Abstract
We demonstrated a microfluidic device for rapidly generating complex mixtures of 32 stock reagents in a 5-nl reactor. This “formulation chip” is fully automated and allows thousands of experiments to be performed in a single day with minimal reagent consumption. It was applied to systematically study the phase behavior of the protein xylanase over a large and complex chemical space. For each chemical formulation that demonstrated a pronounced effect on solubility, the protein phase behavior was completely mapped in the chip, generating a set of empirical phase diagrams. This ab initio phase information was used to devise a rational crystallization screen that resulted in 72-fold improvement in successful crystallization hits compared with conventional sparse matrix screens. This formulations tool allows a physics-based approach to protein crystallization that may prove useful in structural genomics efforts.
The application of x-ray crystallography to the determination of protein structure with atomic resolution was a triumph of structural biology in the 20th century. Since the first solution of the structure of myoglobin in 1958 (1), >23,000 different structures have been deposited in the Protein Data Bank, and their role in relating structure to function in biology has been profound. As structure determination efforts continue to move past the most tractable crystallization targets (typically small soluble proteins) and focus instead on more challenging macromolecules, such as large protein complexes and membrane proteins (2), the need to better understand and explore the crystallization process has become urgent. That is because once high-quality crystals are in hand, advances in x-ray sources, computer codes, and related technology have made it relatively straightforward to obtain the structure. Determining the appropriate crystallization conditions has become one of the most significant remaining bottlenecks to structure determination (3).
Understanding the phase behavior of proteins is an essential part of the crystallization process. The growth of crystals from a protein solution requires the existence of a nontrivial phase diagram, which allows the protein state to be manipulated between at least two thermodynamic phases: soluble and precipitated. The processes of crystal nucleation and growth arise on the boundary between these two phases and are governed by subtle effects in physical chemistry. There are a variety of schemes that manipulate the kinetics of the crystallization process, and all take advantage of generic features of these phase diagrams (4). However, in practice, the phase behavior of very few proteins has been studied in detail (5-12), and solubility information for a specific protein is rarely available for crystallization and optimization experiments (13, 14).
Furthermore, it is often an arduous process to find the right combination of chemicals that yields appropriate phase behavior for a given protein. Every protein is different, and even a modest subset of stock precipitating solutions comprises a vast chemical phase space that must be explored. The large amounts of sample required make systematic exploration by conventional techniques infeasible, and screening is typically directed toward an incomplete factorial or sparse-matrix approach, a brute-force process requiring large numbers of experiments (15, 16). There have been numerous attempts to rationalize this procedure, for example, by using computational approaches to predict phase behavior (17, 18) or by trying to correlate measurements of osmotic second virial coefficients (19, 20) with crystallization conditions. Practical limitations have thus far prevented these techniques from being generally applicable.
Here we describe a microfluidic formulation device that allows for the combinatorial mixing of 16 buffers and 16 precipitation agents with a purified protein sample. The ability of the formulation chip to access a vast number of chemical conditions and to accurately dispense and mix fluids on the picoliter scale makes detailed characterization of macromolecule phase behavior both possible and practical. We used this device to screen 4,300 different solubility conditions of the model protein Endo-1,4-β-xylanase from Trichoderma reesei. Xylanase is a 21-kDa member of the gluconase enzyme family. For those conditions that exhibited nontrivial phase behavior (i.e., precipitation), a full phase diagram was generated. From this thorough characterization of the phase behavior, we designed a rational crystallization screen for xylanase. Comparison of this screen to four commercially available sparse matrix screens showed an increase of nearly two orders of magnitude in crystallization success and allowed new insight into the physics of crystallization.
Materials and Methods
Sample Preparation and Crystallization Protocols. Endo-1,4-β-xylanase (xylanase) from T. reesei (Hampton Research, Riverside, CA) was prepared in deionized water from stock (36 mg/ml protein/43% wt/vol glycerol/0.18 M sodium/potassium phosphate, pH 7.0) by repeated buffer exchange at 4°C using a centrifugal filter with a molecular mass cutoff of 10,000 Da (Micron Separations, Westboro, MA). Protein concentration was measured by absorption at 280 nm and adjusted to a stock concentration of 120 mg/ml. Ten-microliter aliquots were flash frozen in liquid nitrogen and stored at -80°C. To avoid sample-sample variations, a single sample preparation was used for all solubility screening, phase-space mapping, and corresponding crystallization experiments. Batch crystallization trials were actively mixed by repeated aspiration and incubated under paraffin oil. Crystallization trials were inspected daily for a period of 2 weeks. Observed crystals were confirmed to be protein crystals by staining (IZIT dye, Hampton Research) and were recorded as crystallization hits.
Experimental Setup and Data Collection. Automation of metering, mixing, and data acquisition allows for thousands of solubility experiments to be executed without the need for user intervention. In each solubility experiment, a unique mixture of the 32 reagents and the protein sample is produced in the chip; details of chip fabrication are in Supporting Text, which is published as supporting information on the PNAS web site. All device control and data acquisition were implemented by using a custom software driver developed in labview (National Instruments, Austin, TX). Mixing recipes were generated by using a spreadsheet program and translated into valve actuation sequences by the software driver. Off-chip solenoid valves (Fluidigm, South San Francisco, CA), controlled by using a digital input-output card (DIO-32HS; National Instruments), were used to generate square-wave pressure signals at the device control ports. A frame-grabber card (Imagenation PXC200A, CyberOptics, Minneapolis) was used to automate image acquisition from a charge-coupled device camera.
Absorption and Precipitation Measurements. Absorption measurements were taken to determine the concentration of bromophenol blue sodium salt (absorption peak at 590 nm) in the mixing ring. A 9-μm-high segment of the mixing ring (≈300 × 80 μm) having rectangular cross section was illuminated with a 590-nm diode (AND180HYP; Newark Electronics, Chicago) and imaged through a stereoscope (SMZ 1500; Nikon) onto a charge-coupled device camera. Pixel intensities were averaged and compared with an identical adjacent reference channel containing the undiluted dye (2 mM bromophenol blue sodium salt/100 mM Tris·HCl, pH 8.0). In some experiments, glycerol was added to the injected dye to vary the viscosity. Dye concentrations were determined by using the Beer-Lambert relation and used to calculate the injected volume.
Precipitation of the protein was automatically detected by imaging a portion of the mixing ring, calculating the standard deviation of the pixel intensities, and comparing this value to the background (no protein added). To ensure even illumination, images were taken at ×112 magnification at a 9-μm-high section of the mixing ring having a rectangular cross section.
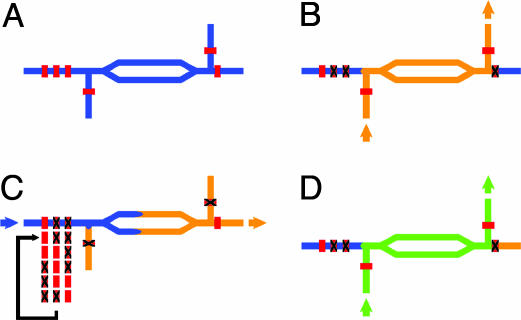
Positive-Displacement Crossinjection Metering. The metering scheme allows for sequential injection of precise sample aliquots from a single microfluidic channel into an array of reaction chambers through a positive displacement crossinjection (PCI) junction. The PCI junction is formed by the combination of a three-valve peristaltic pump (21) and a four-port crossinjection junction with integrated valves on each port (Fig. 1A). At each junction, two sets of valves are actuated to direct the flow either horizontally or vertically. To execute the metering task, the flow is switched vertically through the junction, charging the cross-injector with the sample fluid (Fig. 1B). The flow is then directed horizontally through the junction, and the three valves forming the peristaltic pump are actuated in a five-state sequence to advance the fluid in the horizontal direction (Fig. 1C). Each cycle of the peristaltic pump injects a well defined volume of sample (≈80 pl), determined by the dead volume under the middle valve of the peristaltic pump. The deflection of the valve membranes when not actuated is determined by the pressure difference across the membrane. The volume injected during each cycle therefore may be tuned continuously, allowing for variable positive displacement metering. By repeating the injection sequence, the volume of injected solution may be increased in 80-pl increments, allowing for the dynamic quantized control of the final downstream sample concentration.
Fig. 1.
PCI for robust and programmable high-precision dispensing on chip. These schematic diagrams illustrate the PCI method. (A) A four-port PCI junction. The split-channel architecture creates a large-volume injector region, thereby allowing for an increased number of injections before recharging. Channels/fluids are indicated in blue; valves are red. (B) Charging of the injector region of the PCI junction. Junction valves are actuated to direct the flow vertically through the junction, filling the injector region with orange fluid. (C) Precise positive displacement metering by actuation of peristaltic pump. A peristaltic pump is created by three consecutive valves; the pumping sequence is shown. Pumping the blue fluid injects a metered volume of the orange fluid into the next section of the chip (not shown). (D) The PCI junction is sequentially charged with different solutions to create complex multicomponent mixtures; in this case, the second injection is with a green fluid.
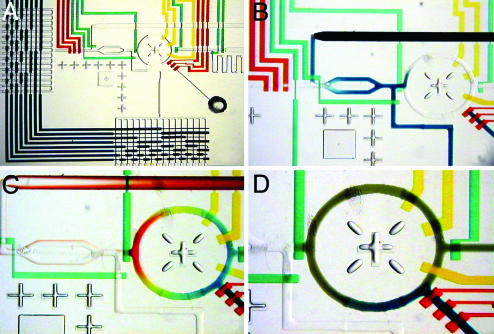
The active region of the microfluidic formulation chip that implements this scheme and allows for the arbitrary combinatorial mixing of 16 stock reagents into one of 16 buffer solutions is shown in Fig. 2A. Two 16-solution multiplexer arrays, actuated by eight control lines, allow for the selection of buffers (Fig. 2A Left) and reagents (bottom, Fig. 2 A). A PCI junction, formed by a three-valve peristaltic injection pump (red) and crossinjection valves (center green), dispenses directly into a 5-nl ring reactor. Once the reactor has been flushed, a reagent line is selected, and the crossinjection sequence is executed. The extended split channel region increases the volume of the crossinjection junction, thereby allowing for up to 15 injections between flushing steps. The maximum number of consecutive injections that may be executed before the junction needs to be refreshed depends on the Taylor dispersion (22) of the injected fluid as it is pumped down the channel and is therefore a function of the viscosity. Fig. 2B shows the injection of four slugs, each having a volume of 80 pl, into the ring reactor. Arbitrary combinations of 16 reagents may be produced in the reactor by sequential flushing and injection steps. Fig. 2C shows a color gradient formed from injections of water and blue, green, yellow, and red dyes. In screening applications that require the interrogation of a precious sample against many premixed reagent formulations, the crossinjection flushing step is wasteful and is circumvented by the addition of a separate sample injection site (Fig. 2D). After the ring is filled with the desired reagents, they are mixed by actuating a rotary peristaltic pump (23). The on-chip peristaltic pumps were pneumatically actuated at 100 Hz, resulting in a maximum flow velocity of ≈2 cm/sec. At these flow rates, complete mixing of aqueous reagents was achieved in <3 sec, and solutions with viscosities of ≈100 cP (1 P = 0.1 Pa·sec) were mixed in 6 sec. A real-time video illustrating the mixing and metering process is included as Movie 1, which is published as supporting information on the PNAS web site.
Fig. 2.
Combinatorial mixing using a microfluidic formulator. These optical micrographs show the manipulation of food dyes with the formulator chip. In all images, the diameter of the mixing ring is 1.5 mm. (A) Integration of multiplexer (dark blue), peristaltic pumps (red), rotary mixer (yellow), and PCI junction (center, green) components for on-chip combinatorial formulation. (B) Injection of ≈250 pl (four injection cycles) of blue dye into rotary mixer. (C) Color gradient formed by consecutive injections into mixing ring (eight injections blue, eight injections green, eight injections yellow, and eight injections red). (D) Pumping around ring for 3 sec results in complete mixing of dye. Blue dye is then added to mixture through sample injection inlet (bottom right).
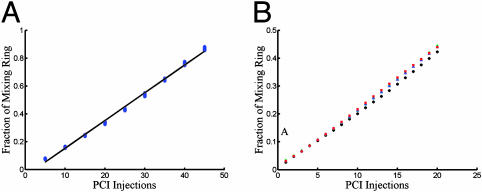
Device Characterization. The precision of metering was evaluated by injecting variable amounts of dye (bromophenol blue sodium salt; Sigma) into a reactor, mixing, and performing absorption measurements. A set of 900 sequential titration experiments (Fig. 3A) shows the metering to be both precise and reproducible, with a slope of 83.4 pl per injection cycle and a coefficient of correlation of 0.996. The standard deviation of the injected slug volume was determined to be ≈0.6 pl. Although positive displacement metering ensures that the injected volume is robust to changes in the fluid viscosity, the viscosity of the working fluid does reduce the bandwidth of the injector. It was found that for a solution having a viscosity of 400 cP, the frequency response of the injector began to roll off at 10 Hz. When operating at an injection frequency of 5 Hz, all solutions having viscosities <400 cP produced equal injection volumes. Because the metering mechanism is completely mechanical, there is no dependence on the pH or ionic strength of the injected fluid. Additionally, because the fluid is not dispensed from the chip, there is no phase interface and therefore little dependence on surface tension. The metering technique is therefore truly robust to the physical properties of the injected fluid. Titration experiments with fluids of varying glycerol concentration show the injection volume to vary by <5% over a viscosity range of 1-400 cP without modification to the injection sequence (Fig. 3B).
Fig. 3.
Precise and robust microfluidic metering. Absorption spectroscopy is used to measure the replaced fraction of fluid in the ring reactor from PCI injections. The total ring volume is nominally 5 nl. (A) Precision and reproducibility of PCI injections. Each of the nine clusters represents 100 identical injection sequences; the standard deviation of the clusters corresponds to an injection error of ≈0.6 pl. (B) Absorption measurements of four sets of 20 injection and mixing sequences show the metering to be robust to the viscosity of the injected fluid. Fluids contain varying amounts of glycerol and have viscosities ranging from 1 to 400 cP.
Results and Discussion
Although small-scale characterization of protein solubility by a precrystallization solubility assay has been reported (15, 24, 25), this technique has not been widely adopted, because the large required sample volumes make it unsuitable for targets that cannot be expressed and purified in large quantities. Microfabricated dispensers have been used to reduce sample consumption in cases where the sequential addition of reagents to a levitated drop of microliter volume is sufficient to explore a restricted chemical space (25). Although microfluidic devices have been previously used to screen crystallization conditions using free interface diffusion (19) and microbatch formats (20), they have not been applied to systematically measure phase behavior (26, 27).
Thorough characterization of protein solubility behavior requires accessing a vast chemical space through the combinatorial mixing of a limited number of stock reagents. The conventional reagents used in crystallization exhibit a large variation in physical properties, such as viscosity, surface tension, ionic strength, and pH. This variation presents a formidable challenge for fluid-handling systems that must allow for arbitrary fluid combinations and proportioning. We developed a PCI metering method that overcomes this obstacle, allowing for variable dispensing to be dynamically programmed by the user in 80-pl increments with <5% variation over a broad range of fluid properties. By combining this method with microfluidic mixing (23) and multiplexing elements (28), large-scale combinatorial screening has been achieved on chip for the first time. The flexibility, precision, and small volume requirements of this device make feasible the systematic mapping of crystallization phase space.
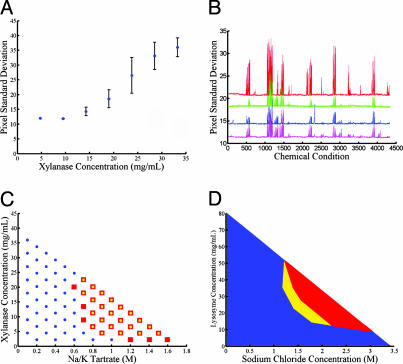
To demonstrate the utility of ab initio protein solubility characterization, we used the formulator chip to exhaustively explore the solubility behavior of a commercially available crystallization standard, Endo-1,4-β-xylanase (xylanase) from T. reesei (Hampton Research) (29, 30). Trial solutions were imaged in the chip's reactor ring, and the standard deviation of the intensity of the imaged pixels was used as a metric of precipitation; this allowed for distinction between precipitated and soluble conditions and provided a rough quantitative measure of the degree of precipitation. Beyond the precipitation limit, the pixel standard deviation increases linearly with the protein concentration and therefore is proportional to the concentration of precipitated protein present in the solution (Fig. 4A). A video of on-chip protein titration and precipitation is included as Movie 2, which is published as supporting information on the PNAS web site.
Fig. 4.
Automated exploration of protein solubility using the microfluidic formulator. (A) Precipitation measurements at varying concentrations of xylanase in 0.6 M potassium phosphate with 0.1 M Tris·HCl, pH 6.5. The standard deviation of pixel intensities provides a quantitative metric of protein precipitation in the ring reactor. Below the precipitation limit, the standard deviation shows constant background level with low variation. Above 12 mg/ml, the solution is in the precipitation regime, and the pixel standard deviation exhibits an approximately linear dependence on protein concentration. All points represent the mean of five identical experiments, with error bars indicating standard deviation of measurements. (B) Solubility fingerprints of xylanase over ≈4,300 chemical conditions. Each data series represents a separate fingerprinting experiment using the same basis of chemical conditions. The top solubility fingerprint (red) is generated by using a sample having elevated protein concentration (90 mg/ml) and exhibits both higher signal-to-noise ratio and additional peaks not present in the other data series (70 mg/ml). The two center solubility fingerprints were generated sequentially on a single device (first green, then blue) with the same loaded sample, demonstrating the stability of the protein over the time of the experiment (≈40 h). The bottom solubility fingerprint (pink) was generated on a separate device by using the same protein sample as the blue fingerprint, showing reproducibility across devices. (C) Comparison of xylanase phase mapping done on chip and in microbatch experiments. Conditions that gave rise to precipitation in microbatch format and in chip are represented by red squares and overlaid yellow circles, respectively. Conditions that did not produce precipitation in either format are shown as blue circles. (D) Reversibility of precipitation and solubility hysteresis for lysozyme observed by outward and return titrations from the origin. The red region shows the area of phase space in which precipitation was observed in both outward and return titrations. The yellow region is the area of hysteresis in which the protein was soluble for the outward titrations but precipitated in the return titrations. The blue region is the area in which the protein was soluble for both the outward and return titrations.
A coarse search of solubility space was performed initially, in which the protein sample was mixed with a large number of chemical formulations to identify conditions that produce nontrivial phase behavior. This generates a solubility fingerprint of the crystallization target in which each precipitation peak indicates a chemical formulation that exerts a pronounced effect on solubility. Fingerprints were generated for xylanase in four independent runs, each of which consists of ≈4,000 titration experiments (Fig. 4B). These fingerprints are highly reproducible and are characteristic of the protein studied. For example, although sodium chloride is a strong precipitating agent for chicken egg white lysozyme, it does not produce a precipitation peak in the solubility fingerprint of xylanase over the pH range studied.
Each solubility fingerprint was generated over a period of 20 h and consumed ≈8 μl of protein sample. Chemical formulations were created by flushing the ring with one of 16 buffers, injecting a precipitating agent (salt or polymer), diluting the ring with water, and then mixing. Protein sample was then introduced at a variety of concentrations and mixed before data acquisition. When a polymer was used as the major precipitating agent, a small amount of salt was also introduced as an additive. Experiments in Fig. 4B were grouped by the identity of the major precipitating agent so that each peak represents the effect of this reagent over a range of pH values and concentrations. The large width of these peaks indicates a high level of experimental redundancy, suggesting that a more efficient search could be conducted using fewer related chemical conditions.
The solubility fingerprint of xylanase reveals five salts (sodium citrate, dipotassium phosphate, ammonium sulfate, and sodium/potassium tartrate) as likely crystallizing agents. A high molecular-weight polymer (polyethylene glycol 8000) in combination with various salt additives was also identified to be a strong precipitating agent at high pH values. The high isoelectric point of xylanase suggests that the reduced effectiveness of this precipitant at low pH values is due to two-body electrostatic repulsion. A smaller molecular-weight polymer (polyethylene glycol 3350) was found to be a much weaker precipitating agent and was not investigated further in phase-space mapping experiments.
The formulator chip was then used to measure full-phase diagrams for the 24 most promising precipitation conditions. Each phase diagram consisted of a grid of 72 separate mixing experiments over all accessible protein and precipitant concentrations. All 24 phase spaces were generated sequentially on a single device by using <3 μl of protein sample (≈100 nl per phase space) and are included in Fig. 6, which is published as supporting information on the PNAS web site. A comparison of precipitation phase spaces measured for xylanase in chip (5-nl reactions) and in microbatch format under paraffin oil (5-μl reactions) shows good agreement in detecting the precipitation boundary (Fig. 4C). Because measurements of precipitation are made immediately after mixing (within 3 sec), the locus of points that separate the precipitated and soluble regions of the graph generates a precipitation curve that is distinct from the thermodynamic solubility curve.
Conditions that reside just below the precipitated region may be in a metastable state conducive to crystallization, so that a detailed knowledge of protein solubility behavior provides an empirical basis for the design of maximum likelihood crystallization trials. The 24 phase spaces generated for xylanase were used to rationally design an optimal crystallization screen consisting of 48 reagents. The formulations and protein concentrations used in the optimized screen are in Table 1, which is published as supporting information on the PNAS web site. A single-batch crystallization trial using the optimal screen was set by combining relative amounts of protein and precipitant stock so that the final condition was located on the boundary of the precipitation region. The efficiency of this screen was evaluated by comparison with standard commercially available sparse matrix screens (Crystal Screens I and II, Hampton Research; Wizards I and II; Emerald Biostructures, Bainbridge Island, WA). Two batch crystallization trials of 48 unique conditions were prepared for each of the four sparse matrix screens for a total of 384 individual assays
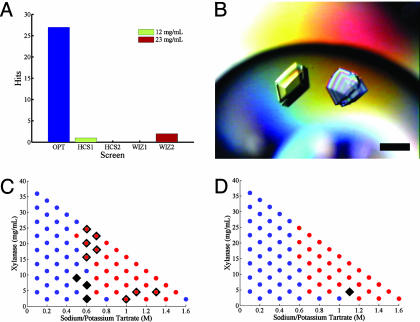
A list of the successful conditions from the rationally designed crystallization screen and from the sparse matrix screens is provided as Table 2, which is published as supporting information on the PNAS web site. Twenty-seven crystallization conditions were observed in the optimal screen compared with a total of three crystallization conditions in the eight standard sparse matrix screens. The use of ab initio solubility information therefore resulted in a 72-fold enrichment in crystallization success (Fig. 5A). A surprising result was that xylanase crystals were observed in the optimal screen for all of the major salt and polymer precipitants identified in the solubility fingerprint. This striking finding suggests that achieving optimal levels of supersaturation is more important in the crystallization of xylanase than is the broad sampling of chemical space. In cases such as this, systematic screening for crystallization using a reduced chemical space may prove more effective than sparse matrix strategies. Additionally, crystallization conditions were identified in the rationally designed screen that gave large single three-dimensional crystals suitable for diffraction studies without further optimization (Fig. 5B). In contrast, only flat plate clusters were observed in the standard sparse matrix screens.
Fig. 5.
Comparison of xylanase microbatch crystallization experiments using an optimal crystallization screen based on solubility phase spaces to commercially available sparse matrix screens. (A) Histogram showing number of successful crystallization conditions identified with sparse matrix screens (each at protein concentrations of 12 and 23 mg/ml) and optimal screen. (B) Polarized micrograph of large single crystals grown directly from optimal screen (16% polyethylene glycol 8000/65 mM sodium chloride/65 mM Tris·HCl, pH 8.2/42 mg/ml xylanase). (Bar, 200 μm.) (C and D) Comparison of phase-space behavior and crystallization variability of the original protein sample (C) and the second protein sample (D) in microbatch format. Conditions that gave rise to crystallization are shown as black triangles. Conditions that gave rise to immediate precipitation are represented as overlaid red circles. Blue circles represent conditions in which the protein was soluble and did not produce crystals.
To evaluate the influence of lot variability on these crystallization results, crystallization trials based on the optimal screen were repeated by using a new protein sample obtained from the same vendor and prepared identically as the original sample. Fourteen of 17 polyethylene glycol conditions that gave crystals in the original experiment were reproduced by using the second sample, compared with 1 of 10 for the salt-based conditions. To determine whether the variable crystallization behavior observed in salt-based conditions was due to variations in phase-space behavior, a complete phase space of one chemical formulation (sodium/potassium tartrate, Tris·HCl, pH 8.5) was measured in microbatch format for both samples (Fig. 5 C and D). It was discovered that, although both samples exhibited similar phase-space behavior, they produced different crystallization results. Eleven conditions produced crystals in the original sample compared with only one successful condition in the second sample. The reason for this difference in behavior is unclear but may be due to variable degrees of proteolysis or trace amounts of chemical contaminants introduced during purification or concentration steps. It is interesting that some crystallization conditions (e.g., those based on polyethylene glycol 8000) are more robust to batch-dependent perturbations than others.
Another application of protein solubility phase-space mapping is in transporting successful crystallization conditions from one experimental format to another. The successful crystallization of a protein is determined both by the established thermodynamic variables and the kinetic trajectory of an experiment. For this reason, experiments conducted with different crystallization kinetics (e.g., hanging-drop vapor diffusion, microbatch, and free-interface diffusion) and the same precipitating agents will not necessarily produce similar results. For example, the hydroxylase domain of a cytochrome p450 alkaline hydroxylase (Mutant 139-3 of BM-3) did not produce crystals in initial hanging-drop trials but was found to crystallize readily by microfluidic free interface diffusion (26) (1 part protein 20 mg/ml/1 part 30% m/v polyethylene glycol 8000/0.2 M sodium acetate/0.1 M Tris·HCl, pH 7.0). However, this condition was unsuccessful when set in hanging-drop vapor diffusion format, resulting only in amorphous precipitate. The microfluidic formulator was used to generate a phase space at constant buffer and salt concentration (100 mM Tris·HCl, pH 7.3/200 mM sodium acetate) with polyethylene glycol concentration and protein concentration as variables. Two hanging-drop experiments were designed to equilibrate near the solubility limit determined from the phase-space map. One condition (8 μl of 35 mg/ml protein sample mixed with 6.7 μl of 10% polyethylene glycol/100 mM sodium acetate/50 mM Tris·HCl, pH 7.3, and equilibrated at 20°C against 1 mM 20% polyethylene glycol/200 mM sodium acetate/100 mM Tris·HCl, pH 7.3) produced crystals within 3 days. This success demonstrates the usefulness of solubility mapping in transporting conditions across crystallization formats.
Finally, we used the formulator to make a direct observation of the supersaturation region of chicken egg white lysozyme. The concentrations of salt and lysosyme were manipulated while keeping the buffer concentration constant to evolve the chemical state of the mixing ring radially out from the origin and then back again. Measurements of precipitation were taken at ≈1-min intervals. The addition of a family of such radial titrations was used to generate two phase-space diagrams for chicken egg white lysosyme: one for the outward and one for the return titrations (Fig. 4D). The first observation of protein precipitation appears at higher salt and protein concentration during the outward trajectory than on the return path, thereby exhibiting solubility hysteresis. The intersection of the soluble region of the outward phase space with the precipitated region of the return path phase space provides a direct observation of a metastable regime in which the aggregate phase is thermodynamically stable but not observed at short times. The observation of the reversible formation of a protein aggregate may be used to distinguish between denatured and well folded protein aggregates. Additionally identified metastable regions in phase space provide likely candidates for crystal seeding and growth experiments.
Conclusion
We have shown that complex sample processing at the nanoliter scale allows for a practical implementation of automated protein solubility characterization. Ab initio solubility information obtained through systematic protein phase-space mapping provides a physical basis for the design of optimal crystallization screens, giving rise to dramatic enrichment in crystallization success. It must be noted that chemical conditions and phase behavior are not the only variables that can be adjusted in the search for good crystals; it is often equally important to tune the properties of the protein by creating point mutants with or truncated constructs. However, the path to crystallization always includes extensive chemical screening with precious protein sample, and for this step it appears that microfluidic formulation devices can play an important role. Beyond applications in protein crystallization, the formulation capability of this device should find diverse applications in areas such as combinatorial chemistry, chemical synthesis, and cell culture studies.
Supplementary Material
Acknowledgments
We thank Sebastian Maerkl and James Berger for helpful conversations and insights. This work was supported by the National Science Foundation, the Defense Advanced Research Planning Agency, and the Army Research Office. C.L.H. was partially funded by the Natural Sciences and Engineering Research Council (Canada) (Julie Payette Fellowship).
Abbreviation: PCI, positive displacement crossinjection.
References
- 1.Kendrew, J. C., Bodo, G., Dintzis, H. M., Parrish, R. G., Wyckoff, H. & Phillips, D. C. (1958) Nature 181, 662-666. [DOI] [PubMed] [Google Scholar]
- 2.Loll, P. J. (2003) J. Struct. Biol. 142, 144-153. [DOI] [PubMed] [Google Scholar]
- 3.Chayen, N. E. & Saridakis, E. (2002) Acta Crystallogr. D 58, 921-927. [DOI] [PubMed] [Google Scholar]
- 4.Luft, J. R. & DeTitta, G. (1997) Macromol. Crystallogr. A. 276, 110-131. [Google Scholar]
- 5.Rosenbaum, D. F. & Zukoski, C. F. (1996) J. Cryst. Growth 169, 752-758. [Google Scholar]
- 6.Ataka, M. (1993) Phase Transitions 45, 205-219. [Google Scholar]
- 7.Carbonnaux, C., Rieskautt, M. & Ducruix, A. (1995) Protein Sci. 4, 2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikol, V. & Giege, R. (1989) J. Cryst. Growth 97, 324-332. [Google Scholar]
- 9.Howard, S. B., Twigg, P. J., Baird, J. K. & Meehan, E. J. (1988) J. Cryst. Growth 90, 94-104. [Google Scholar]
- 10.Kam, Z., Shore, H. B. & Feher, G. (1978) J. Mol. Biol. 123, 539-555. [DOI] [PubMed] [Google Scholar]
- 11.Muschol, M. & Rosenberger, F. (1997) J. Chem. Phys. 107, 1953-1962. [Google Scholar]
- 12.Forsythe, E. L., Judge, R. A. & Pusey, M. L. (1999) J. Chem. Eng. Data 44, 637-640. [Google Scholar]
- 13.Saridakis, E. E. G., Stewart, P. D. S., Lloyd, L. F. & Blow, D. M. (1994) Acta Crystallogr. D 50, 293-297. [DOI] [PubMed] [Google Scholar]
- 14.Saridakis, E. & Chayen, N. E. (2003) Biophys. J. 84, 1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter, C. W., Baldwin, E. T. & Frick, L. (1988) J. Cryst. Growth 90, 60-73. [Google Scholar]
- 16.Jancarik, J. & Kim, S. H. (1991) J. Appl. Crystallogr. 24, 409-411. [Google Scholar]
- 17.tenWolde, P. R. & Frenkel, D. (1997) Science 277, 1975-1978. [DOI] [PubMed] [Google Scholar]
- 18.DeLucas, L. J., Bray, T. L., Nagy, L., McCombs, D., Chernov, N., Hamrick, D., Cosenza, L., Belgovskiy, A., Stoops, B. & Chait, A. (2003) J. Struct. Biol. 142, 188-206. [DOI] [PubMed] [Google Scholar]
- 19.George, A. & Wilson, W. W. (1994) Acta Crystallogr. D 50, 361-365. [DOI] [PubMed] [Google Scholar]
- 20.Guo, B., Kao, S., McDonald, H., Asanov, A., Combs, L. L. & Wilson, W. W. (1999) J. Cryst. Growth 196, 424-433. [Google Scholar]
- 21.Unger, M. A., Chou, H. P., Thorsen, T., Scherer, A. & Quake, S. R. (2000) Science 288, 113-116. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, G. (1953) Proc. R. Soc. London Ser. A 219, 186-203. [Google Scholar]
- 23.Chu, H., Unger, M. A. & Quake, S. R. (2001) Biomed. Microdev. 3, 323-330. [Google Scholar]
- 24.Stura, E. A., Nemerow, G. R. & Wilson, I. A. (1992) J. Cryst. Growth 122, 273-285. [Google Scholar]
- 25.Santesson, S., Cedergren-Zeppezauer, E. S., Johansson, T., Laurell, T., Nilsson, J. & Nilsson, S. (2003) Anal. Chem. 75, 1733-1740. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, C. L., Skordalakes, E., Berger, J. M. & Quake, S. R. (2002) Proc. Natl. Acad. Sci. USA 99, 16531-16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng, B., Roach, L. S. & Ismagilov, R. F. (2003) J. Am. Chem. Soc. 125, 11170-11171. [DOI] [PubMed] [Google Scholar]
- 28.Thorsen, T., Maerkl, S. J. & Quake, S. R. (2002) Science 298, 580-584. [DOI] [PubMed] [Google Scholar]
- 29.Torronen, A., Harkki, A. & Rouvinen, J. (1994) EMBO J. 13, 2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torronen, A. & Rouvinen, J. (1995) Biochemistry 34, 847-856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.