurring upon the reduction of Complex I and its operation. The amplitude of the NADH-induced responses did not change in case of NBD-Acg in the temperature range of 15–45 °C. However, the rate of the response, calculated from the maximal steepness of the curve, changed drastically (Fig. 9A,B). Shown for a comparison the temperature dependence of this rate in the presence of the electron acceptor DQ has a bell shape with the maximum at approximately 25–30 °C (Fig. 9B), which is typical for the enzyme activity, whereas the rate in the absence of an electron acceptor grows exponentially with the temperature (Fig. 9B). In the case of Annine 6+, the amplitude of the responses starts to drop at the temperature over 30 °C (Fig. 9C) what could be due to concentration quenching if several Annine 6+ molecules are involved. Due to complicated temperature dependence, it is difficult to treat the data of Annine6+ response rate at a higher temperature; however, the rate in the presence of DQ grows significantly slower than the rate in its absence (Fig. 9D).
Fig. 9.
Temperature dependence of NADH-induced fluorescence changes of Annine 6+ and NBD-acetogenin bound to solubilized Complex I. A. Responses of 0.1 μM NBD-Acg on 10 μM NADH addition in the absence of DQ from 15 to 45 °C (right to left). B. Temperature dependence of the rate of NBD-Acg responses to NADH in the absence of DQ (empty circles) and in its presence (grey circles). C. Responses of 0.6 μM Annine 6+ on 10 μM NADH addition in the absense of DQ from 15 to 45 °C (right to left, the dotted lines correspond to the highest temperatures). D. Temperature dependence of the rate of Anine 6+ responses to NADH in the absence of DQ.
The data on temperature dependence of NADH-induced NBD-Acg responses is presented as Arrhenius plot. The plot displays the break at 23 °C (Fig. 10); calculated activation energy is 130 kJ*mol−1 and 50 kJ*mol−1 at temperatures below and higher 23 °C, correspondingly.
Fig. 10.
Arrhenius plot of the rate of NADH-induced fluorescence responses of NBD-Acg bound to solubilized Complex I.
3.6. Dependence of NADH-induced NBD-Acg and Annine 6+ fluorescence responses on the ubiquinone reductase activity of Complex I
The effect of the enzyme activity on NADH-induced dyes responses was tested by using mutated Complex I. Two mutants with low ubiquinone reductase activity were studied: NuoCD R274A [34], and NuoM E144A [33]. The decrease of these mutants activity was due to mutations at different positions. In NuoCD R274A the mutation was introduced upstream of the ubiquinone-binding site in the hydrophilic NuoCD subunit, by the single amino acid replacement making the redox potential of FeS cluster N2 more negative [34] and, therefore, the electron transfer to ubiquinone is hampered. In NuoM E144A the mutation was made downstream from the ubiquinone-binding site, at the distance of approximately by 60 Å, also by the single amino acid replacement in the membrane NuoM subunit; this mutation blocks the ubiquinone reduction and proton translocation [33]. In the absence of added ubiquinone, without turnover, the rate of NADH-induced fluorescence response of Annine6+ bound to mutated Complex I was strongly decreased (Fig. 11A) and it correlates with the quinone reductase activity of these variants (NADH:ubiquinone oxidase activity: 100% wt, 22% NuoCD R274A [34], 12% NuoM E144A [33]; maximal rate of NADH-induced Annine6+ response: 100% wt, 15% NuoCD R274A, 5% NuoM E144A). The effect of the mutation is less in the case of NADH-induced fluorescence responses of NBD-Acg because the wild type enzyme was already inhibited by the dye, nevertheless, the initial rate of these responses was about 50% of wild type rate for both mutants (Fig. 11B). A significant deceleration of NADH-induced dye responses by the mutations in the absence of turnover indicates that the dyes do not respond to the reduction of FeS and FMN but their fluorescence changes reflect the events occurring upon ubiquinone reduction and/or release.
Fig. 11.
Dependence of Annine 6+ (A) and NBD-Acg (B) NADH-induced responses on the activity of solubilized Complex I variants without added DQ. 10 μM NADH was added at zero time. Wild type, blue; NuoCD R274A, green; NuoM E144A, red. Conditions as specified in Fig. 4.
3.7. Alternative reduction of Complex I
To study the role of NADH in observed fluorescence responses the effect of strong reductant dithionite (−0.44 > Em > −0.66 V [56]) was tested. Dithionite could be used in Annine 6+ assay because it does not interact with this dye (Fig. 5A) in contrast to NBD-Acg. Reduction of Complex I by dithionite under anaerobic conditions resulted in Annine 6+ fluorescence changes similar to that upon the addition of NADH, regardless whether the latter was added under aerobic or anaerobic conditions (Fig. 12). The reductant with the higher redox potential (Em = −0.33 V), dithiothreitol, caused only negligible fluorescence changes of Annine 6+ bound to Complex I (not shown). The response to dithionite was fully reversible and can be followed by NADH response and vice versa. The rate of the response to dithionite was much faster than that to NADH (Fig. 12) and cannot be resolved by the used experimental setup.
Fig. 12.
Response of Annine 6+ fluorescence on reduction of solubilized Complex I by NADH and dithionite. 100 μM dithionite added under anaerobic conditions at zero time, red curve; 10 μM NADH added under aerobic and anaerobic conditions at zero time, blue and green. Annine 6+ concentration 0.6 μM.
It should be noted that after the enzyme is oxidized it converts again into the initial form: subsequent additions of NADH or dithionite resulted in the same fluorescence responses. Longer exposure of Complex I to NADH without an electron acceptor under aerobic conditions caused the enzyme destruction.
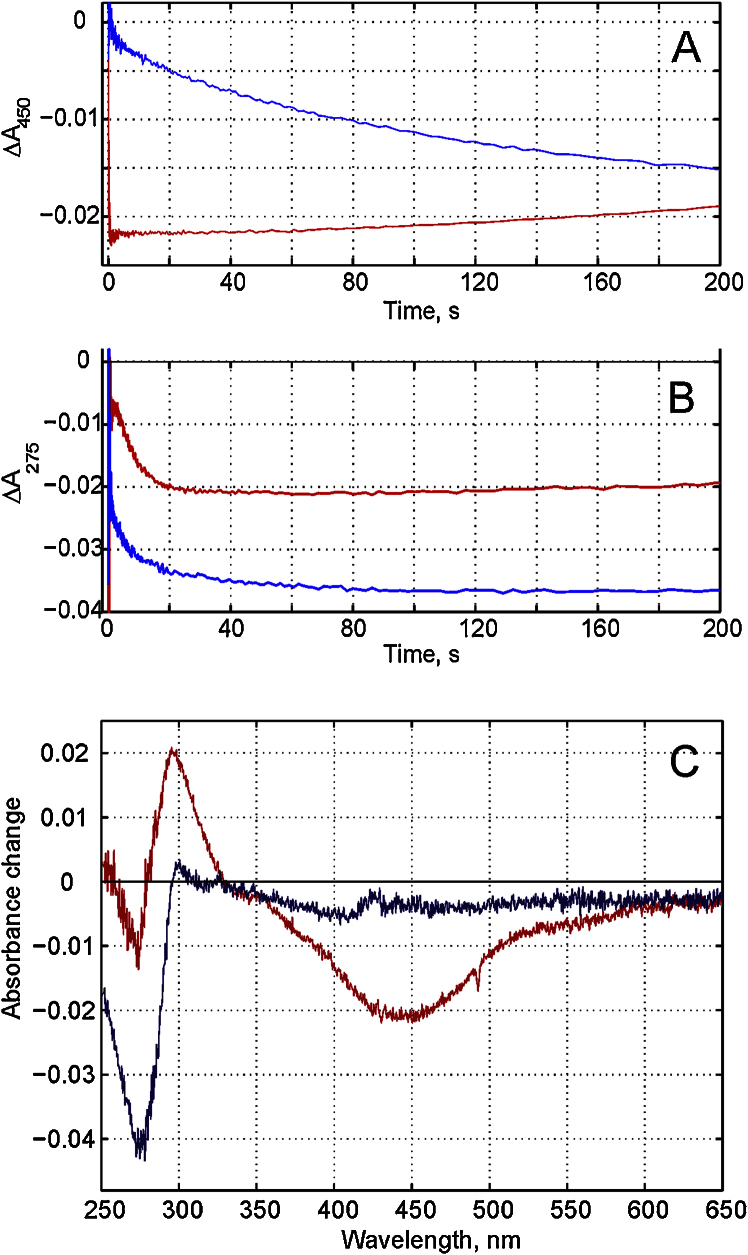
3.8. Redox spectra of Complex I upon its reduction
To study the nature of the compound reduced by dithionite or NADH and possibly responsible for the Annine 6+ response we obtained the kinetics of absorbance changes upon Complex I reduction in UV-Vis range in the absence of added ubiquinone. The sets of redox spectra of Complex I were taken for 300 s after addition of dithionite or NADH under anaerobic conditions. Previously we have shown that the reduction of FeS clusters of Complex I intraprotein redox chain by dithionite is a slow process in the absence of mediators [57]. Indeed, the kinetics at 450 nm, where most of Complex I redox centers absorb, indicated slow, in the time scale of minutes, reduction of the intraprotein redox chain (Fig. 13A, blue line). However, the kinetics at 275 nm, in ubiquinone band, was fast (Fig. 13B, blue line). In contrast, the reduction of the intraprotein redox chain by NADH is very fast process (Fig. 13A, red line), it is not resolved here since it takes less than a couple of milliseconds [4, 6], but the kinetics in the ubiquinone band is much slower (Fig. 13B, red line). The spectra taken at 3 s after NADH or dithionite addition are shown in Fig. 13C. By this time the reduction of the intraprotein redox chain is complete with NADH but negligible with dithionite. On the other hand, the absorption at ubiquinone band is close to its saturated level with dithionite, whereas it is less than a half with NADH. Comparison the kinetics of Annine 6+ response and absorption changes clearly shows that the reduction of Complex I intraprotein redox chain has no effect on Annine 6+ response.
Fig. 13.
Kinetics of solubilized Complex I reduction by addition of 35 μM dithionite, blue lines, or 40 μM NADH, red lines, under anaerobic conditions. A. Absorption changes at 450 nm. B. Absorption changes at 275 nm. C. Redox spectra of Complex I taken at 3 s after addition of dithionite or NADH. Complex I at concentration of 0.5 mg/ml was solubilized in buffer containing 40 mM Hepes-KOH, pH 7.0 and 0.5 mM MgSO4.
3.9. Fast measurements of Complex I activation and NADH-induced response of Annine 6+ bound to Complex I
Following NADH:ubiquinone reductase activity by optic spectroscopy upon fast mixing of Complex I and NADH showed a significant initial delay [30]. We reproduced these experiment and monitored NADH-induced fluorescence response of Annine 6+ bound to Complex I using stopped-flow approach for the better time resolution. NADH oxidation kinetics could be fitted with an exponential curve with the time constant of 1.2 s. The track of fluorescence response has substantial sigmoidal character and it can be fitted by a logistic curve with the breakpoint at 2.1 s. The juxtaposition of kinetics of these processes is shown in Fig. 14.
Fig. 14.
Juxtaposition of NADH:DQ oxidoreductase activity of Complex I monitored by NADH absorbtion at 340 nm (blue line) and fluorescent response of Annine 6+ on NADH addition to Complex I (green line). The curves were obtained by stopped-flow apparatus (see Materials and Methods). The sample containing 60 μg/ml Complex I equilibrated with 50 μM DQ with or without Annine 6+ (1.2 μM) was fast mixed with equal volume of buffer containing 80 μM NADH. The kinetics of NADH oxidation was fitted by an exponential curve with time constant 1.2 s (red line). The kinetics of Annine+ fluorescence response was fitted by a logistic curve with breakpoint at 2.1 s (red line).
4. Discussion
4.1. Interaction of fluorescent dyes with Complex I
Here we report that two fluorescent dyes, Annine 6+ and NBD-Acg, are capable of binding specifically to Complex I and responding to events occurring in the enzyme upon NADH oxidation. The fluorescence of both dyes is strongly quenched by ubiquinone and much less by ubiquinol. A similar effect on particular lipophilic fluorescent probes by ubiquinone was observed earlier; the quenching property of ubiquinone was used to study its localization and dynamics in the mitochondrial membrane [50] and model bilayers [51] by means of fluorescent anthroylstearate or pyrene derivatives [54] and the development of ubiquinone-quantum dot bioconjugates for in vitro and intracellular Complex I sensing [52] and reactive oxygen species imaging [53]. This discriminative ubiquinone/ubiquinol response is also a characteristic feature of the probes used in this study; we showed that the fluorescence of the probes corresponds the reduction level of quinone in the lipophilic phase (Results, section III). In the absence of added quinone, NADH-induced fluorescence responses of Annine 6+ and NBD-Acg report the reduction of the bound ubiquinone and conformational changes within the protein as a result of an increase in hydrophobicity in the chromophore environment. In the presence of added ubiquinone these responses also reflect Complex I activity. Such properties make both probes, Annine 6+ and NBD-Acg, useful tools to study Complex I.
4.2. NADH-induced fluorescence response reflects resting-to-active (R/A) transition of Complex I
Previously we have shown that oxidized Complex I exists in de-active, resting state, and it converts to the active state upon turnover. Here we present the results of a study aimed to shed light on this transition. NADH-induced fluorescence responses of both dyes regardless whether DQ was added exhibit similar non-linear kinetics with a lag phase typical for Complex I R/A transition determined by the rate of NADH:ubiquinone oxidoreductase activity [30]. Comparison of the kinetics of NADH oxidation and Annine6+ fluorescence response obtained by stopped-flow approach (Fig. 14) cannot be straightforward since the former is exponential and the latter sigmoidal suggesting a number of sequential reactions. Fluorescence response kinetics can be fitted by a logistic curve with the breakpoint at 2.1 s; the value of this empiric parameter indicates that two processes, activation of NADH oxidation (characteristic time of 1.2 s) and Annine 6+ response, fall in a similar time range.
4.3. Tight- and loosely- bound ubiquinone
Monitoring redox spectra of Complex I after NADH addition showed fast (unresolved in this setup) reduction of FMN and FeS clusters whereas the bound quinone(s) reduction occurred slowly, in the same time range as the fluorescence changes. The fast reduction of the bound quinone, as well as fast fluorescence response, both faster than 1 s (Fig. 13), were observed upon the addition of the hydrophilic low-potential reductant dithionite despite the fact that an interaction of dithionite with the catalytic center of Complex I proceeds slowly, in a minute time scale (Fig. 13). Therefore, electrons from dithionite are not delivered to the quinone via the intraprotein redox chain, it remains oxidized at least for 3 s (Fig. 13C). This means that quinone bound to Complex I is solvent accessible. Dithionite-induced fast fluorescence changes of Annine 6+ indicate that the dye responds to this quinone reduction. However, high-resolution structural data revealed that the hydrophilic head of the quinone molecule binds in the deep end of a narrow cavity inside the protein; the entrance of the cavity is most probably sealed by ubiquinone hydrophobic tail that may not allow any solvent into the cavity [58]. This finding combined with previously reported data on low-potential ubiquinone in Complex I [10, 11] makes plausible the presence of tightly-bound ubiquinone molecules in purified Complex I. The data presented here suggest the presence of loosely-bound quinone, which is not deeply buried in the protein and can be directly reduced by dithionite. If this is the case, the quinone content in the purified Complex I preparation should be 2 quinone molecules per FMN. Previously we reported the ratio Q:FMN = 1.3 in the samples of purified Complex I from E. coli; however, i) the amount of ubiquinone could be underestimated by incomplete extraction of the tightly-bound quinone and ii) the population of the loosely-bound ubiquinone in Complex I may be lower than equimolar.
4.4. The timing of events in the initial stage of Complex I turnover
Taking together the data on the electron transfer upon Complex I reduction by NADH obtained earlier [4, 6, 10] and results presented here allowed us to derive the approximate timing of the events (Fig. 15). Electrons from NADH are delivered to the intraprotein redox chain in approximately 0.1 ms; the full reduction of FeS chain occurs in millisecond time range [4, 5, 6]. The reduction rate of the tightly-bound quinone is not yet known; due to the method limitation, we could observe only its partial reduction with a high time resolution. However, the time frames can be set: reduction of the tightly-bound ubiquinone falls within ≈1 ms (freeze-quench approach [4]) and ≈15 ms (stopped-flow approach [10]). Then the activation stage proceeds (1–2 s obtained by stopped-flow approach [30]). We suggest that in this stage Complex I undergoes the conformational changes that enable the reduction of the loosely-bound quinone and conversion of the enzyme to the fully active state. The loosely-bound quinone is released, and the turnover number approaches its inherent value of ≈300 s−1.
Fig. 15.
Approximate timing of the events occurring in Complex I upon its reduction by NADH.
4.5. Indication of conformational changes upon the activation stage
The reason for the delay in the reduction of loosely-bound quinone could be the long distance between loosely and tightly-bound quinones, which impedes electron transport. Conformational changes should enable the ubiquinone dynamics and provide efficient electron tunneling. The fluorescence of the probes used here is dependent on the hydrophobicity of the dye-binding site which may be altered upon conformational changes and the reduction level of the bound ubiquinone(s); it is hard to determine the contribution of these two parameters in the redox-dependent fluorescence responses. However, the temperature dependence of the responses allowed us to make some conclusions. NBD-Acg is presumably located deep in the protein in the suggested acetogenin binding site; therefore, it should be subject to conformational changes. The temperature dependence of NADH-induced fluorescence responses of NBD-Acg, is indeed consistent with known conformational changes within the protein. The Arrhenius plot of this data obtained with NDB-Acg showed breakpoint at 23 °C and two values of activation energy of 50 and 130 kJ*mol−1, above and below the breakpoint, correspondingly, were obtained. The lower value of activation energy is comparable to that reported for the A/D transition of the mitochondrial Complex I from frog muscle, 66 kJ*mol−1, and carp muscle, 67 kJ*mol−1, [59]; such transition in mitochondrial Complex I is realized via conformational changes [27, 29, 60]. The higher activation energy obtained at lower temperatures could be determined by the detergent properties: the viscosity in DDM micelles increases and affects Complex I dynamics.
4.6. Mapping the site of conformational changes
Since NBD-Acg, which binds Complex I in almost equimolar proportion, should reside like acetogenin, the conformational changes can be localized between the hydrophobic and hydrophilic domains, in accordance with the data obtained by acetogenin photoaffinity labeling of mitochondrial Complex I [41, 42, 43, 44]. Two fragments of the ND1 (NuoH in E. coli, Nqo8 in T. thermophilus) subunit were labeled by acetogenin. One of them composes TMS 4th and 5th and part of the loop between 5th and 6th TMSs (site A) and the other includes 6th TMS and the second part of the loop between 5th and 6th TMSs (site B) [42, 44]. Acetogenin binding to the site B was suppressed by exogenous short-chain ubiquinone [42] suggesting a proximity of an entrance for pool ubiquinone. Recent studies of the acetogenin-binding site in bovine Complex I also mapped Asp160 in the 49 kDa subunit (Asp139 in Nqo4 in T. thermophilus, Asp 329 in NuoCD in E. coli) [43] which is located in the putative quinone binding cavity [2]. The 4th TMS in the NuoH subunit involved in acetogenin binding is the nearest neighbor of NuoA subunit, particularly its 2nd TMS (Fig. 16) and plausibly takes part in energy transduction to the transporter subunits of Complex I. It is possible to assume that the observed conformational changes upon Complex I reduction result in closer contact between the NuoH and NuoA subunits and, thus, provide more efficient connection between the ubiquinone redox transitions and the proton translocation machinery in the membrane fragment that results in Complex I activation.
Fig. 16.
Acetogenin binding sites in Complex I. Fragment of the T. thermophilus enzyme (PDB entry: 4HEA) is shown. The Nqo8 (NuoH in E. coli) subunit is shown in green, Nqo7 (NuoA in E. coli), in yellow. Two fragments of Nqo8 corresponding to that in the mitochondrial Complex I labeled by acetogenin are shown in red. Acetogenin binding sites were suggested to reside in the third matrix loop. An approximate position of the ubiquinone channel is designated by pale red. At the entry of the channel the Asp139 which is also shown to participate in acetogenin binding. In subunit Nqo7 loop Ser46 homologous to Cys39 involved in active/de-active transition in mitochondrial enzyme is shown.
In should be noted that our results on the localization of conformational changes in Complex I upon its activation are in line with studies of the A/D transition of bovine Complex I by specific labeling. The selective fluorescence labeling identified the transition-dependent exposure of Cys39 in the ND3 (Ser46 in Nqo7 in T. thermophilus, Ser52 in NuoA) subunit loop facing to the junction of hydrophilic and membrane domains [27] and resided close to acetogenin binding site (Fig. 16).
Declarations
Author contribution statement
Nikolai Belevich: Performed the experiments; analyzed and interpreted the data; contributed reagents, materials, analysis tools or data; wrote the paper.
Galina Belevich, Zhiyong Chen: Performed the experiments.
Subhash C. Sinha: Conceived and designed the experiments; contributed reagents, materials, analysis tools or data; wrote the paper.
Marina Verkhovskaya: Conceived and designed the experiments; performed the experiments; analyzed and interpreted the data; wrote the paper.
Competing interest statement
The authors declare no conflict of interest.
Funding statement
This work was supported by Biocentrum Helsinki (7919530), the Sigrid Juselius Foundation (4702217), and the Magnus Ehrnrooth Foundation (96133).
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Prof. Mårten Wikström for insightful comments. We thank Eija Haasanen for purification of Complex I.
References
- 1.Efremov R.G., Sazanov L.A. The coupling mechanism of respiratory complex I—a structural and evolutionary perspective. Biochim. Biophys. Acta. 2012;1817:1785–1795. doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Baradaran R., Berrisford J.M., Minhas G.S., Sazanov L.A. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sazanov L.A., Baradaran R., Efremov R.G., Berrisford J.M., Minhas G. A long road towards the structure of respiratory complex I, a giant molecular proton pump. Biochem. Soc. Trans. 2013;41:1265–1271. doi: 10.1042/BST20130193. [DOI] [PubMed] [Google Scholar]
- 4.Verkhovskaya M.L., Belevich N., Euro L., Wikström M., Verkhovsky M.I. Real-time electron transfer in respiratory complex I. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belevich N.P., Verkhovskaya M.L., Verkhovsky M.I. Chapter 4. Electron transfer in respiratory complexes resolved by an ultra-fast freeze-quench approach. Methods Enzymol. 2009;456:75–93. doi: 10.1016/S0076-6879(08)04404-2. [DOI] [PubMed] [Google Scholar]
- 6.Belevich N., Belevich G., Verkhovskaya M. Real-time optical studies of respiratory Complex I turnover. Biochim. Biophys. Acta. 2014;1837:1973–1980. doi: 10.1016/j.bbabio.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Verkhovskaya M., Bloch D.A. Energy-converting respiratory Complex I: on the way to the molecular mechanism of the proton pump. Int. J. Biochem. Cell Biol. 2013;45:491–511. doi: 10.1016/j.biocel.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Hirst J. Mitochondrial Complex I. Ann. Rev. Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 9.Wikström M., Sharma V., Kaila V.R., Hosler J.P., Hummer G. New perspectives on proton pumping in cellular respiration. Chem. Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 10.Verkhovsky M., Bloch D.A., Verkhovskaya M. Tightly-bound ubiquinone in the Escherichia coli respiratory Complex I. Biochim. Biophys. Acta. 2012;1817:1550–1556. doi: 10.1016/j.bbabio.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Verkhovskaya M., Wikström M. Oxidoreduction properties of bound ubiquinone in Complex I from Escherichia coli. Biochim. Biophys. Acta. 2014;1837:246–250. doi: 10.1016/j.bbabio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Hunte C., Zickermann V., Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 13.Zickermann V., Wirth C., Nasiri H., Siegmund K., Schwalbe H., Hunte C., Brandt U. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 14.Ohnishi S.T., Salerno J.C., Ohnishi T. Possible roles of two quinone molecules in direct and indirect proton pumps of bovine heart NADH-quinone oxidoreductase (complex I) Biochim. Biophys. Acta. 2010;1797:1891–1893. doi: 10.1016/j.bbabio.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich T., Hellwig P. Redox-induced conformational changes within the Escherichia coli NADH ubiquinone oxidoreductase (complex I): an analysis by mutagenesis and FT-IR spectroscopy. Biochim. Biophys. Acta. 2010;1797:659–663. doi: 10.1016/j.bbabio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Hellwig P., Kriegel S., Friedrich T. Infrared spectroscopic studies on reaction induced conformational changes in the NADH ubiquinone oxidoreductase (complex I) Biochim. Biophys. Acta. 2016;1857:922–927. doi: 10.1016/j.bbabio.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Marshall D., Fisher N., Grigic L., Zickermann V., Brandt U., Shannon R.J., Hirst J., Lawrence R., Rich P.R. ATR-FTIR redox difference spectroscopy of Yarrowia lipolytica and bovine complex I. Biochemistry. 2006;45:5458–5467. doi: 10.1021/bi052561e. [DOI] [PubMed] [Google Scholar]
- 18.Berrisford J.M., Thompson C.J., Sazanov L.A. Chemical and NADH-induced, ROS-dependent, cross-linking between subunits of complex I from Escherichia coli and Thermus thermophilus. Biochemistry. 2008;47:10262–10270. doi: 10.1021/bi801160u. [DOI] [PubMed] [Google Scholar]
- 19.Berrisford J.M., Sazanov L.A. Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 2009;284:29773–29783. doi: 10.1074/jbc.M109.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pohl T., Spatzal T., Aksoyoglu M., Schleicher E., Rostas A.M., Lay H., Glessner U., Boudon C., Hellwig P., Weber S., Friedrich T. Spin labeling of the Escherichia coli NADH ubiquinone oxidoreductase (complex I) Biochim. Biophys. Acta. 2010;1797:1894–1900. doi: 10.1016/j.bbabio.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Shiraishi Y., Murai M., Sakiyama N., Ifuku K., Miyoshi H. Fenpyroximate binds to the interface between PSST and 49 kDa subunits in mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry. 2012;51:1953–1963. doi: 10.1021/bi300047h. [DOI] [PubMed] [Google Scholar]
- 22.Murai M., Miyoshi H. Chemical modifications of respiratory complex I for structural and functional studies. J. Bioenerg. Biomembr. 2014;46:313–321. doi: 10.1007/s10863-014-9562-z. [DOI] [PubMed] [Google Scholar]
- 23.Minakami S., Estabrook R.W., Schindler F.J. Hydrogen transfer between reduced diphosphopyridine nucleotide dehydrogenase and the respiratory chain. 2. Initial lag in oxidation of reduced diphosphopyridine nucleotide. J. Biol. Chem. 1964;239:2049–2054. [PubMed] [Google Scholar]
- 24.Gavrikova E.V., Vinogradov A.D. Active/de-active state transition of the mitochondrial complex I as revealed by specific sulfhydryl group labeling. FEBS Lett. 1999;455:36–40. doi: 10.1016/s0014-5793(99)00850-9. [DOI] [PubMed] [Google Scholar]
- 25.Gostimskaya I.S., Cecchini G., Vinogradov A.D. Topography and chemical reactivity of the active-inactive transition-sensitive SH-group in the mitochondrial NADH: ubiquinone oxidoreductase (Complex I) Biochim. Biophys. Acta. 2006;1757:1155–1161. doi: 10.1016/j.bbabio.2006.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grivennikova V.G., Kapustin A.N., Vinogradov A.D. Catalytic activity of NADH-ubiquinone oxidoreductase (Complex I) in intact mitochondria − Evidence for the slow active/inactive transition. J. Biol. Chem. 2001;276:9038–9044. doi: 10.1074/jbc.M009661200. [DOI] [PubMed] [Google Scholar]
- 27.Galkin A., Meyer B., Wittig I., Karas M., Schägger H., Vinogradov A., Brandt U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 2008;283:20907–20913. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babot M., Birch A., Labarbuta P., Galkin A. Characterisation of the active/de-active transition of mitochondrial complex I. Biochim. Biophys. Acta. 2014;1837:1083–1092. doi: 10.1016/j.bbabio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babot M., Labarbuta P., Birch A., Kee S., Fuszard M., Botting C.H., Wittig I., Heide H., Galkin A. ND3, ND1 and 39 kDa subunits are more exposed in the de-active form of bovine mitochondrial complex I. Biochim. Biophys. Acta. 2014;1837:929–939. doi: 10.1016/j.bbabio.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belevich N., Verkhovskaya M. Resting state of respiratory Complex I from Escherichia coli. FEBS Lett. 2016 doi: 10.1002/1873-3468.12199. [DOI] [PubMed] [Google Scholar]
- 31.Sinha S.C., Chen Z., Huang Z.Z., Nakamaru-Ogiso E., Pietraszkiewicz H., Edelstein M., Valeriote F. Alteration of the bis-tetrahydrofuran core stereochemistries in asimicin can affect the cytotoxicity. J. Med. Chem. 2008;51:7045–7048. doi: 10.1021/jm801028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z.Y., Sinha S.C. Synthesis of 10 stereochemically distinct bis-tetrahydrofuran intermediates for creating a library of 64 asimicin stereoisomers. Tetrahedron Lett. 2009;50:6398–6401. [Google Scholar]
- 33.Euro L., Belevich G., Verkhovsky M.I., Wikström M., Verkhovskaya M. Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I) Biochim. Biophys. Acta. 2008;1777:1166–1172. doi: 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Belevich G., Euro L., Wikström M., Verkhovskaya M. Role of the conserved arginine 274 and histidine 224 and 228 residues in the NuoCD subunit of complex I from Escherichia coli. Biochemistry. 2007;46:526–533. doi: 10.1021/bi062062t. [DOI] [PubMed] [Google Scholar]
- 35.Euro L., Belevich G., Wikström M., Verkhovskaya M. High affinity cation-binding sites in Complex I from Escherichia coli. Biochim. Biophys. Acta. 2009;1787:1024–1028. doi: 10.1016/j.bbabio.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Verkhovskaya M., Knuuti J., Wikström M. Role of Ca2+ in structure and function of Complex I from Escherichia coli. Biochim. Biophys. Acta. 2011;1807:36–41. doi: 10.1016/j.bbabio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Bloch D.A., Jasaitis A., Verkhovsky M.I. Elevated proton leak of the intermediate OH in cytochrome c oxidase. Biophys. J. 2009;96:4733–4742. doi: 10.1016/j.bpj.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belevich Y.V.B.N.P., Verkhovskaya M.L., Baykov A.A., Bogachev A.V. Identification of the coupling step in Na+-translocating NADH:quinone oxidoreductase from real-time kinetics of electron transfer. Biochim. Biophys. Acta. 2016;1857:141–149. doi: 10.1016/j.bbabio.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Morgan J.E., Verkhovsky M.I., Puustinen A., Wikström M. Identification of a peroxy intermediate in Cytochrome bo(3) of Escherichia coli. Biochemistry. 1995;34:15633–15637. doi: 10.1021/bi00048a005. [DOI] [PubMed] [Google Scholar]
- 40.Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochim. Biophys. Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 41.Sekiguchi K., Murai M., Miyoshi H. Exploring the binding site of acetogenin in the ND1 subunit of bovine mitochondrial complex I. Biochim. Biophys. Acta. 2009;1787:1106–1111. doi: 10.1016/j.bbabio.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Kakutani N., Murai M., Sakiyama N., Miyoshi H. Exploring the binding site of delta(lac)-acetogenin in bovine heart mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry. 2010;49:4794–4803. doi: 10.1021/bi100454b. [DOI] [PubMed] [Google Scholar]
- 43.Masuya T., Murai M., Morisaka H., Miyoshi H. Pinpoint chemical modification of Asp160 in the 49 kDa subunit of bovine mitochondrial complex I via a combination of ligand-directed tosyl chemistry and click chemistry. Biochemistry. 2014;53:7816–7823. doi: 10.1021/bi501342w. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi S., Abe M., Yamamoto S., Murai M., Miyoshi H. Bis-THF motif of acetogenin binds to the third matrix-side loop of ND1 subunit in mitochondrial NADH-ubiquinone oxidoreductase. Biochim. Biophys. Acta. 2011;1807:1170–1176. doi: 10.1016/j.bbabio.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Fromherz P., Hübener G., Kuhn B., Hinner M.J. ANNINE-6plus, a voltage-sensitive dye with good solubility, strong membrane binding and high sensitivity. Eur. Biophys. J. 2008;37:509–514. doi: 10.1007/s00249-007-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hübener G., Lambacher A., Fromherz P. Anellated hemicyanine dyes with large symmetrical solvatochromism of absorption and fluorescence. J. Phys. Chem. B. 2003;107:7896–7902. [Google Scholar]
- 47.Flickinger B., Berghofer T., Hohenberger P., Eing C., Frey W. Transmembrane potential measurements on plant cells using the voltage-sensitive dye ANNINE-6. Protoplasma. 2010;247:3–12. doi: 10.1007/s00709-010-0131-y. [DOI] [PubMed] [Google Scholar]
- 48.Habeck M., Cirri E., Katz A., Karlish S.J., Apell H.J. Investigation of electrogenic partial reactions in detergent-solubilized Na, K-ATPase. Biochemistry. 2009;48:9147–9155. doi: 10.1021/bi901148k. [DOI] [PubMed] [Google Scholar]
- 49.Mares L.J., Garcia A., Rasmussen H.H., Cornelius F., Mahmmoud Y.A., Berlin J.R., Lev B., Allen T.W., Clarke R.J. Identification of Electric-Field-Dependent Steps in the Na+,K+-Pump Cycle. Biophys. J. 2014;107:1352–1363. doi: 10.1016/j.bpj.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chance B., Erecinska M., Radda G.K. 12-(9-Anthroyl)stearic acid, a fluorescent-probe for ubiquinone region of mitochondrial-membrane. Eur. J. Biochem. 1975;54:521–529. doi: 10.1111/j.1432-1033.1975.tb04165.x. [DOI] [PubMed] [Google Scholar]
- 51.Fato R., Battino M., Castelli G.P., Lenaz G. Measurement of the lateral diffusion-coefficients of ubiquinones in lipid vesicles by fluorescence quenching of 12-(9-anthroyl)stearate. FEBS Lett. 1985;179:238–242. doi: 10.1016/0014-5793(85)80526-3. [DOI] [PubMed] [Google Scholar]
- 52.Ma W., Qin L.X., Liu F.T., Gu Z., Wang J., Pan Z.G., James T.D., Long Y.T. Ubiquinone-quantum dot bioconjugates for in vitro and intracellular complex I sensing. Sci. Rep. 2013;3:1537. doi: 10.1038/srep01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin L.X., Ma W., Li D.W., Li Y., Chen X.Y., Kraatz H.B., James T.D., Long Y.T. Coenzyme Q functionalized CdTe/ZnS quantum dots for reactive oxygen species (ROS) imaging. Chemistry—A Eur. J. 2011;17:5262–5271. doi: 10.1002/chem.201003749. [DOI] [PubMed] [Google Scholar]
- 54.Lenaz G., Fato R., Di Bernardo S., Jarreta D., Costa A., Genova M.L., Castelli G.P. Localization and mobility of coenzyme Q in lipid bilayers and membranes. Biofactors. 1999;9:87–93. doi: 10.1002/biof.5520090202. [DOI] [PubMed] [Google Scholar]
- 55.White J.A., Pliquett U., Blackmore P.F., Joshi R.P., Schoenbach K.H., Kolb J.F. Plasma membrane charging of Jurkat cells by nanosecond pulsed electric fields. Eur. Biophys. J. Biophys. Lett. 2011;40:947–957. doi: 10.1007/s00249-011-0710-7. [DOI] [PubMed] [Google Scholar]
- 56.Mayhew S.G. The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur. J. Biochem. 1978;85:535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- 57.Knuuti J., Belevich G., Sharma V., Bloch D.A., Verkhovskaya M. A single amino acid residue controls ROS production in the respiratory Complex I from Escherichia coli. Mol. Microbiol. 2013;90:1190–1200. doi: 10.1111/mmi.12424. [DOI] [PubMed] [Google Scholar]
- 58.Berrisford J.M., Baradaran R., Sazanov L.A. Structure of bacterial respiratory complex I. Biochim. Biophys. Acta. 2016;1851:892–901. doi: 10.1016/j.bbabio.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Maklashina E., Kotlyar A.B., Cecchini G. Active/de-active transition of respiratory complex I in bacteria, fungi, and animals. Biochim. Biophys. Acta. 2003;1606:95–103. doi: 10.1016/s0005-2728(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 60.Ciano M., Fuszard M., Heide H., Botting C.H., Galkin A. Conformation-specific crosslinking of mitochondrial complex I. FEBS Lett. 2013;587:867–872. doi: 10.1016/j.febslet.2013.02.039. [DOI] [PubMed] [Google Scholar]