Abstract
Bovine insulin has long been known to self-assemble in vitro into amyloid fibrils. We have observed a further higher-order self-association of the protein into spherical structures, with diameters typically around 50 μm but ranging from 10 to 150 μm. In a polarizing light microscope, these structures exhibit a “Maltese-cross” extinction pattern typical of spherulites. Spherical structures of a similar size distribution can be observed in the environmental scanning electron microscope, which also reveals the presence of significant amounts of water in the structures. The spherulites contain a large quantity of well defined amyloid fibrils, suggesting that they are formed at least in part as a consequence of the self-assembly of preformed fibrils. Similar structures also have been observed in the tissues of patients suffering from amyloid disorders. The ability of amyloid fibrils to form such higher-order assemblies supports the hypothesis that they represent a generic form of polypeptide structure with properties that are analogous to those of classical synthetic polymers.
Spherulite formation is a very common form of self-assembly observed during polymer crystallization (1, 2), particularly from synthetic polymer melts. It also is a commonly observed phenomenon associated with crystallization in many other types of systems including metals (2). In addition, in the biphasic region of lyotropic liquid crystalline polymers, when the isotropic and anisotropic phases coexist, the anisotropic phase often forms spherulitic drops within an isotropic matrix (3). This paper describes the observation and preliminary structural study of spherulites formed through the self-assembly of amyloid fibrils, thread-like assemblies most commonly associated with protein deposition disorders such as Alzheimer's disease and type II diabetes.
The basic structural characteristics of spherulites have largely been inferred from polarized light microscopy. Under the polarizing microscope, spherulites show a typical “Maltese cross” pattern of light extinction. The process by which a spherulite self-assembles is best documented for synthetic polymers such as polyethylene (PE), although some details are still being elucidated for this system even after 50 years of research (1). For flexible polymer chains such as those of PE, the basic building block in the assembly process is a structure in which the polymer chain folds back on itself repeatedly in a rather regular pattern, forming lamellar crystals, which have a well defined thickness, whose value is determined by the conditions under which they form. Typically, the lamellae are ≈10 nm thick (along the chain length); laterally, however, their length is on the order of micrometers, with the top and bottom surfaces known as fold surfaces, because this is where the chain folding occurs. This arrangement, however, is never perfect, because the chains in the lamellae do not all fold in exactly the same way, resulting in “rough” fold surfaces. Furthermore, some chains are only anchored in the lamellae at one end, protruding from the surface at the other end, resulting in so-called cilia (1).
The lamellae of PE are observed to form particularly readily, as melts of this polymer are cooled. The presence of cilia, however, prevents the lamellae from lying parallel to each other, resulting in the formation of structures resembling a wheat sheaf. In effect, the lamellae exhibit a splay form, usually at angles of ≈20°. As they assemble, “subsidiary” lamellae can form in the spaces between the so-called “dominant” lamellae at angles that are similar to those observed between the dominant lamellae. Furthermore, any areas that are still vacant can be taken up by polymer chains not in lamellae. In time, this structure extends outwards, resulting in an approximately spherical overall shape (1). The lamellae lie in a radial orientation, and as the polymer chains lie perpendicular to the fold surfaces of the lamellae, the chains are oriented tangentially in the spherulite.
The formation of higher-order semicrystalline assemblies, including spherulites, has been observed in many other systems, including natural polymers. In carbohydrates, for example, synthetic cellulose (4), chitin and chitosan (5, 6), and amylose (7) readily form spherulites in vitro, and the morphology of the starch granules formed in vivo has been described as spherulitic (8). Chitin and cellulose are known to form crystallites, rod-like particles formed by the lateral association of chitin molecules (9, 10). In spherulites, the crystallites are suggested to lie along the radius (radial or positive spherulites) or along the circumference (tangential or negative spherulites) (4, 6). Several other systems also are known to form spherulites: sodium adducts of DNA (11), proteins such as lysozyme (12, 13) and carboxypeptidase (14) under crystallization conditions, tubules from bacteriochlorophyll protein (15), and fibers formed from hemoglobin S, a mutational variant of the human protein leading to sickle cell anemia (16).
The characteristic Maltese-cross pattern of spherulites when viewed under a polarizing microscope has been observed in brain sections from human patients with a particular strain of Creutzfeldt-Jakob disease (CJD) (17) and from a rat model of Alzheimer's disease (18). Such tissue samples have been shown to contain amyloid fibrils, large thread-like assemblies of peptide or protein molecules, containing multiple copies of a single polypeptide chain in highly ordered arrays that are rich in β-sheet structure. The fibrils are typically long (often >1 μm) and unbranched, with diameters generally between 6 and 12 nm (19). All amyloid fibrils possess a common underlying structure irrespective of the polypeptide precursor and give rise to a “cross-β” x-ray fiber diffraction pattern indicating that the β-strands lie perpendicular to the fibril axis (19). Formation of these structures is best known through their association with many types of pathological conditions including Alzheimer's disease, CJD, type II diabetes, and a wide range of other systemic and neurological disorders (20). Recent evidence suggests, however, that the ability to form amyloid fibrils is a common and perhaps generic property of polypeptide chains, because similar structures can be formed in vitro from a variety of peptides and proteins (21, 22).
Amyloid plaques associated with Alzheimer's disease, Gerstmann-Strausler-Scheinker disease, and Down's syndrome have recently been found to contain fibrils that are aligned in a radial fashion (23). Maltese-cross extinction patterns, indicative of spherulitic structures, also have been observed in amyloid deposits associated with mammary tumors in dogs (24, 25), and in amyloid tumors, localized nodular masses of amyloid deposits not linked to systemic amyloidoses, observed in the jejunum, part of the small intestine (26). The possible significance of spherulitic deposits in the development of these diseases has not, however, been established. The formation of spherulites in solutions containing amyloid fibrils in vitro has been reported for a pathogenic Ig light chain (27), for several different synthetic peptides (28), including a helix-turn-helix peptide (29), for gels of β-lactoglobulin (30, 31), and for peptide analogues of silkmoth eggshell proteins, chorion (32). Furthermore, spherical aggregates have been observed in solutions of the nonpathogenic protein α-L-iduronidase (33), in gels formed from solutions of a synthetic peptide derived from the β-sheet domain of platelet factor 4 (34) and in solutions of the 40-residue Aβ peptide implicated in Alzheimer's disease, where the spherical aggregates were termed “β-amy balls” (35). Although in these three cases the spherical aggregates could be spherulitic in nature, the lack of optical birefringence data prevents a definitive characterization at this stage.
Insulin is a highly flexible polypeptide system with which to study the formation and properties of amyloid fibrils. It readily self-assembles after incubation in solutions at pH 2.0 and temperatures above 30°C, forming viscous solutions that contain fibrils with an average diameter of ≈8 nm (36-38). In this paper, we describe the characterization of spherulites formed in such solutions by optical microscopy and environmental scanning electron microscopy (ESEM).
Materials and Methods
Insulin Solutions and Incubation Conditions. Bovine insulin and all other chemicals were of analytical grade or better and were obtained from Sigma-Aldrich and used without further purification. Solutions were made up by weighing out the required amount of insulin and dissolving the dry powder in distilled and deionized water adjusted to pH 2.0 with HCl. The pH was then checked and, if necessary, adjusted with solutions of HCl or NaOH. It was found by UV absorption at 280 nm that insulin concentrations obtained by this method were precise to within 10%. Solutions were incubated in glass vials at temperatures of 37°C or 65°C for up to 24 h. Under these conditions, chemical modification of bovine insulin is limited to a degree of deamidation (39).
For Fourier transform infrared (FTIR) measurements, bovine insulin was first dissolved in D2O at pH 2.0, left for several hours, and freeze-dried. This procedure was repeated three times. Solutions of the deuterated material were made up in D2O, and the pH was adjusted to a meter reading of 2.0 with DCl. No correction was made for any isotope effects. These solutions were then incubated as described above.
Optical Microscopy and ESEM. After incubation, aliquots of insulin solutions were removed from the glass vials by using Pasteur pipettes and put onto microscope slides. The slides were then studied by using an optical microscope (Axioplan, Zeiss) without coverslips and at magnifications of up to ×50. The polarizer and analyzer were in a fixed position (east-west and north-south on the images, as shown). A 551-nm retardation plate was inserted at 45° to the polarizers, with the slow axis running northeast-southwest and the fast axis running northwest-southeast. Digital images were taken, and scale bars were obtained by taking images of a calibration slide with the same settings on the microscope and the digital camera.
For ESEM, aliquots of incubated solutions were taken and placed on copper stubs inside the microscope chamber of an Electroscan ESEM 2010 (FEI UK, Cambridge, U.K.). The samples were left to equilibrate at 2°C (temperature controlled by a Peltier device under the copper stub). A few drops of distilled and deionized water were placed around the sample, and the chamber was sealed and evacuated initially to ≈6 Torr. The chamber was flooded repeatedly with water vapor, and the pressure was reduced slowly to ≈5 Torr. Further decreases in pressure led to evaporation of the water and drying of the samples; conversely, increasing the pressure resulted in the deposition of larger quantities of water on the sample.
Transmission Electron Microscopy (TEM). Copper 400 mesh grids (Agar Scientific, Stansted, U.K.) were coated with Formvar and carbon film. Insulin solutions were sonicated at 30-40 kHz in an Ultrawave bath (Fisher Scientific) for 60-90 s. Insulin solutions were subsequently diluted 10- to 100-fold, and 3.5-μl aliquots were placed on the grids. After 30 s, two drops containing 15 μl of 2% uranyl acetate (Agar Scientific) were then placed on the grid and left for 10-15 s. Excess water was removed with filter paper, and the grid was then left to air-dry. Imaging was done by using an electron microscope (Technai 20, FEI) with an acceleration voltage of 120 kV and magnifications typically around ×26,000.
FTIR Spectra and Deconvolution Procedures. Solutions of insulin were centrifuged after incubation, the supernatant was removed, and the pellet was resuspended in D2O pH 2.0. This procedure was repeated three times, after which the pellet was placed between CaF2 windows with a 50-μm Mylar spacer, and 64 spectra were collected and averaged by using a FTIR spectrometer (Equinox 55, Bruker, Billerica, MA). Spectra of D2O at pH 2.0 and spectra of water vapor also were collected and used for background and baseline corrections. Spectra were fitted to mixtures of Gaussian and Lorentzian peaks, with peak positions obtained from second-derivative spectra.
Thioflavin-T (ThT) Measurements. Aliquots (5 μl) of insulin solutions after incubation were added to 995 μl of a 10 mM Na2H/NaH2PO4, 150 mM NaCl, and 50 μM ThT solution and stirred for 30 s. By using a fluorescence spectrophotometer (Cary Eclipse, Varian), fluorescence emission intensity was measured for 30 s at 482 nm (10-nm slit width), exciting at 440 nm (5-nm slit width). Values were compared with those of the ThT solution and of samples taken from the insulin solution before any incubation.
Results
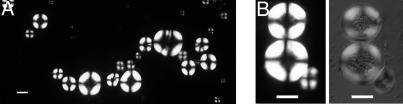
Prolonged incubation of 1 mM solutions of insulin at pH 2.0 and 37°C or 65°C resulted in the formation of a precipitate that collected at the bottom of the vials within a few hours. Optical microscopy revealed that the precipitate consisted largely of spherical particles ranging in size from 10 to 150 μm, with the most common size being ≈50 μm. Between crossed polarizers, a clear Maltese-cross extinction pattern could be observed (Fig. 1 A and B), indicating that the structures present had a spherulitic morphology. The spherulite centers contained substantial quantities of protein (as indicated by differential interference contrast microscopy in Fig. 1B Right), but the lack of birefringence indicated that the polypeptide chains in these regions are unlikely to be oriented in any regular manner. This nonbirefringent core is present even if the insulin solutions are filtered through 0.22-μm filters before incubation, indicating that the core is unlikely to be any external contaminant or preformed aggregate.
Fig. 1.
Insulin spherulites observed by polarized optical microscopy. (A) Representative polarized optical microscopy images of insulin spherulites formed by incubating solutions containing 1 mM insulin at pH 2.0 and 65°C for several hours. A distribution of spherulite sizes is clearly observable. (B Left) Insulin spherulites, as viewed between crossed polars. (B Right) Differential interference contrast microscopy shows that the central region of the spherulites is made up of material that is not oriented in a regular manner. (Scale bar, 50 μm.)
As samples of the solutions were air-dried under the optical microscope, the spherulites were observed to shrink and deform; ultimately, the Maltese-cross pattern disappeared, leaving only patches of weakly birefringent material. Rehydration of the sample did not reestablish the Maltese-cross extinction pattern, indicating a permanent disruption of the spherulite structure.
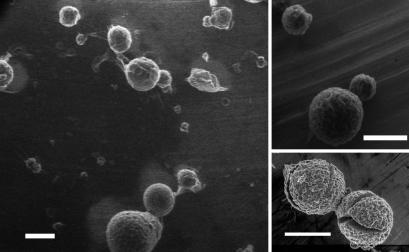
The spherulites also were imaged by using ESEM, a technique that allows the investigation of wet and insulating samples without the need for extensive sample preparation (e.g., fixing or Au-coating). As such, it is an ideal method for studying biological samples as well as other “soft” or insulating materials (40). Furthermore, the sample chamber can be flooded with water vapor, allowing control over the wetness of the sample by controlling the pressure in the sample chamber and the sample temperature (40). By maintaining the sample temperature at 2°C and the chamber pressure at ≈5 Torr, the vapor pressure of water at this temperature, the hydration state of the sample could be controlled. At higher pressures, water condensed onto the samples, as a result of which no spherulites could be discerned. Conversely, at pressures below the vapor pressure of water at 2°C, the evaporation of water from the sample could be readily controlled, ultimately revealing spheres with radii similar to those observed for the spherulites in microscopy (Fig. 2). Further reductions in the chamber pressure resulted in the spherulites shrinking in size, presumably because of, at least in part, the removal of water from their interiors, ultimately resulting in the appearance of cracks on the surface (Fig. 3). Increasing the pressure inside the chamber at this stage resulted in the deposition of a layer of water on the spherulites, and surface cracks were often seen to close. The structures did not anneal completely, however, because a second reduction in chamber pressure and concomitant loss of water resulted in the reopening of the same cracks, often accompanied by new ones (data not shown).
Fig. 2.
Insulin spherulites recorded by ESEM. Images were obtained from solutions incubated at pH 2.0 and 65°C for several hours. Note the rough surface of the spherulites and the variation in their size. Some fractured spherulites also are shown. (Scale bar, 50 μm.)
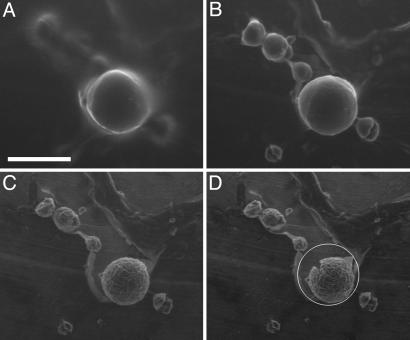
Fig. 3.
Changes in spherulite morphology on drying. ESEM images of a group of spherulites as the pressure is reduced from 5.4 to 4.8 Torr. (A) At 5.4 Torr, a spherulite becomes visible, surrounded by water. Smaller spherulites are apparent but ill defined. (B) At 5.3 Torr, most of the surface water has evaporated, although the spherulite surface is still smooth, possibly covered with water. The smaller spherulites, several of which have fractured into smaller pieces, are now clearly visible. (C) At 4.9 Torr, water has been removed from the spherulites; the surfaces are rough, and further shrinkage has occurred. (D) At 4.8 Torr, the sample is now drier still, and the larger spherulite has developed cracks. The white circle indicates the original size of the spherulite observed in A. (Scale bar, 100 μm.)
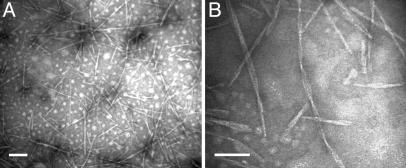
It is well known that, under the conditions described here (low pH and high temperature), insulin forms amyloid fibrils (36-38). Direct application of solutions containing spherulites to grids resulted in deposits that were too thick to be investigated by TEM. Dilution and sonication of spherulite solutions, however, resulted in preparations that could be investigated by TEM, presumably resulting from substantial disruption of the spherulites. Grids thus prepared revealed the presence of large numbers of structures that are typical of amyloid fibrils (Fig. 4).
Fig. 4.
Amyloid fibrils formed from bovine insulin. (A) Solutions containing spherulites were sonicated before application to TEM grids. (B) Many fibrils appear to be twisted when examined at higher magnification. (Scale bar, 100 nm.)
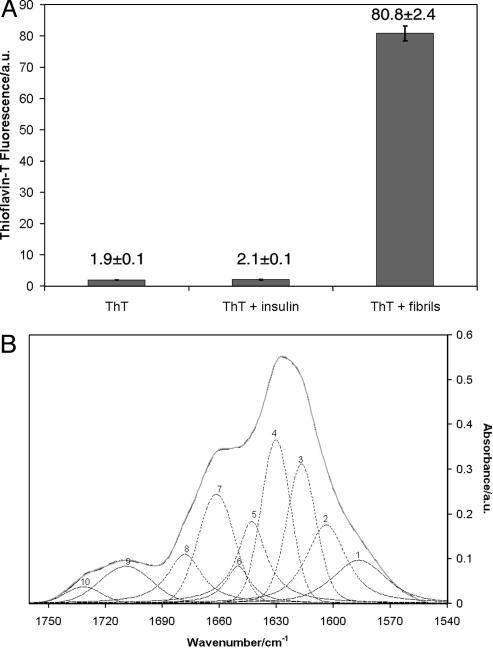
Aliquots of nondiluted and nonsonicated solutions containing spherulites were added to solutions of ThT (Fig. 5A). The enhanced fluorescence relative to solutions of ThT and to solutions of ThT containing freshly dissolved insulin indicated the presence of amyloid structures (41). By measuring the concentration of protein in the supernatant after centrifugation, it was found that 90 ± 3% of the protein in samples incubated for 24 h at 65°C had been converted into fibrillar or spherulitic structures. It was further shown by FTIR (Fig. 5B) that the pellets resulting from centrifugation of incubated deuterated insulin solutions contained significant quantities of β-sheet (53 ± 1%) and turn (24 ± 1%) structures. Because turns are most commonly found linking other elements of secondary structure (42), this result implies that up to 77% of the material in the centrifuged pellets is involved in β-structure; this finding is similar to that reported by Bouchard et al. (38), who observed that amyloid fibrils of insulin contained as much as 90% of the protein in a β-sheet state. The remaining 23% of the protein structure in the present study was either α-helical (17 ± 1%) or random-coil (6 ± 0.5%). Because FTIR characterizes all of the material in the pellet, i.e., all of the aggregate species, these data would be consistent with a model in which amyloid fibrils form the birefringent regions of the spherulites, with additional, perhaps non-β-sheet material forming the core region of the spherulites visible in the polarized optical microscope images (Fig. 1B).
Fig. 5.
ThT fluorescence and FTIR spectra of insulin solutions containing spherulites. (A) Fluorescence of a solution of ThT only, of ThT with added insulin, before (ThT + insulin) and after (ThT + fibrils) incubation at 65°C for several hours. Three different solutions were used in each experiment. For clarity, the value and error of each measurement are shown on the graph. (B) FTIR spectra of pellets formed by centrifugation of solutions of insulin that were incubated at 65°C. The secondary structure content, based on fitted peak areas, is α-helix 17 ± 1%, β-sheet 53 ± 1%, random-coil 6 ± 0.5%, turns 24 ± 1%. Assignments are based on published procedures (49-51).
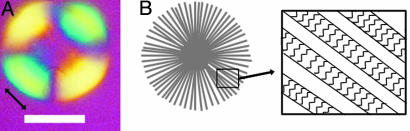
To reveal the orientation of the fibrils within the spherulites, a full-wave retardation plate was inserted at a 45° angle to the polarizers. The four quadrants of the spherulite are now colored (Fig. 6A). The appearance of the color blue indicates that the fast optical axis of the spherulites is aligned with the fast optical axis of the wave retardation plate (43). Amyloid fibrils of insulin and other proteins have been shown to have their slow optical axis parallel to the fibril axis (i.e., they are positively birefringent) (43-45). Thus, as in the plaques associated with Alzheimer's disease, Gerstmann-Strausler-Scheinker disease, and Down's syndrome (23), the fibrils are arranged radially in the spherulites, as shown in schematic form in Fig. 6B.
Fig. 6.
Schematic structure of insulin spherulites. (A) Optical microscopy image of a spherulite recorded after the insertion of a wave plate. The double-headed arrow indicates the orientation of the fast optical axis of the wave plate. (Scale bar, 50 μm.) (B) Schematic representation of the proposed structure of the spherulites. The expanded picture shows individual fibrils illustrating that the β-strands that assemble to form sheets are oriented perpendicular to the fibril axis. A theoretical wave plate, superimposed onto this picture in the same orientation as in A, would result in the fast axis of the fibrils, the protein backbone, being perpendicular to the fast axis of the wave plate, resulting in the yellow color seen in A.
Discussion
Under conditions where insulin is known to form amyloid fibrils, we have observed the formation of spherical aggregates on a length scale of 10-150 μm, very significantly larger than that characteristic of the fibrils. In addition, the aggregates show a Maltese-cross extinction pattern when observed in the polarizing microscope, indicating that they have a spherulitic structure. The core of each spherulite appears to be nonbirefringent, but differential interference contrast microscopy reveals the presence of protein molecules that are probably less regularly packed. Remarkably, species similar to these spherulites were observed >50 years ago in the light microscope and called “spherites” (36). A combination of TEM, FTIR, and ThT fluorescence data has revealed the presence of amyloid fibrils in the spherulites, suggesting that the noncore regions of these particles consist of these species oriented in a radial fashion. The spherical structures also could be observed by ESEM and appear similar to those observed by SEM for aggregates of peptides from the β-sheet domain of human platelet factor 4 (34), structures formed in i-poly(4-methyl pentene-1) melts (1), and some forms of amylose (7). Further observations of these structures by optical microscopy and ESEM show that they are unstable when dried, resulting in their deformation and, ultimately, in the loss of the Maltese-cross extinction pattern.
We have demonstrated here that spherulites can readily assemble in insulin samples in which amyloid formation is occurring. The mechanism by which bovine insulin forms the observed spherulites is currently under investigation.
Because the literature indicates that spherulites could be prevalent in many other amyloid-containing systems (17, 18, 23-35), it is possible that the ability to form such structures is a property common to all, or at least most, amyloid fibril systems. From the data presented here, and from the wider protein literature, it appears that once biological control of the native structure of a protein has been lost for whatever reason, the assembly of proteins into organized structures can proceed via a pathway not dissimilar to that of typical synthetic polymers. These observations would suggest that the laws of polymer physics could be applied to these systems, and an apparently common behavior is seen in misfolded protein systems, just as the ability to form amyloid fibrils has been described as a generic property of proteins (22). This seemingly common ability can therefore explain the occurrence of spherulitic structures in the body, once the local concentration of amyloid fibrils is sufficiently high; a similar concentration effect has been observed in the formation of higher-order assemblies by synthetic peptides (46). Although the importance of such large aggregates on human health has not yet been established, it is possible, as has been suggested for amyloid fibrils themselves (47), that the assembly process acts to sequester the amyloid aggregates and their precursors into localized regions and, hence, to help protect the organism from their toxic properties.
The results of the present study therefore provide strong evidence that the structures formed by polypeptide chains, when released from the constraints of their functional biological environments, are characteristic of synthetic polymers. A full understanding of the assembly process that leads from amyloid fibrils to spherulites is likely to lead not just to advances in our understanding of neurodegeneration and other amyloid diseases, but also to a fundamental understanding of the assembly characteristics of amyloid fibrils that is likely to be important for their exploitation as novel biomaterials and in nanotechnology (21, 48).
Acknowledgments
We thank Dr. Neville Boden for helpful discussions and Dr. Jesús Zurdo for help with the FTIR spectra and fitting. This work was funded by the Engineering and Physical Sciences Research Council (A.F.M. and M.R.H.K.), the Royal Society (C.E.M.), and a Wellcome Trust Programme Grant (to C.M.D.).
Abbreviations: ESEM, environmental scanning electron microscopy; FTIR, Fourier transform infrared; TEM, transmission electron microscopy; ThT, thioflavin T.
References
- 1.Bassett, D. C. (2003) J. Macromol. Sci. B Phys. 42, 227-256. [Google Scholar]
- 2.Magill, J. H. (2001) J. Mater. Sci. 36, 3143-3164. [Google Scholar]
- 3.Donald, A. M. & Windle, A. H. (1992) Liquid Crystalline Polymers (Cambridge Univ. Press, Cambridge, U.K.).
- 4.Kobayashi, S., Hobson, L. J., Sakamoto, J., Kimura, S., Sugiyama, J., Imai, T. & Itoh, T. (2000) Biomacromol. 1, 168-173. [DOI] [PubMed] [Google Scholar]
- 5.Murray, S. B. & Neville, A. C. (1998) Int. J. Biol. Macromol. 22, 137-144. [DOI] [PubMed] [Google Scholar]
- 6.Murray, S. B. & Neville, A. C. (1997) Int. J. Biol. Macromol. 20, 123-130. [DOI] [PubMed] [Google Scholar]
- 7.Ring, S. G., Miles, M. J., Morris, V. J., Turner, R. & Colonna, P. (1987) Int. J. Biol. Macromol. 9, 158-160. [Google Scholar]
- 8.Marchant, J. L. & Blanshard, J. M. V. (1980) Stärke 7, 223-226. [Google Scholar]
- 9.Revol, J.-F., Bradford, H., Giasson, J., Marchessault, R. H. & Gray, D. G. (1992) Int. J. Biol. Macromol. 14, 170-172. [DOI] [PubMed] [Google Scholar]
- 10.Revol, J.-F. & Marchessault, R. H. (1993) Int. J. Biol. Macromol. 15, 329-335. [DOI] [PubMed] [Google Scholar]
- 11.Rill, R. L. (1986) Proc. Natl. Acad. Sci. USA 83, 342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka, S., Yamamoto, M., Ito, K., Hayakawa, R. & Ataka, M. (1997) Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 56, R67-R69. [Google Scholar]
- 13.Chow, P. S., Zhang, J., Liu, X. Y. & Tan, R. B. H. (2002) Int. J. Mod. Phys. B 16, 354-358. [Google Scholar]
- 14.Coleman, J. E., Allan, B. J. & Vallee, B. L. (1960) Science 131, 350-352. [DOI] [PubMed] [Google Scholar]
- 15.Olson, R. A. (1970) Science 169, 81-82. [DOI] [PubMed] [Google Scholar]
- 16.Hofrichter, J. (1986) J. Mol. Biol. 189, 553-571. [DOI] [PubMed] [Google Scholar]
- 17.Manuelidis, L., Fritch, W. & Xi, Y.-G. (1997) Science 277, 94-98. [DOI] [PubMed] [Google Scholar]
- 18.Snow, A. D., Sekiguchi, R., Nochlin, D., Fraser, P., Kimata, K., Mizutani, A., Arai, M., Schreier, W. A. & Morgan, D. G. (1994) Neuron 12, 219-234. [DOI] [PubMed] [Google Scholar]
- 19.Sunde, M. & Blake, C. C. F. (1997) Adv. Protein Chem. 50, 123-159. [DOI] [PubMed] [Google Scholar]
- 20.Pepys, M. B. (1996) in Oxford Textbook of Medicine, eds. Weatherall, D. J., Ledingham, J. G. G. & Warrell, D. A. (Oxford Univ. Press, Oxford), Vol. 2, pp. 1512-1524. [Google Scholar]
- 21.MacPhee, C. E. & Dobson, C. M. (2000) J. Am. Chem. Soc. 122, 12707-12713. [Google Scholar]
- 22.Dobson, C. M. (2003) Nature 426, 884-890. [DOI] [PubMed] [Google Scholar]
- 23.Jin, L.-W., Claborn, K. A., Kurimoto, M., Geday, M. A., Maezawa, I., Sohraby, F., Estrada, M., Kaminsky, W. & Kahr, B. (2003) Proc. Natl. Acad. Sci. USA 100, 15294-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vos, J. H. & Gruys, E. (1985) Vet. Pathol. 22, 347-354. [DOI] [PubMed] [Google Scholar]
- 25.Taniyama, H., Kitamura, A., Kagawa, Y., Hirayama, K., Yoshino, T. & Kamiya, S. (2000) Vet. Pathol. 37, 104-107. [DOI] [PubMed] [Google Scholar]
- 26.Acebo, E., Mayorga, M. & Val-Bernal, J. F. (1999) Pathology 31, 8-11. [DOI] [PubMed] [Google Scholar]
- 27.Raffen, R., Dieckman, L. J., Szpunar, M., Wunschl, C., Pokkuluri, P. R., Dave, P., Wilkins-Stevens, D., Cai, X., Schiffer, M. & Stevens, F. J. (1999) Protein Sci. 8, 509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aggeli, A., Bell, M., Carrick, L. M., Fishwick, C. W. G., Harding, R., Mawer, P. J., Radford, S. E., Strong, A. E. & Boden, N. (2003) J. Am. Chem. Soc. 125, 9619-9628. [DOI] [PubMed] [Google Scholar]
- 29.Fezoui, Y., Hartley, D. M., Walsh, D. M., Selkoe, D. J., Osterhout, J. J. & Teplow, D. B. (2000) Nat. Struct. Biol. 7, 1095-1099. [DOI] [PubMed] [Google Scholar]
- 30.Sagis, L. M. C., Veerman, C., Ganzevles, R., Ramaekers, M., Bolder, S. G. & van der Linden, E. (2002) Food Hydrocoll. 16, 207-213. [Google Scholar]
- 31.Bromley, E. H. C., Krebs, M. R. H. & Donald, A. M. (2004) Faraday Discuss. 128, in press. [DOI] [PubMed]
- 32.Hamodrakas, S. J., Hoenger, A. & Iconomidou, V. A. (2004) J. Struct. Biol. 145, 226-235. [DOI] [PubMed] [Google Scholar]
- 33.Ruth, L., Eisenberg, D. & Neufeld, E. F. (2000) Acta Crystallogr. D 56, 524-528. [DOI] [PubMed] [Google Scholar]
- 34.Lockwood, N. A., van Tankeren, R. & Mayo, K. H. (2002) Biomacromolecules 3, 1225-1232. [DOI] [PubMed] [Google Scholar]
- 35.Westlind-Danielsson, A. & Arnerup, G. (2001) Biochemistry 40, 14736-14743. [DOI] [PubMed] [Google Scholar]
- 36.Waugh, D. F. (1946) J. Am. Chem. Soc. 68, 247-250. [Google Scholar]
- 37.Nielsen, L., Khurana, R., Coats, A., Frokjaer, S., Brange, J., Vyas, S., Uversky, V. N. & Fink, A. L. (2001) Biochemistry 40, 6036-6046. [DOI] [PubMed] [Google Scholar]
- 38.Bouchard, M., Zurdo, J., Nettleton, E. J., Dobson, C. M. & Robinson, C. V. (2000) Protein Sci. 9, 1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson, M. R. & Dobson, C. M. (2003) Protein Sci. 12, 2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donald, A. M. (2003) Nat. Mater. 2, 511-515. [DOI] [PubMed] [Google Scholar]
- 41.Naiki, H., Higuchi, K., Hosokawa, M. & Takeda, T. (1989) Anal. Biochem. 177, 244-249. [DOI] [PubMed] [Google Scholar]
- 42.Branden, C. & Tooze, J. (1991) Introduction to Protein Structure (Garland, New York).
- 43.Schmitt, F. O. (1944) in Medical Physics, ed. Glasser, O. (Year Book Publishers, Chicago), pp. 1586-1591.
- 44.Ambrose, E. J. & Elliott, A. (1951) Proc. R. Soc. London 208, 75-90. [Google Scholar]
- 45.Senti, F. R., Copley, M. J. & Nutting, G. C. (1945) J. Phys. Chem. 49, 192-211. [Google Scholar]
- 46.Aggeli, A., Fytas, G., Vlassopoulos, D., McLeish, T. C. B., Mawer, P. J. & Boden, N. (2001) Biomacromolecules 2, 378-388. [DOI] [PubMed] [Google Scholar]
- 47.Lansbury, P. T. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3342-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheibel, T., Parthasarathy, R., Sawicki, G., Liu, X.-M., Jaeger, H. & Lindquist, S. L. (2003) Proc. Natl. Acad. Sci. USA 100, 4527-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Surewicz, W. K., Mantsch, H. H. & Chapman, D. (1993) Biochemistry 32, 389-394. [DOI] [PubMed] [Google Scholar]
- 50.Susi, H. & Byler, D. M. (1986) Methods Enzymol. 130, 290-311. [DOI] [PubMed] [Google Scholar]
- 51.Krimm, S. & Bandekar, J. (1986) Adv. Protein Chem. 38, 181-364. [DOI] [PubMed] [Google Scholar]