Abstract
Background
Acetyl-CoA-derived chemicals are suitable for multiple applications in many industries. The bio-production of these chemicals has become imperative owing to the economic and environmental problems. However, acetate overflow is the major drawback for acetyl-CoA-derived chemicals production. Approaches for overcoming acetate overflow may be beneficial for the production of acetyl-CoA-derived chemicals.
Results
In this study, a transcriptional regulator iclR was knocked out in E.coli BL21(DE3) to overcome acetate overflow and improve the chemicals production. Two important acetyl-CoA-derived chemicals, phloroglucinol (PG) and 3-hydroxypropionate (3HP) were used to evaluate it. It is revealed that knockout of iclR significantly increased expressions of aceBAK operon. The cell yields and glucose utilization efficiencies were higher than those of control strains. The acetate concentrations were decreased by more than 50% and the productions of PG and 3HP were increased more than twice in iclR mutants. The effects of iclR knockout on cell physiology, cell metabolism and production of acetyl-CoA-derived chemicals were similar to those of arcA knockout in our previous study. However, the arcA-iclR double mutants couldn’t gain higher productions of PG and 3HP. The mechanisms are unclear and needed to be resolved in future.
Conclusions
Knockout of iclR significantly increased gene expression of aceBAK operon and concomitantly activated glyoxylate pathway. This genetic modification may be a good way to overcome acetate overflow, and improve the production of a wide range of acetyl-CoA-derived chemicals.
Keywords: Acetyl-CoA, Escherichia coli, iclR, arcA, 3-hydroxypropionate, Phloroglucinol
Background
Acetyl-CoA-derived chemicals, including polyhydroxyalkanoates, isoprenoids, polyketides, lipids and butyrate, are suitable for multiple applications in food additive, medicine, agriculture, cosmetic, and chemical industries [1–3]. The bio-production of acetyl-CoA-derived chemicals has become imperative owing to the diminishing petroleum reserves and growing environmental concerns [4]. To date, Escherichia coli remains one of the most widely used hosts in recombinant bioprocesses because of its low manufacturing cost and easy manipulation [5, 6]. The bio-production of acetyl-CoA-derived chemicals through engineering Escherichia coli has made significant progresses in recent years. However, it is worth mentioning that acetyl-CoA can be converted to acetate through the acetate kinase/phosphate acetyl transferase (AckA-Pta) pathway when cells grow rapidly on glucose abundant conditions, also known as ‘acetate overflow’ [7–9] (Fig. 1). Acetate excretion affects the cell density, biomass accumulation and macromolecule biosynthesis even at concentrations as low as 0.5 g/l [10, 11]. Mutations in both ackA and pta have been reported a strong reduction of acetate production. However, this is at the expense of the cell growth rate and is accompanied by an increase in the production of other by-products such as formate and lactate [12]. So, decreasing the undesirable conversion from acetyl-CoA to acetate may be beneficial for acetyl-CoA-derived chemicals production. As predicted, our previous study has revealed that knockout of arcA in E.coli BL21(DE3) exhibited surprising efficacy of overcoming acetate formation, and the productions of acetyl-CoA-derived chemicals PG and 3HP were improved by 2.25-fold and 2.08-fold, respectively [13].
Fig. 1.
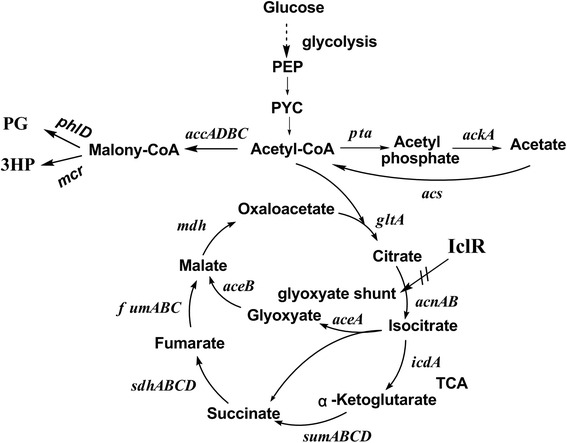
The main carbon metabolic pathways in recombinant E.coli strains
Acetate overflow is observed in all Escherichia coli strains but the extent can differ greatly between the various strains [7, 14]. E.coli BL21 strains showed lower acetate yields as compared to K12 strains [15, 16]. C13-Constrained metabolic flux analysis revealed that the lower acetate yields in BL21 were caused by the higher activation of glyoxylate pathway and acetate assimilation pathway [15, 17]. IclR (isocitrate lyase regulator) is a local regulator, repressing the gene expression of aceBAK operon, which codes for the glyoxylate pathway enzymes malate synthase (AceB), isocitrate lyase (AceA) and isocitrate dehydrogenase kinase/phosphatase(AceK) [18–20] (Fig. 1). Decreasing the gene expression of iclR may activate glyoxylate pathway and decrease acetate formation. If not, lots of acetyl-CoA would be transformed to acetate through AckA-Pta pathway, which in turn starve the precursor for the acetyl-CoA-derived chemicals production and hurt the yields.
In this study, the iclR gene of E.coli BL21(DE3) was knocked out, and the resultant strain was used to produce two acetyl-CoA-derived chemicals, phloroglucinol (PG) and 3-hydroxypropionate (3HP) which are both important bulk chemicals [21, 22]. The effects of iclR deletion on gene expression of aceBAK operon, the cell physiology and metabolism were also determined and discussed. As arcA deletion also had positive effects on PG and 3HP production, the cell physiology and metabolism and production of acetyl-CoA-derived chemicals in E.coli BL21(DE3) arcA-iclR double mutant were also investigated in this study.
Methods
Strains construction
The oligonucleotide primers used in this study were listed in Table 1, and the recombinant plasmids and strains used in this study were listed in Table 2. The iclR mutants were constructed using P1 phage transduction from BL21(DE3) and arcA single mutant Q1949 as previously described [23]. The donor strain of E.coli BW25113 iclR::kan (JW3978) was purchased from the Keio collection [24]. The kanamycin genes were removed by a temperature-sensitive plasmid of pCP20, and the resultant strains were named as Q2280 and Q2138, respectively . The four subunits of accA, accD and accBC from E.coli were cloned into pACYCDuet-1, resulting in plasmid pA-accADBC. The phlD gene from P. fluorescens Pf5 (ATCC BAA -477) and mar gene from E.coli were cloned into pET30a, resulting in plasmid pET-phlDmar. [13, 22] The plasmids pA-accADBC and pET-phlDmar were transformed into E.coli BL21(DE3), arcA mutant Q1949, iclR mutant Q2280 and arcA-iclR double mutant Q2138 to generate PG-producing strains Q1944, Q1963, Q2283 and Q2176. The mcr-N and mcr-C (N940V/ K1106W/ S1114R) from Chloroflexus aurantiacus were cloned into pETDuet-1, resulting in plasmid pMCR-N-C [25]. The plasmids pA-accADBC and pMCR-N-C were also transformed into E.coli BL21(DE3), Q1949, Q2280 and Q2138 to generate 3HP -producing strains Q2204, Q2284, Q2285 and Q2286.
Table 1.
Primers used in this study
| Primer name | Sequence |
|---|---|
| Strain construction | |
| ID-iclR_F | TACGAAATGCCGGATCGTTG |
| ID-iclR_R | TCTTGTTTATCAAGAGTGTC |
| pKD13-Kan_R | GGTGAGATGACAGGAGATCC |
| RT-PCR Reaction | |
| 16S rRNA_F | TGGTCTGAGAGGATGACCAG |
| 16S rRNA_R | TGCTTCTTCTGCGGGTAACG |
| aceB_F | ATGACTGAACAGGCAACAAC |
| aceB_R | CGAATGGAAGCT GTTTCCGA |
| aceA_F | ATGAAAACCCGTACACAACA |
| aceA_R | CGAGGCTGTTGATGTAGCCT |
| aceK_F | CGTGGCCTGGAATTATTGAT |
| aceK_R | GTTAGTAATGCAGCGCAGTT |
Table 2.
Plasmids and strains used in this study
| Plasmids and strains | Description | Source |
|---|---|---|
| Plasmids | ||
| pETDuet-1 | Ampr oripBR322 lacI q T 7 p | Novagen |
| pACYCDuet-1 | Cmr oriP15A lacI q T 7 p | Novagen |
| pCP20 | Cmr, Ampr pSC101 ori CI857 | Novagen |
| pA-accADBC | repp15A CmR lacI PT7 accA PT7 accD PT7 accBC | [22] |
| pET-phlDmar | reppBR322 kanR lacI PT7 phlD PT7 mar | [22] |
| pMCR-N-C | reppBR322 AmpR lacI PT7 His 6 -mcr 1-549 PT7 His 6 -mcr 550-1219(N940V/ K1106W/ S1114R) | [25] |
| Strains | ||
| E. coli DH5α | F- supE44 ΔlacU169 (ϕ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| E. coli BL21(DE3) | F- ompT gal dcm lon hsdSB (rB- mB-) λ(DE3) | Invitrogen |
| JW3978 | E. coli BW25113 iclR::kan | Keio collection |
| Q1949 | E. coli BL21(DE3)△arcA | [13] |
| Q2133 | E. coli BL21(DE3) iclR::kan | This study |
| Q2280 | E. coli BL21(DE3)△iclR | This study |
| Q2135 | E. coli BL21(DE3)△arcA iclR::kan | This study |
| Q2138 | E. coli BL21(DE3)△arcA△iclR | This study |
| Q1944 | E. coli BL21(DE3)/pA-accADBC/pET-phlDmar | This study |
| Q1963 | Q1949/pA-accADBC/pET-phlDmar | This study |
| Q2283 | Q2280/pA-accADBC/pET-phlDmar | This study |
| Q2176 | Q2138/pA-accADBC/pET-phlDmar | This study |
| Q2204 | E. coli BL21(DE3)/pA-accADBC/pMCR-N-C | This study |
| Q2284 | Q1949/pA-accADBC/pMCR-N-C | This study |
| Q2285 | Q2280/pA-accADBC/pMCR-N-C | This study |
| Q2286 | Q2138/pA-accADBC/pMCR-N-C | This study |
Gene expression by RT-PCR
Total RNAs from BL21(DE3) and Q2280 were isolated using Bacteria RNA Kit (omega) according to the manufacturer’s recommendations. The quantity and purity of RNAs were determined by optical density measurements at 260nm and 280nm by 1% agarose gel electrophoresis. Reverse-transcription (RT)-PCR reactions were carried out in a PCR thermocycler using TransScrip® One-step gDNA removal and cDNA synthesis kit. The reaction mix was first incubated 10min at 25°C and then incubated 30 min at 42°C for reverse transcription. Inactivated gDNA remover by heating 5 s at 85°C. Gene expression levels of aceBAK operon in BL21(DE3) and iclR mutant Q2280 were analyzed by PCR amplification with primers in Table 1. The gene transcription levels of 16S rRNA were used as inner reference for normalization.
Shake-flask cultivation of the recombinant E.coli strains
Shake-flask experiments were carried out in triplicate in 250 ml flask containing 50 ml fermentation medium. The strains were grown overnight at 37°C with shaking in LB broth, and then 1:50 diluted into 50 ml fermentation medium. When OD600 of the culture reached about 0.6, IPTG was added to a final concentration of 0.1 mM/l and further incubated at 30°C. 10 mg/l Biotin and 20mM/l NaHCO3 were needed for 3HP production. The fermentation medium for PG production contains 9.8 g/l K2HPO4•3H2O, 2.1 g/l citric acid•H2O, 0.3 g/l ferric ammonium citrate, 3.0 g/l (NH4)2SO4, 0.2 g/l MgSO4•7H2O, 20 g/l glucose and 1000 × trace mental (3.7 g/l (NH4)6Mo7O24•4H2O, 2.9 g/l ZnSO4•7H2O, 24.7 g/l H3BO3, 2.5 g/l CuSO4•5H2O, 15.8 g/l MnCl2•4H2O). The fermentation medium for 3HP production contains 14 g/l K2HPO4•3H2O, 5.2 g/l KH2PO4, 1 g/l NaCl, 1 g/l NH4Cl, 0.25 g/l MgSO4•7H2O, 0.2 g/l yeast extract, 20 g/l glucose. The OD600 of the culture and the concentrations of residual glucose were measured during the whole fermentation course.
Analytical methods
The cell concentration was measured by the optical density of the culture at 600 nm, and the value of density was converted to cell dry weight (CDW) based on that one unit of OD600 was equivalent to 0.36 g/l CDW. The concentration of residual glucose was quantified by using an SBA-40D Biological Sensing Analyzer. The concentration of PG in the fermentation supernatant was quantified using the colorimetric reaction at OD446 between cinnamaldehyde and PG. [26] The concentrations of 3HP and acetate in medium were determined by HPLC as described previously [13, 27].
Results
Knockout of iclR increased gene expression of aceBAK operon
IclR is a transcriptional regulator that regulates gene expression of aceBAK operon [18, 20]. In order to investigate the effect of iclR deletion on gene expression of aceBAK operon, the total RNAs of wild strain BL21(DE3) and iclR mutant strain Q2280 were extracted to carry out the RT-PCR reaction. The gene transcription levels of 16S rRNA were used as inner reference for normalization. As shown in Fig. 2, the mRNA levels of aceB, aceA and aceK in iclR mutant Q2280 were much higher than those of the wild-type strain BL21(DE3), while the mRNA levels of 16S rRNA were very similar in these two strains, suggesting that iclR deletion could up-regulate the gene expression of aceBAK operon.
Fig. 2.
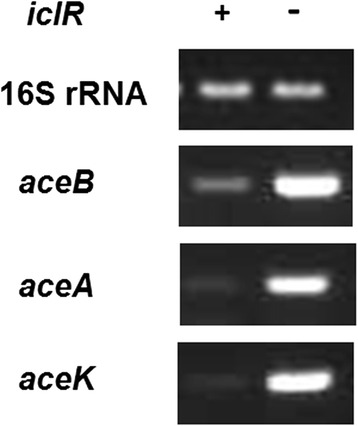
Genes transcription of aceBAK operon in E.coli BL21(DE3) and iclR mutant by RT-PCR. Constitutively, gene transcriptional level of 16S rRNA was used as inner reference
Effects of iclR knockout on cell physiology and metabolism
The iclR mutants were constructed using P1 phage transduction and the recombinant plasmids were transformed for PG and 3HP production. During the cultivation process, the OD600 and residual glucose concentrations of the control strains (Q1944 and Q2204) and iclR mutants (Q2283 and Q2285) were measured to investigate the effects of iclR deletion on cell growth and glucose utilization. As shown in Fig. 3 and Fig. 4, the iclR mutants showed similar cell dry weight (CDW) as compared to their corresponding control strains. After fermentation, the cell dry weight of PG-producing strains Q1944 and Q2283 were 2.05 ± 0.06 g/l and 2.08 ± 0.09 g/l, while 3HP-producing strains Q2204 and Q2285 accumulated 2.64 ± 0.13 g/l and 2.76 ± 0.22 g/l CDW, respectively. These results drew a conclusion that deletion of iclR had no obvious effect on cell growth during the bio-production of PG and 3HP. Meanwhile, the glucose consumptions and cell yields were also compared. The glucose consumptions of iclR mutants obviously decreased since IPTG induction. Finally, the residual glucose concentrations of iclR mutants Q2283 and Q2285 were significantly higher than their corresponding control strains (Fig. 3). Knockout of iclR
Fig. 3.
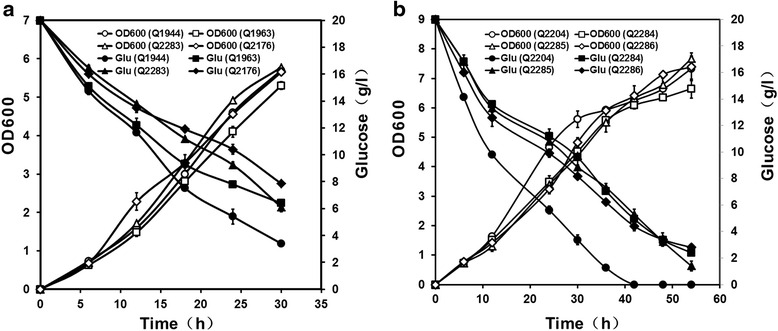
Cell growth and glucose consumption of recombinant E.coli strains during the process of cultivation in shaking flasks. a The OD600 and residual glucose concentrations of PG-producing strains Q1944, Q1963, Q2283 and Q2176 in their cultures. b The OD600 and residual glucose concentrations of 3HP-producing strains Q2204, Q2284, Q2285 and Q2286 in their cultures
Fig. 4.
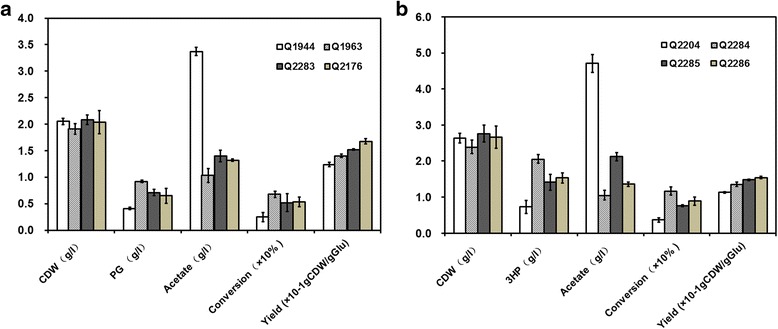
The effects of arcA and iclR knockout on the cell phycology and metabolism of recombinant E.coli strains during the process of cultivation in shaking flasks. a The cell dry weight, PG productions, acetate concentrations, glucose conversion efficiencies and cell yields of PG-producing strains. b The cell dry weight, 3HP productions, acetate concentrations, glucose conversion efficiencies and cell yields of 3HP-producing strains
decreased the glucose consumptions without impairing the cell dry weight, therefore, iclR mutants possessed higher cell yields. It is speculated that the genetic modification of iclR knockout could improve the glucose utilization efficiency and reduce the manufacturing cost in industrial applications.
Acetate is the major undesirable metabolite in the production of acetyl-CoA-derived chemicals. After cultivation, the control strain Q1944 and Q2204 produced 3.14 ± 0.13 g/l and 5.86 ± 0.05 g/l acetate, while the iclR mutants Q2283 and Q2285 produced 1.41 ± 0.01 g/l and 2.12 ± 0.13 g/l acetate, decreased by 55% and 64%, respectively. In conclusion, iclR deletion showed dramatic effects on glucose utilization efficiency and acetate formation during the process of PG and 3HP production. However, the cell growth was very similar between the control strains and iclR mutants.
Knockout of iclR improved PG and 3HP production
The effects of iclR deletion on PG and 3HP production were also investigated (Fig. 4). The PG production of iclR mutant Q2283 was 0.71 ± 0.06 g/l, showing 1.7-time higher than that of the control strain Q1944. The glucose conversion efficiency to PG was increased by 2.1-fold, from 2.47% to 5.19%. In 3HP production, the iclR mutant Q2285 produced 1.41 ± 0.22 g/l 3HP, showing 1.9-time higher than that of the control strain Q2204. The glucose conversion efficiency to 3HP was increased by 2.0-fold, from 3.65% to 7.58%. Knockout of iclR significantly improved the production of acetyl-CoA-derived chemicals and the glucose conversion efficiency.
Effects of arcA and iclR knockouts on cell physiology and metabolism
Summarizing the above results, deletion of arcA or iclR can improve the cell yield and glucose utilization efficiency and repress the acetate formation, so we are curious whether the double knockout of arcA and iclR should further affect the cell physiology and metabolism. The detailed comparison of the cell growth, glucose consumption, cell yield and acetate production in arcA mutants, iclR mutants and arcA-iclR double mutants were also made. As shown in Fig. 3 and Fig. 4, the cell growth was seriously impaired through arcA deletion. While, it was not affected in iclR mutants and arcA-iclR double mutants. The double mutants Q2176 and Q2286 produced 2.03 ± 0.22 g/l and 2.66 ± 0.31 g/l CDW, the same as the control strains of 2.05 ± 0.06 g/l and 2.64 ± 0.12 g/l CDW. In addition, the cell yields of arcA-iclR double mutants were slightly higher than those of the control strains, the arcA single mutants and the iclR single mutants. It was assumed that the combined deletion of arcA and iclR was beneficial for the conversion from glucose to the biomass.
All the mutant strains showed reduced acetate productions as compared to the control strains (Fig. 4). However, the ability to overcome acetate formation of arcA-iclR double mutants was less than that of the arcA single mutants. The regulatory mechanisms remained unclear and needed to be resolved.
Effects of arcA and iclR knockouts on PG and 3HP production
The arcA-iclR double mutants showed lower productions of PG and 3HP than the arcA single mutants. The productions of PG and 3HP in arcA single mutants were increased by 2.25-fold and 2.83-fold than those in the control strains Q1944 and Q2204, while they were only increased by 1.58-fold and 2.15-fold in arcA-iclR double mutants. Consistent with that effects, the arcA-iclR double mutants also showed lower glucose conversion efficiencies than the arcA single mutants. The glucose conversions efficiencies to PG and 3HP in arcA single mutants were 6.76% and 11.7% respectively, while they were only 5.35% and 8.89% in arcA-iclR double mutants.
Taken together, both mutants of arcA and iclR showed positive effects on the productions and glucose conversion efficiencies to PG and 3HP. However the arcA single mutants had the most prominent results, and the iclR single mutants, the arcA-iclR double mutants showed the similar effects on these aspects.
Discussion
The excretion and assimilation of acetate usually undergo a metabolic switch when cells growing on glucose. Acetate is produced by the Pta-AckA pathway in exponential growth phase, and it is converted to acetyl-CoA by the catalysis of acetyl-CoA synthetase (Acs) with the cells convert to stationary phase [28]. As the intracellular enzyme pyroposphatase removes the intermediate pyrophosphate, the reaction from acetate to acetyl-CoA is irreversible. Acetyl-CoA was utilized to generate ATP and metabolic intermediates for the cellular activities and exogenous genes expression via TCA cycle and glyoxylate shunt. So, acetate overflow is not only detrimental to cell growth and macromolecular biosynthesis, it is also a loss of carbon and therefore an economic sink [12]. To date, numerous approaches have been reported to overcome acetate excretion. If some of these approaches could also be used to improve the production of target compounds and decrease the production cost, these studies will have important significance and market value. In this study, the effects of arcA and/or iclR deletion on the production of acetyl-CoA-derived chemicals were determined and discussed. The results revealed that the arcA and/or iclR deletion significantly decreased the acetate concentrations. All the mutants showed higher cell yields and higher glucose utilization efficiencies. More importantly, the productions of PG and 3HP were increased than those of the control strains (Fig. 3 and Fig. 4). So, overcoming acetate overflow through arcA and/or iclR deletion could improve the production of acetyl-CoA-derived chemicals. Acetate overflow is observed in all Escherichia coli strains, however, the extent is very different between the K-12 and B strains, which are the most common E.coli strains for laboratorial and biotechnological applications [6]. BL21 strains showed less acetate accumulations than K-12 strains during the process of high cell-density fermentation with glucose as the sole carbon. A comparison of the genome sequences between BL21 and K12 strains showed that BL21 strains possessed rare codons in arcA and two mutations in the promoter region of iclR [29]. These variations of BL21 resulted in the reduced expressions of arcA and iclR, presumably contributing to the lower acetate accumulation. A previous research revealed that the central metabolic fluxes of BL21(DE3), especially with respect to the TCA fluxes, the glyoxylate pathway and the pentose pathway were similar to that of the K12 strains with arcA and iclR deletion [29]. Meanwhile, the acetate concentration of E. coli MG1655 Δ arcA Δ iclR was decreased to the value of BL21(DE3) [30]. Considering all that, the expression levels of arcA and iclR in BL21 strains were lower than K12 strains due to the variations in rare codons and promoter region, respectively. A previous study considered that the levels of iclR gene and its gene products in E. coli BL21 strains were much lower compared to in K12 strains, probably, the iclR levels were so low that a complete deletion of iclR [14]. However, our results revealed that knockout of iclR in BL21(DE3) could also significantly decrease acetate producitons by more than 50% and improve the PG and 3HP productions by more than twice. Thus, genetic modifications of iclR in K12 and BL21 strains both had obvious effect on overcoming acetate overflow, though the iclR levels were already very low in BL21 strains.
ArcA (anaerobic redox control) is the cytosolic response regulator of the dual-component regulator system ArcAB [31, 32]. It is reported that phosphorylated ArcA can repress genes expression of TCA cycle and glyoxylate shunt, such as gltA, acnAB, icdA, sucABCD, sdhCDAB, fumA, mdh, and aceB [33–35]. Our previous results demonstrated that the acetate concentrations of arcA mutants in BL21(DE3) were decreased by more than 70% and the productions of PG and 3HP were increased by more than twice. The decreased acetate concentrations and increased PG and 3HP productions of arcA mutants could be caused by the following reasons: the higher activity of TCA cycle and glyoxylate pathway; the more metabolic flux through pyruvate dehydrogenase (PDH) complex, which catalyzes pyruvate to acetyl-CoA [13, 32, 36]. IclR (isocitrate lyase regulator) is a local regulator, inhibiting genes expression of aceBAK operon in glyoxylate pathway. Knockout of iclR significantly increased gene expression of aceBAK operon according to the results of RT-PCR (Fig. 2). In glyoxylate pathway, isocitrate is directly split into succinate and glyoxylate without CO2 loss, it may be used to explain the fact that iclR mutants showed higher cell yields than their corresponding control strains. Glyoxylate pathway is the main pathway for acetate assimilation, knockout of iclR can activate the glyoxylate pathway and promote the conversion from acetate to acetyl-CoA [17, 37]. In addtion, the metabolic flux from phosphoenolpyruvic acid (PEP) to acetyl-CoA is obviously increased in iclR mutant strains [29]. So, knockout of iclR can elevate the metabolic flux from acetate to acetyl-CoA and from PEP to acetyl-CoA. On the one hand, acetyl-CoA is metabolized to provide the energy and precursors for exogenous genes expression in recombinant pathways. And on the other hand, acetyl-CoA is the direct substrate for the production of acetyl-CoA-derived chemicals. Therefore, the iclR knockout can decrease the acetate formation and increase the production of acetyl-CoA-derived chemicals.
To summarize, the arcA deletion and iclR deletion were observed to behave similar effects on the cell physiology and metabolism, except the cell growth. The cell growth of arcA mutants was much lower than their corresponding control strains, however it was not impaired in iclR mutants. It still remains unclear what causes this phenomenon. Then, the arcA and iclR double mutants were also constructed to study the effects on the cell physiology, the metabolism and the production of acetyl-CoA-derived chemicals. Surprisingly, the double mutants had no better results for PG and 3HP productions than those of arcA and iclR single mutants. Meanwhile, the glucose conversion efficiencies and the abilities for overcoming acetate formation were also lower than those of the arcA single mutants. These results proved that the effects of polygenic modification on the produciton of target compounds may not be better than the single genic modification, especially by the global regulators. A global regulator usually controls several operons that belong to different groups [36]. Engineering of a global regulator could show pleiotropic phenotypes and variations in metabolic pathways and carbon fluxes. It is hard to determine the certain gene or the single pathway with a global regulator was modified. Furthmore, the results will be more unpredictable and complex when the several regulators are combined modified. In this study, why the effects of arcA-iclR double mutant on overcoming acetate overflow and improving the acetyl-CoA-derived chemicals production were less than the arcA single mutants? A previous study reported that arcA-iclR deletion of E. coli K12 MG1655 had dramatic effects on the cell physiology and metabolism. However, no significant differences of metabolic flux were observed between the wild type BL21(DE3) and the arcA-iclR double mutant [14]. The tiny variations of metabolic flux in BL21(DE3) arcA-iclR double mutant may lead to the less effects on the cell physiology and metabolism. However, the exact mechanisms would be unravelled by determining the distribution of metabolic fluxes in arcA single mutant, iclR single mutant and arcA-iclR double mutant in future.
Conclusions
Acetate overflow is the major drawback for acetyl-CoA-derived chemicals production. Approaches for overcoming acetate overflow may be beneficial for the production of acetyl-CoA-derived chemicals. IclR is a transcriptional regulator that regulates gene expression of aceBAK operon in glyoxylate pathway. In this work, iclR was knocked out in E. coli BL21(DE3) to construct a mutant strain, and the productions of two important acetyl-CoA-derived chemicals, phloroglucinol and 3-hydroxypropionate were used to evaluate it. The results revealed that knockout of iclR significantly improved gene expression of aceBAK operon. The iclR mutants showed higher cell yields and higher glucose utilization efficiencies without sacrificing cell growth. The acetate concentrations were decreased by more than 50%. More importantly, the productions of PG and 3HP were 2-times higher than those of control strains. These phenomena of iclR knockout were similar to those of the global regulator arcA knockout in our previous study. However, the arcA-iclR double mutants could not produce higher productions of PG and 3HP. Considering all that, the genetic modification of iclR would be a good choice to improve the production of a wide range of acetyl-CoA-derived chemicals in industry.
Acknowledgement
The authors would like to thank the financial support from the Chinese Academy of Sciences, Shandong Province and the postdoctoral Science Foundation of China.
Funding
This work was supported by100-Talent Project of CAS (to GZ), Key Program of CAS (ZDRW-ZS-2016-3M), Scientific and Technological Project of Tianjin (14ZCZDSY00047), the 60-class General Financial Grant from the China Postdoctoral Science Foundation (2016M600564) and Taishan Scholars Climbing Program of Shandong (No.tspd20150210).
Availability of data and materials
All data generated and analysed during this study are included in this published article.
Authors’ contributions
ML and MX participated in the design of the study. ML and GZ were responsible for the review of literature and preparation of the manuscript. YD and HL helped to analyze the statistic dates and revise the manuscript. ML constructed the recombinant strains, carried out the RT-PCR experiments. ML, HC and ZZ participated in the fermentation researches. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- 3HP
3-hydroxypropionate
- Ace
Acetate
- DCW
Dry cell weight
- Glu
Glucose
- PEP
Phosphoenolpyruvate
- PG
Phloroglucinol
- RT-PCR
Reverse-transcription polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Contributor Information
Min Liu, Email: liumin@qibebt.ac.cn.
Yamei Ding, Email: elvadym@hotmail.com.
Hailin Chen, Email: chenhailin@qibebt.ac.cn.
Zhe Zhao, Email: zhaozhe16@mails.ucas.ac.cn.
Huizhou Liu, Email: liuhuizhou@qibebt.ac.cn.
Mo Xian, Phone: +86-532-80662767, Email: xianmo1@qibebt.ac.cn.
Guang Zhao, Phone: +86-532-80662767, Email: zhaoguang@qibebt.ac.cn.
References
- 1.Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Ajikumar PK, Xiao W-H, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–4. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–72. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, Li Z, Feng S, Lucas M, Wu G, Li Y, Li C, Jiang G. Biomass energy utilization in rural areas may contribute to alleviating energy crisis and global warming: A case study in a typical agro-village of Shandong. China Renew Sust Energ Rev. 2010;14:3132–9. doi: 10.1016/j.rser.2010.07.052. [DOI] [Google Scholar]
- 5.Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24:530–6. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–22. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 7.Luli GW, Strohl WR. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol. 1990;56:1004–11. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valgepea K, Adamberg K, Nahku R, Lahtvee PJ, Arike L, Vilu R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst Biol. 2010;4:166. doi: 10.1186/1752-0509-4-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Shimizu K. Catabolite regulation analysis of Escherichia coli for acetate overflow mechanism and co-consumption of multiple sugars based on systems biology approach using computer simulation. J Biotechnol. 2013;168:155–73. doi: 10.1016/j.jbiotec.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Farmer WR, Liao JC. Reduction of aerobic acetate production by Escherichia coli. Appl Environ Microbiol. 1997;63:3205–10. doi: 10.1128/aem.63.8.3205-3210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano K, Rischke M, Sato S, Märkl H. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl Microbiol Biotechnol. 1997;48:597–601. doi: 10.1007/s002530051101. [DOI] [PubMed] [Google Scholar]
- 12.De MM, De MS, Soetaert W, Vandamme E. Minimizing acetate formation in E. coli fermentations. J Ind Microbiol Biot. 2007;34:689–700. doi: 10.1007/s10295-007-0244-2. [DOI] [PubMed] [Google Scholar]
- 13.Liu M, Yao L, Mo X, Ding Y, Liu H, Zhao G. Deletion of arcA increased the production of acetyl-CoA-derived chemicals in recombinant Escherichia coli. Biotechnol Lett. 2016;38:97–101. doi: 10.1007/s10529-015-1953-7. [DOI] [PubMed] [Google Scholar]
- 14.Waegeman H, Maertens J, Beauprez J, De Mey M, Soetaert W. Effect of iclR and arcA deletions on physiology and metabolic fluxes in Escherichia coli BL21 (DE3) Biotechnol Lett. 2012;34:329–37. doi: 10.1007/s10529-011-0774-6. [DOI] [PubMed] [Google Scholar]
- 15.Noronha SB, Yeh HJC, Spande TF, Shiloach J. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using 13C-NMR/MS. Biotechnol Bioeng. 2000;68:316–27. [PubMed]
- 16.Phue J-N, Shiloach J. Transcription levels of key metabolic genes are the cause for different glucose utilization pathways in E. coli B (BL21) and E. coli K (JM109). J Biotechnol. 2004;109:21–30. [DOI] [PubMed]
- 17.Phue JN, Noronha SB, Hattacharyya R, Wolfe AJ, Shiloach J. Glucose metabolism at high density growth of E. coli B and E. coli K: differences in metabolic pathways are responsible for efficient glucose utilization in E. coli B as determined by microarrays and Northern blot analyses. Biotechnol Bioeng. 2005;90:805–20. doi: 10.1002/bit.20478. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto K, Ishihama A. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mole Microbiol. 2003;47:183–94. doi: 10.1046/j.1365-2958.2003.03287.x. [DOI] [PubMed] [Google Scholar]
- 19.Gui L, Sunnarborg A, Pan B, Laporte DC. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J Bacteriol. 1996;78:321–4. doi: 10.1128/jb.178.1.321-324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina-Henares AJ, Krell T, Guazzaroni ME, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. Fems Microbio Rev. 2006;30:157–86. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 21.Rathnasingh C, Raj SM, Jo JE, Park S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng. 2009;104:729–39. doi: 10.1002/bit.22429. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Jiang X, Zhang R, Xian M. Improved phloroglucinol production by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2011;91:1545–52. doi: 10.1007/s00253-011-3304-5. [DOI] [PubMed] [Google Scholar]
- 23.Moore SD. Assembling new Escherichia coli strains by transduction using phage P1. In: Strain Engineering. Springer; 2011;765:155–69. doi:10.1007/978-1-61779-197-0_10. [DOI] [PubMed]
- 24.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mole syst biol. 2006;2:8. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Ding Y, Zhang R, Liu H, Xian M, Zhao G. Functional balance between enzymes in malonyl-CoA pathway for 3-hydroxypropionate biosynthesis. Meta Eng. 2016;34:104–11. [DOI] [PubMed]
- 26.Zha W, Rubin-Pitel SB, Zhao H. Exploiting genetic diversity by directed evolution: molecular breeding of type III polyketide synthases improves productivity. Mole BioSys. 2008;4:246–8. doi: 10.1039/b717705d. [DOI] [PubMed] [Google Scholar]
- 27.Rathnasingh C, Raj SM, Lee Y, Catherine C, Ashok S, Park S. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J Biotechnol. 2012;157:633–40. doi: 10.1016/j.jbiotec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Kumari S, Beatty C. Df, Busby S, Simel E, Hovel MG, Wolfe A: Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–9. doi: 10.1128/JB.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquie-Moreno MR, Heijnen JJ, Charlier D, Soetaert W. Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3) BMC Microbiol. 2011;11:70. doi: 10.1186/1471-2180-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waegeman H, De Lausnay S, Beauprez J, Maertens J, De Mey M, Soetaert W. Increasing recombinant protein production in Escherichia coli K12 through metabolic engineering. N Biotechnol. 2013;30:255–61. doi: 10.1016/j.nbt.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Loui C, Chang AC, Lu S. Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol. 2009;9:1–14. doi: 10.1186/1471-2180-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nizam SA, Zhu J, Ho PY, Shimizu K. Effects of arcA and arcB genes knockout on the metabolism in Escherichia coli under aerobic condition. Biochem Eng J. 2009;44:240–50. doi: 10.1016/j.bej.2008.12.017. [DOI] [Google Scholar]
- 33.Iuchi S, Lin EC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–92. [DOI] [PMC free article] [PubMed]
- 34.Chao G, Park SJ. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode…-ketoglutarate. J Bacteriol. 1997;179:4138–42. doi: 10.1128/jb.179.13.4299-4304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–96. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 36.Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol. 2005;187:3171–9. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon SH, Jeong H, Kwon S-K, Kim JF: Genomics, biological features, and biotechnological applications of Escherichia coli B:“Is B for better?!”. In: Systems Biology and Biotechnology of Escherichia coli. Springer; 2009: 1–17. doi:10.1007/978-1-61779-197-0_10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analysed during this study are included in this published article.


