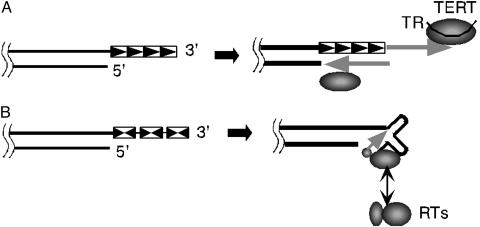
In the past two decades, intense efforts have been devoted to uncovering the mechanisms responsible for the maintenance of telomeres in eukaryotic cells. These efforts have led to the identification of an unusual reverse transcriptase (RT), named telomerase, that uses an integral RNA subunit as template to synthesize a short reiterated sequence at the ends of eukaryotic chromosomes (Fig. 1A) (1). In recent years, the study of eukaryotic telomeres and telomerase has received additional attention because of their established roles in cellular senescence and genome stability (2). Although much less commonly appreciated, linear chromosomes and telomeres are not exclusive to the eukaryotic kingdom; they can be found in a number of bacteria, including Streptomyces, Borrelia, Rhodococcus, etc. (3). In contrast to eukaryotic telomeres, the bacterial versions (at least in some cases) consist of multiple inverted repeats (Fig. 1B). Much of the current knowledge on bacterial telomere maintenance is derived from analyses of linear chromosomes and plasmids in Streptomyces spp. Early studies indicate that replication of these plasmids initiates from an internal origin, resulting in the generation of a leading strand 3′ overhang, and incomplete duplication of the lagging strand (4). Thus, similar to eukaryotic telomeres, a restorative or compensatory mechanism is required to prevent the loss of genetic information. In a series of elegant papers, Cohen and colleagues (5-7) showed that the “patching” of the 5′-recessed ends of Streptomyces plasmids is likely accomplished through a protein-primed mechanism, in which a protein named Tap recognizes a folded structure generated by the 3′ overhang, recruits the Tpg protein, which then serves as primer for the synthesis of DNA on the 5′-recessed strand (Fig. 1B) (5-7). This view of bacterial telomere maintenance would thus not appear to require the participation of an RT.
Fig. 1.
The eukaryotic and bacterial solution to the problem of incomplete end replication. (A) In eukaryotes, telomeres consist of multiple copies of a direct repeat. To compensate for the loss of DNA incurred by incomplete replication of the strand with the 5′ end, eukaryotes use a special RT [telomerase RT (TERT)] to extend the strand with the 3′ end. The template for reverse transcription is embedded in an RNA (TR) that associates stably with TERT. The lengthened 3′ strand can then provide additional template residues for the synthesis of the 5′ strand by DNA polymerases. (B) In bacteria, telomeres consists of multiple inverted repeats. The 3′ overhang generated by incomplete end replication is presumed to fold into a structure that is recognized by a protein named Tap. The Tap protein can associate with Tpg, which is believed to provide the primer for the synthesis of the missing portion of the 5′ strand. The new article by Bao and Cohen (8) suggests that Tap is associated with two additional polypeptides with RT activity.
Tap-associated polypeptides have reverse transcriptase activity.
Or would it? In this issue of PNAS, Bao and Cohen (8) describe that the aforementioned Tap protein is, in fact, associated with RTs. They introduced a His6-tagged Tap into Streptomyces, purified it along with associated proteins, and showed that the purified complex bears substantial RT activity. Further analysis revealed that neither Tap nor Tpg was responsible for the activity. Instead, the RT activity copurified with two polypeptides that were in the telomere complex but that can be separately resolved from Tap by additional chromatographic steps. That these two polypeptides have inherent RT activity was further supported by analysis of purified recombinant proteins. Do these remarkable results imply that there is significant mechanistic similarity between telomere replication in bacterial and eukaryotic cells? This notion, though quite unexpected, could be strengthened by (i) a reverse genetic demonstration that the RT activity is important for telomeric maintenance and (ii) identification of the physiologic substrate of the RTs. [Interestingly, although Bao and Cohen (8) were unable to identify an RNA species in their Tap complex that could specify the synthesis of telomere, they did find such a potential template sequence in the genome.] It is important, however, to keep in mind a key distinction between eukaryotic and bacterial telomere maintenance: the former entails the lengthening of 3′ overhangs to compensate for telomere loss, whereas the latter apparently entails the complete synthesis of the 5′-recessed strand. Nonetheless, the discovery of telomere protein-associated RTs is certainly striking and would undoubtedly stimulate continued investigation aimed at uncovering the physiologic template and function of these RTs.
Perhaps equally striking are the identities of the two polypeptides that were shown to have inherent RT activity; they turn out to be DNA polymerase I and DNA topoisomerase I. Whereas RT activity has been reported for several DNA polymerase I proteins (e.g., those from Thermus aquaticus and Thermus thermophilus), this activity is clearly not conserved in all family members (9, 10). In this regard, a careful comparative analysis of DNA polymerase I proteins with and without RT activity may reveal features of the enzyme responsible for substrate recognition and utilization. On the other hand, as the authors point out, the occurrence of RT activity innate to a topoisomerase protein is truly unprecedented. The Streptomyces topoisomerase I is homologous to all other bacterial DNA topoisomerase I (which are type IA enzymes) (11), but belongs to a distinct subfamily that also includes the enzymes from, among others, Mycobacterium and Deinococcus. Unique to this subfamily of DNA topoisomerase I are two ≈30- to 40-aa insertions. Bao and Cohen (8) surmised that two highly conserved Asp-Asp pairs located in one of the insertions might be equivalent to the metal-binding Asp-Asp pairs in RTs (despite the complete absence of sequence similarity elsewhere) and be essential for activity. Indeed, a recombinantly expressed Streptomyces coelicolor topoisomerase I mutant protein that bears substitutions of the Asp-Asp pairs was found to be capable of DNA supercoil relaxation, but not reverse transcription. This observation provides very strong support for the notion that the RT activity detected in the topoisomerase I preparation is innate to the protein.
Could there be some distant evolutionary relationship between this topoisomerase subfamily and other RTs? This idea is contradicted by the absence of other RT signatures in the topoisomerase protein. It seems more plausible that the acquisition of RT activity by Streptomyces topoisomerase I was a more recent event that involved the addition and modification of the two aforementioned insertions. Homology modeling suggests that the basic architecture of type IA DNA topoisomerase I is retained by the Streptomyces protein with the exception of these two insertions, which are located in conserved domain II, and in close proximity to each other. Intriguingly, one of the insertions lies adjacent to a previously identified nucleotide-binding site on the homologous Escherichia coli topoisomerase I, hinting at its potential contact with nucleotides or nucleic acids (12). Clearly, many questions relating to the RT activity of Streptomyces topoisomerase are worthy of further study. For example, do other members of this topoisomerase subfamily also possess RT activity? What are the mechanistic bases for this unusual RT? Is the RT activity required for telomere replication? What is the physiologic advantage, if any, of having a bifunctional topoisomerase/RT? Thus, in summary, the new article by Bao and Cohen (8) not only raises interesting questions concerning the role of RTs in bacterial telomere maintenance but also provokes speculations about the mechanisms, evolution, and function of an ingenious protein that possesses an unprecedented combination of enzyme activities.
See companion article on page 14361.
References
- 1.Blackburn, E. (2000) Nat. Struct. Biol 7, 847-850. [DOI] [PubMed] [Google Scholar]
- 2.Granger, M., Wright, W. & Shay, J. (2002) Crit. Rev. Oncol. Hematol. 41, 29-40. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. (1996) Trends Genet. 12, 192-196. [DOI] [PubMed] [Google Scholar]
- 4.Chang, P. & Cohen, S. (1994) Science 265, 952-954. [DOI] [PubMed] [Google Scholar]
- 5.Qin, Z. & Cohen, S. (1998) Mol. Microbiol. 28, 893-903. [DOI] [PubMed] [Google Scholar]
- 6.Bao, K. & Cohen, S. (2001) Genes Dev. 15, 1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao, K. & Cohen, S. (2003) Genes Dev. 17, 774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao, K. & Cohen, S. N. (2004) Proc. Natl. Acad. Sci. USA. 101, 14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shandilya, H., Griffiths, K., Flynn, E., Astatke, M., Shih, P., Lee, J., Gerard, G., Gibbs, M. & Bergquist, P. (2004) Extremophiles 8, 243-251. [DOI] [PubMed] [Google Scholar]
- 10.Myers, T. & Gelfand, D. (1991) Biochemistry 30, 7661-7666. [DOI] [PubMed] [Google Scholar]
- 11.Wang, J. (2002) Nat. Rev. Mol. Cell Biol. 3, 430-440. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg, H., Changela, A. & Mondragon, A. (1999) Nat. Struct. Biol 6, 961-968. [DOI] [PubMed] [Google Scholar]